NR4A Expression by Human Marginal Zone B-Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Cell Sorting of Human Blood MZ and Precursor-Like MZ B-Cells, RNA Isolation, and Sequencing
2.3. Validation of the Cross-Reactivity of the PE-Conjugated Human Anti-Mouse Nurr77 (NR4A1) Monoclonal Antibody (mAb)
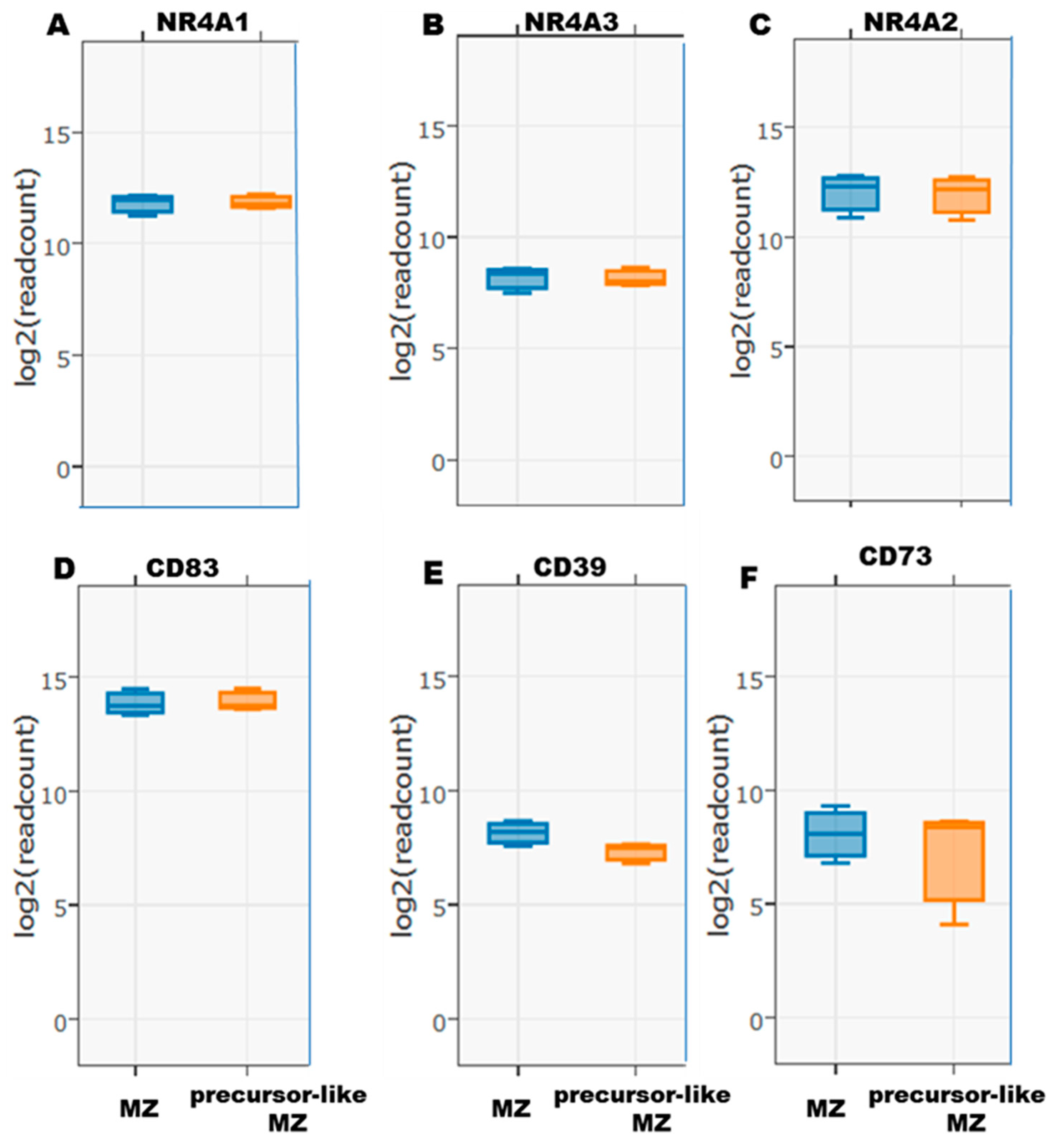
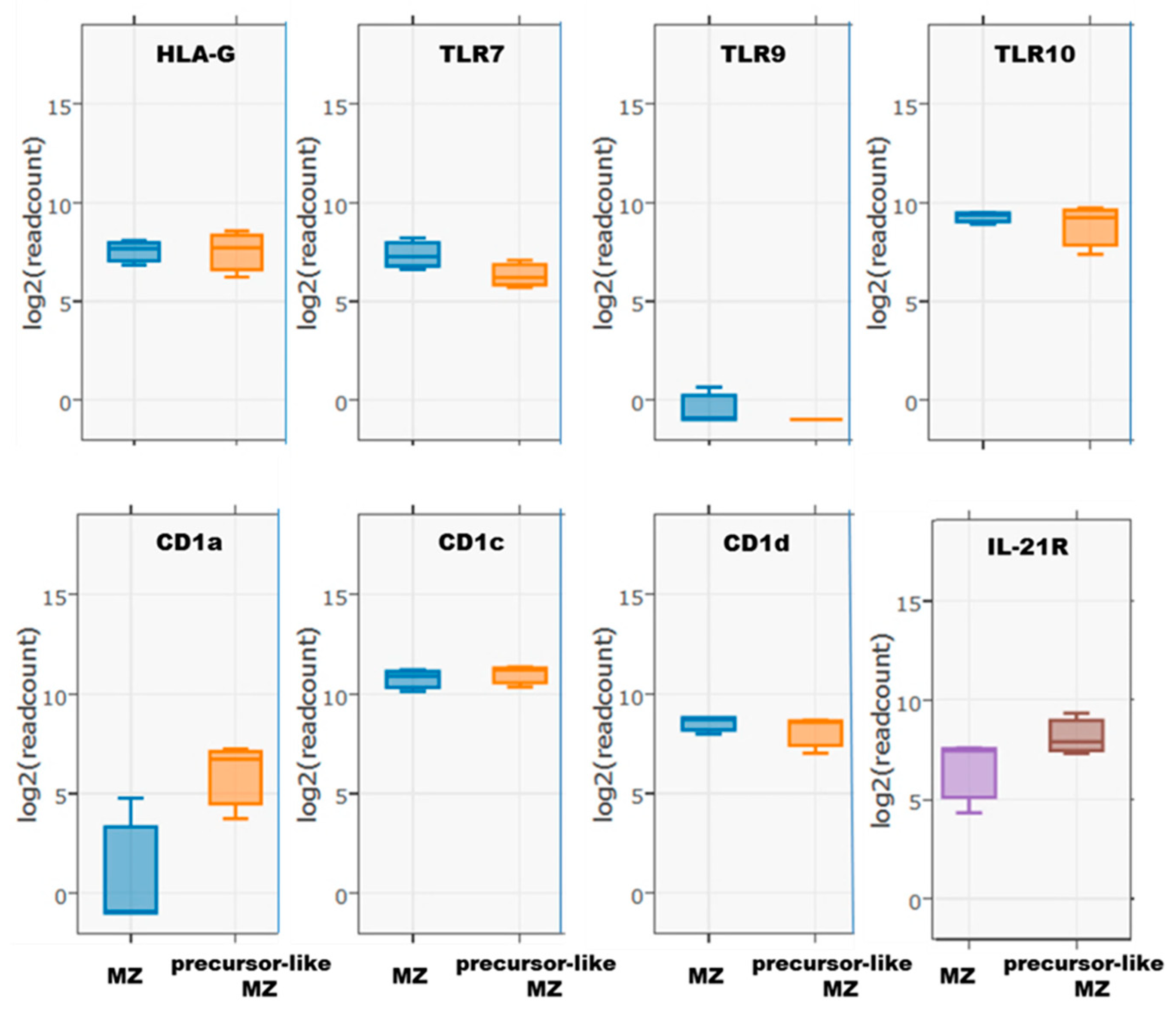
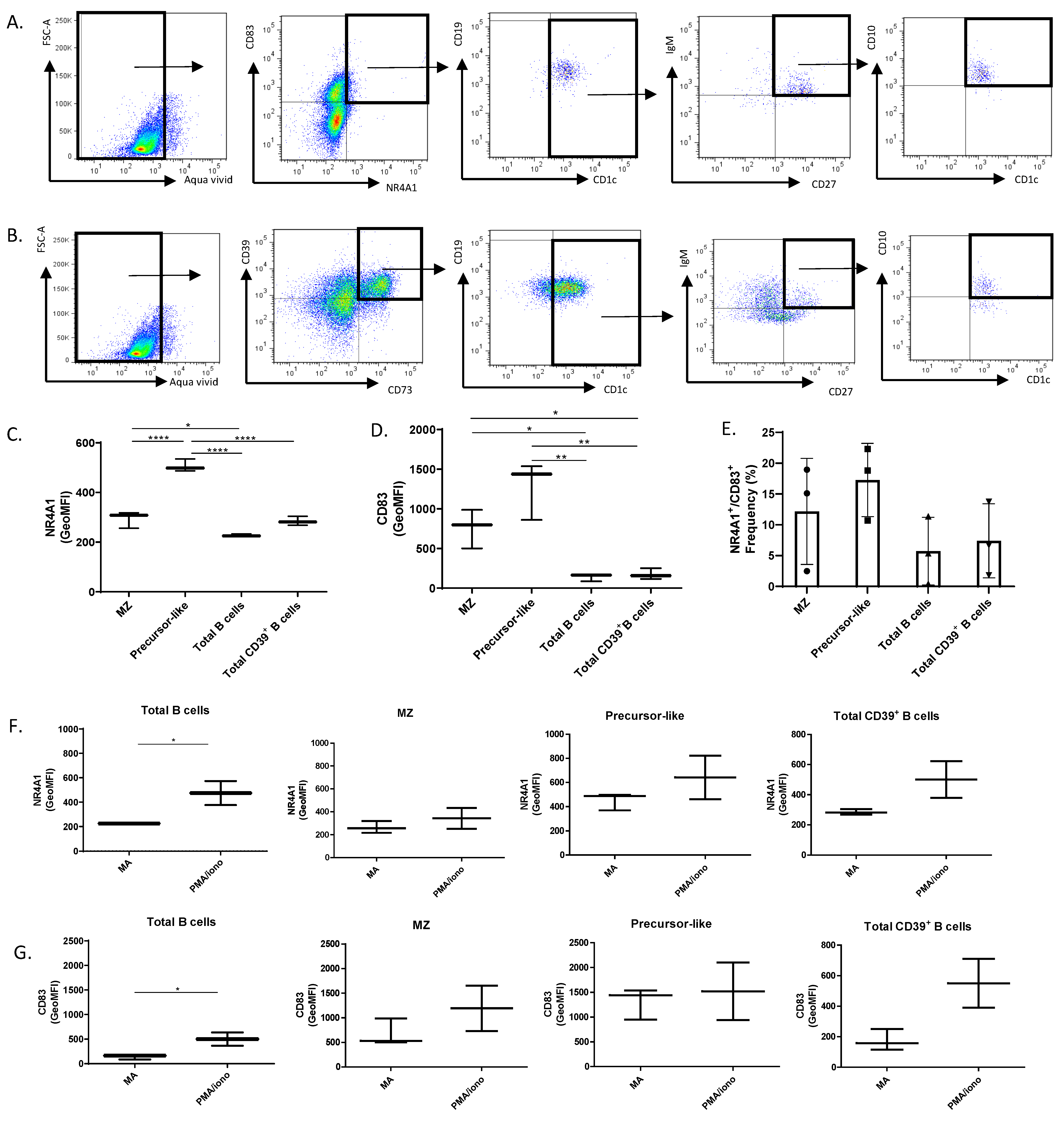
2.4. Flow-Cytometry Characterization of NR4A1, NR4A3, CD83, CD39, and CD73 Expression by Human Peripheral Blood Total, MZ, and Precursor-Like MZ B-Cells
2.5. Flow-Cytometry Characterization of NR4A1, NR4A3, CD83, CD39, and CD73 Expression by Human Tonsillar B-Cells
2.6. Functional Assays
2.7. Statistical Analyses
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fontaine, J.; Chagnon-Choquet, J.; Valcke, H.S.; Poudrier, J.; Roger, M. High expression levels of B lymphocyte stimulator (BLyS) by dendritic cells correlate with HIV-related B-cell disease progression in humans. Blood 2011, 117, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Chagnon-Choquet, J.; Fontaine, J.; Poudrier, J.; Roger, M. IL-10 and lymphotoxin-α expression profiles within marginal zone-like B-cell populations are associated with control of HIV-1 disease progression. PLoS ONE 2014, 9, e101949. [Google Scholar] [CrossRef] [PubMed]
- Sabourin-Poirier, C.; Fourcade, L.; Chagnon-Choquet, J.; Labbé, A.-C.; Alary, M.; Guédou, F.; Poudrier, J.; Roger, M. Blood B Lymphocyte Stimulator (BLyS)/BAFF levels may reflect natural immunity to HIV in highly exposed uninfected Beninese Commercial Sex Workers. Sci. Rep. 2016, 6, 32318. [Google Scholar] [CrossRef] [PubMed]
- Mauri, C.; Bosma, A. Immune regulatory function of B Cells. Annu. Rev. Immunol. 2012, 30, 221–241. [Google Scholar] [CrossRef] [PubMed]
- Iizuka-Koga, M.; Nakatsukasa, H.; Ito, M.; Akanuma, T.; Lu, Q.; Yoshimura, A. Induction and maintenance of regulatory T cells by transcription factors and epigenetic modifications. J. Autoimmun. 2017, 83, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Ashouri, J.F.; Weiss, A. Endogenous Nur77 is a specific indicator of antigen receptor signaling in human T and B cells. J. Immunol. 2016, 198, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, T.; Kashiwagi, I.; Yoshida, R.; Fukaya, T.; Morita, R.; Kimura, A.; Ichinose, H.; Metzger, D.; Chambon, P.; Yoshimura, A. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat. Immunol. 2013, 14, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Bandukwala, H.S.; Rao, A. ‘Nurr’ishing Treg cells: Nr4a transcription factors control Foxp3 expression. Nat. Immunol. 2013, 14, 201–203. [Google Scholar] [CrossRef] [PubMed]
- Breloer, M.; Kretschmer, B.; Lüthje, K.; Ehrlich, S.; Ritter, U.; Bickert, T.; Steeg, C.; Fillatreau, S.; Hoehlig, K.; Lampropoulou, V.; et al. CD83 is a regular of murine B cell function in vivo. Eur. J. Immunol. 2007, 37, 634–648. [Google Scholar] [CrossRef]
- Breloer, M.; Fleischer, B. CD83 regulates lymphocyte maturation, activation and homeostasis. Trends Immunol. 2008, 29, 186–194. [Google Scholar] [CrossRef]
- Duren, R.P.; Boudreaux, S.P.; Conneely, O.M. Genome wide mapping of NR4A binding reveals cooperativity with ETS factors to promote epigenetic activation of distal enhancers in acute myeloid leukemia cells. PLoS ONE 2016, 11, e0150450. [Google Scholar] [CrossRef] [PubMed]
- Figueiró, F.; Muller, L.; Funk, S.; Jackson, E.K.; Battastini, A.M.O.; Whiteside, T.L. Phenotypic and functional characteristics of CD39high human regulatory B cells (Breg). Oncoimmunology 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.-S.; Whiteside, T.L.; Schuler, P.J.; Saze, Z.; Cheng, D.; Jackson, E.K. Adenosine production by human B cells and B cell-mediated suppression of activated T cells. Blood 2013, 122, 9–18. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- van Rhijn, I.; Godfrey, D.I.; Rossjohn, J.; Moody, D.B. Lipid and small-molecule display by CD1 and MR1. Nat. Rev. Immunol. 2015, 15, 643–654. [Google Scholar] [CrossRef] [PubMed]
- Allard, B.; Longhi, M.S.; Robson, S.C.; Stagg, J. The ectonucleotidases CD39 and CD73: Novel checkpoint inhibitor targets. Immunol. Rev. 2017, 276, 121–144. [Google Scholar] [CrossRef] [PubMed]
- Bahar, B.; Crean, D.; McMorrow, J.P.; Cummins, E.P.; Mohan, H.; Murphy, E.P. Adenosine modulates NR4A orphan nuclear receptors to attenuate hyperinflammatory responses in monocytic cells. J. Immunol. 2015, 195, 1436–1448. [Google Scholar]
- Mauri, C.; Menon, M. Human regulatory B cells in health and disease: Therapeutic potential. J. Clin. Investig. 2017, 127, 772–779. [Google Scholar] [CrossRef]
- Amodio, G.; de Albuquerque, R.S.; Gregori, S. New insights into HLA-G mediated tolerance. Tissue Antigens 2014, 84, 255–263. [Google Scholar] [CrossRef]
- Abdollahi-Roodsaz, S.; Dinarello, C.A.; O’Neill, L.; Netea, M.G.; Joosten, L.A.B. Toll-like receptors and chronic inflammation in rheumatic diseases: New developments. Nat. Rev. Rheumatol. 2016, 12, 344–357. [Google Scholar] [CrossRef]
- Guan, Y.; Hess, N.J.; Tapping, R.I.; Jiang, S.; Li, X. TLR10 is a B cell intrinsic suppressor of adaptive immune responses. J. Immunol. 2016, 198, 699–707. [Google Scholar]
- Oleinika, K.; Rosser, E.C.; Matei, D.E.; Nistala, K.; Bosma, A.; Drozdov, I.; Mauri, C. CD1d-dependent immune suppression mediated by regulatory B cells through modulations of iNKT cells. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Yoshizaki, A.; Miyagaki, T.; Dilillo, D.J.; Matsushita, T.; Horikawa, M.; Kountikov, E.I.; Spolski, R.; Poe, J.C.; Leonard, W.J.; Tedder, T.F. Regulatory B cells control T-cell autoimmunity through IL-21-dependent cognate interactions. Nature 2012, 491, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Blair, P.A.; Norena, L.Y.; Flores-Borja, F.; Rawlings, D.J.; Isenberg, D.A.; Ehrenstein, M.R.; Mauri, C. CD19+CD24hiCD38hi B cells exhibit regulatory capacity in healthy individuals but are functionally impaired in systemic lupus erythematosus patients. Immunity 2009. [Google Scholar] [CrossRef] [PubMed]
- Cerutti, A.; Cols, M.; Puga, I. Marginal zone B cells: Virtues of innate-like antibody-producing lymphocytes. Nat. Rev. Immunol. 2013, 13, 118–132. [Google Scholar] [CrossRef] [PubMed]
- Bates, J.M.; Flanagan, K.; Mo, L.; Ota, N.; Ding, J.; Ho, S.; Liu, S.; Roose-Girma, M.; Warming, S.; Diehl, L. Dendritic cell CD83 homotypic interactions regulate inflammation and promote mucosal homeostasis. Mucosal Immunol. 2015, 8, 414–428. [Google Scholar] [CrossRef] [PubMed]


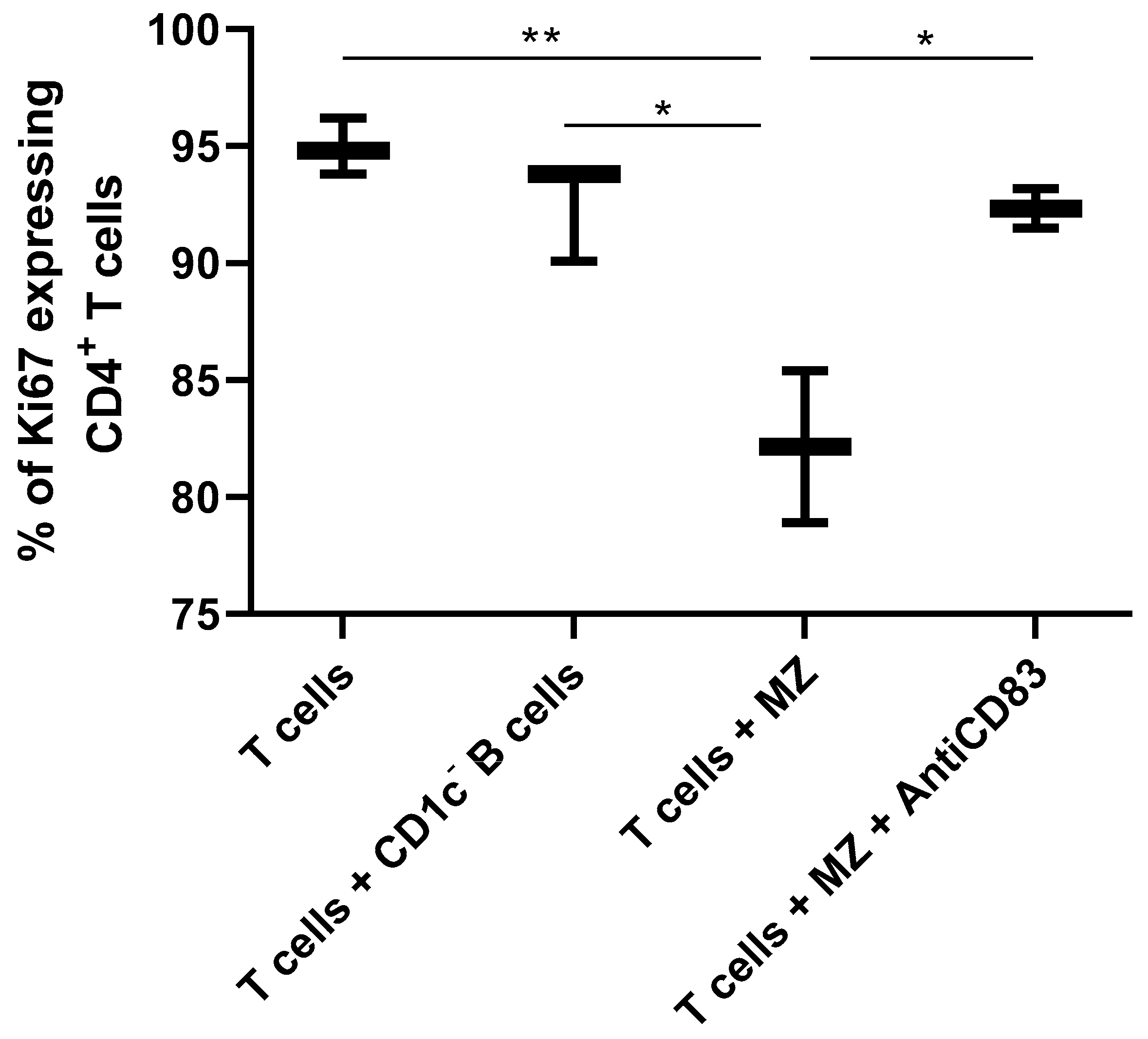
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doyon-Laliberté, K.; Chagnon-Choquet, J.; Byrns, M.; Aranguren, M.; Memmi, M.; Chrobak, P.; Stagg, J.; Poudrier, J.; Roger, M. NR4A Expression by Human Marginal Zone B-Cells. Antibodies 2019, 8, 50. https://doi.org/10.3390/antib8040050
Doyon-Laliberté K, Chagnon-Choquet J, Byrns M, Aranguren M, Memmi M, Chrobak P, Stagg J, Poudrier J, Roger M. NR4A Expression by Human Marginal Zone B-Cells. Antibodies. 2019; 8(4):50. https://doi.org/10.3390/antib8040050
Chicago/Turabian StyleDoyon-Laliberté, Kim, Josiane Chagnon-Choquet, Michelle Byrns, Matheus Aranguren, Meriam Memmi, Pavel Chrobak, John Stagg, Johanne Poudrier, and Michel Roger. 2019. "NR4A Expression by Human Marginal Zone B-Cells" Antibodies 8, no. 4: 50. https://doi.org/10.3390/antib8040050
APA StyleDoyon-Laliberté, K., Chagnon-Choquet, J., Byrns, M., Aranguren, M., Memmi, M., Chrobak, P., Stagg, J., Poudrier, J., & Roger, M. (2019). NR4A Expression by Human Marginal Zone B-Cells. Antibodies, 8(4), 50. https://doi.org/10.3390/antib8040050




