Relative Contribution of Framework and CDR Regions in Antibody Variable Domains to Multimerisation of Fv- and scFv-Containing Bispecific Antibodies
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Plasmid Construction
2.3. Antibody Expression
2.4. Octet Quantification Assay
2.5. Immobilised Metal Affinity Chromatography (IMAC)
2.6. Protein G and Protein A HPLC Purification
2.7. SDS-PAGE
2.8. ELISA
2.9. Size Exclusion Chromatography (SEC)
2.10. Differential Scanning Fluorimetry
3. Results
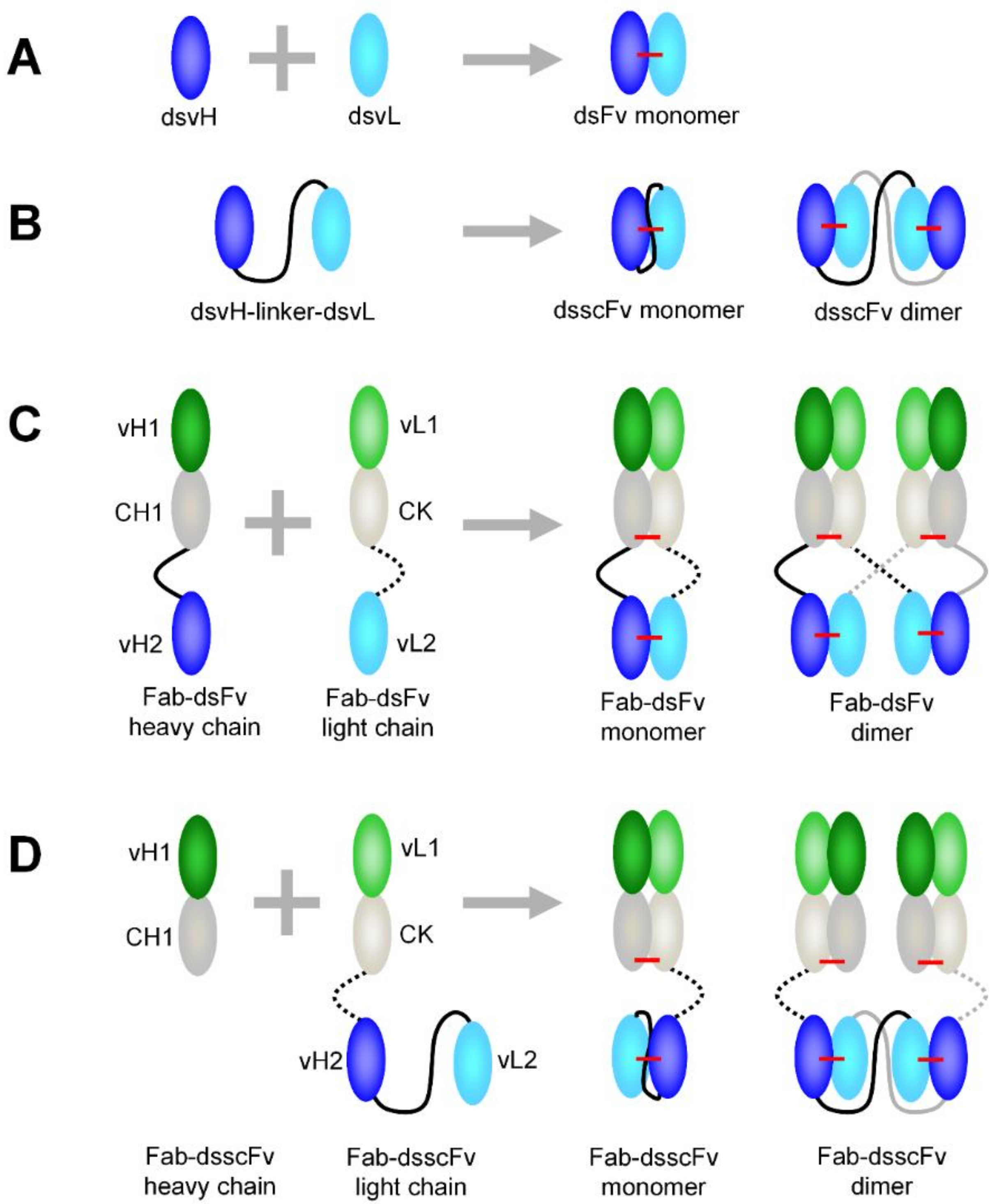
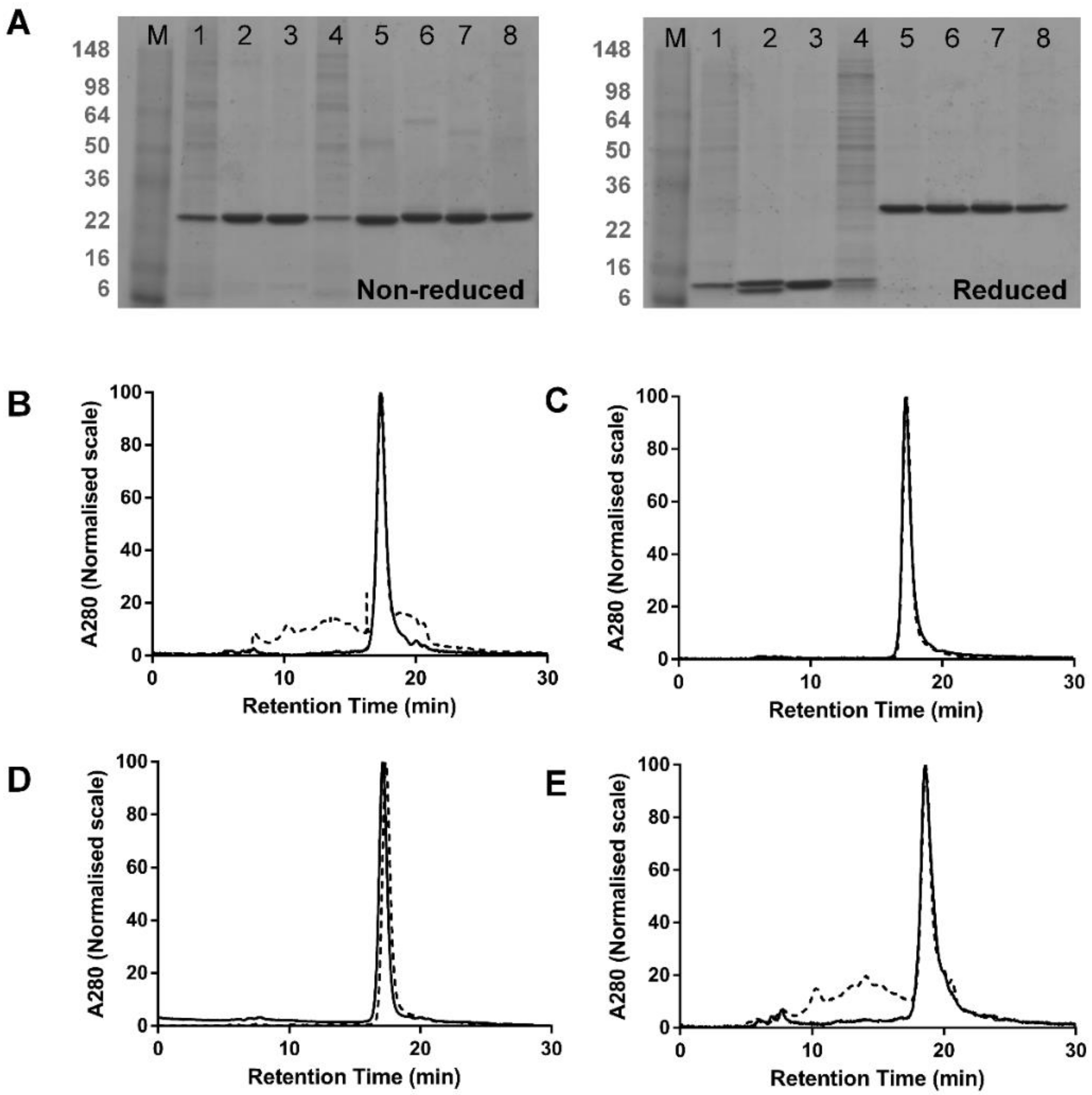
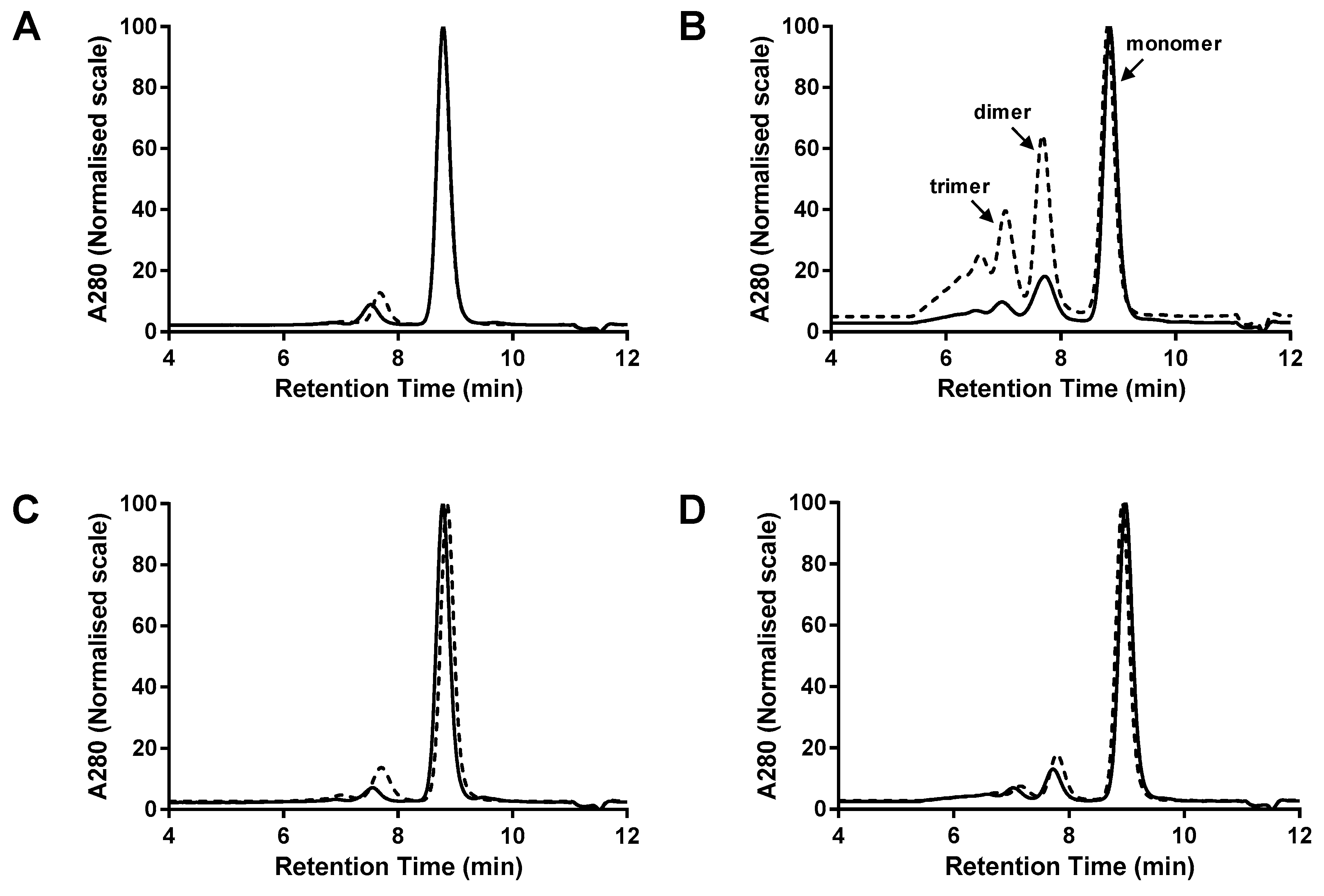
3.1. Comparison of dsFv and dsscFv
3.2. Comparison of Fab-dsFv and Fab-dsscFv
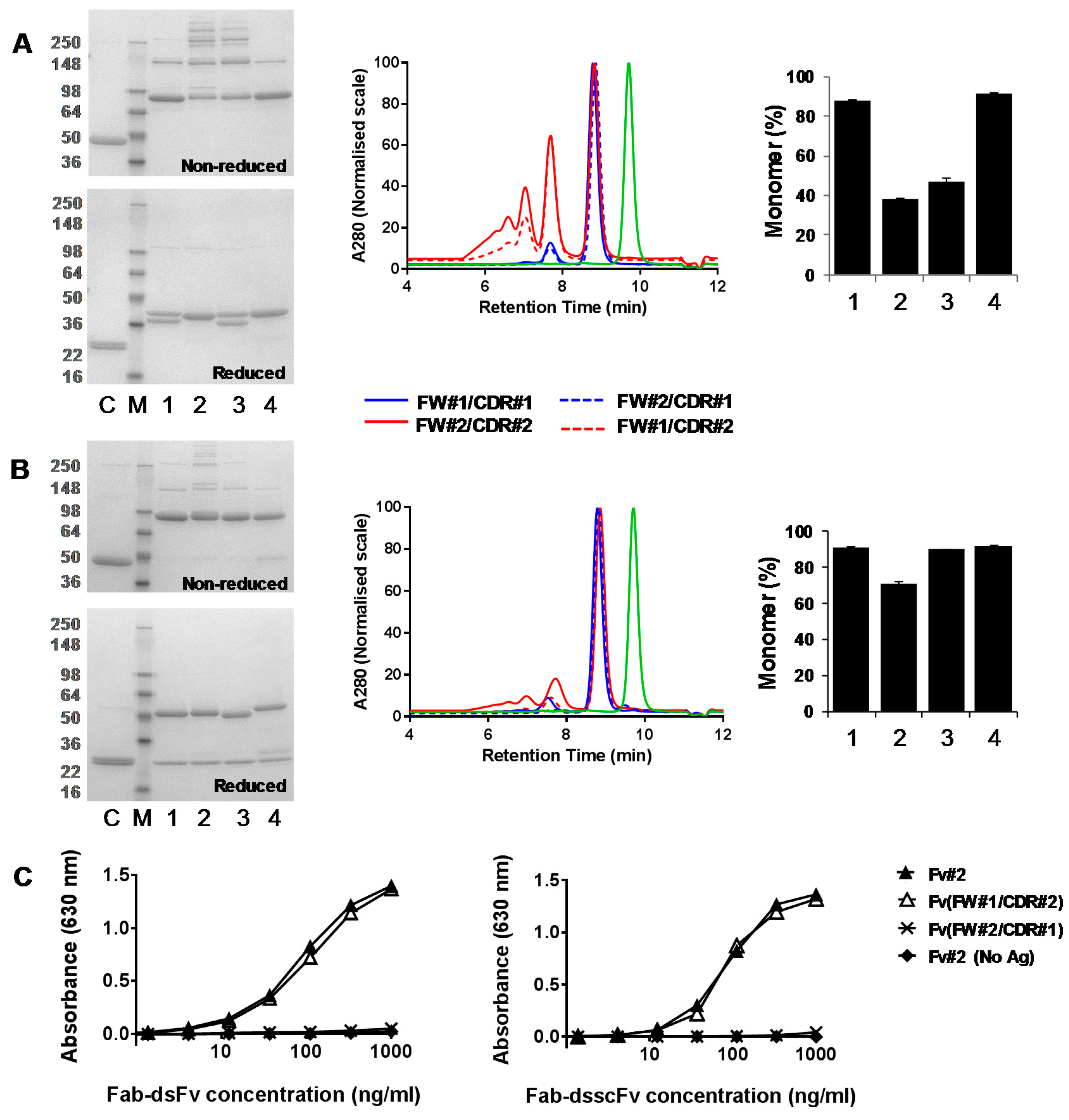
3.3. Analysis of Fab-dsFv and Fab-dsscFv with Framework/CDR-‘Swapped’ Fv
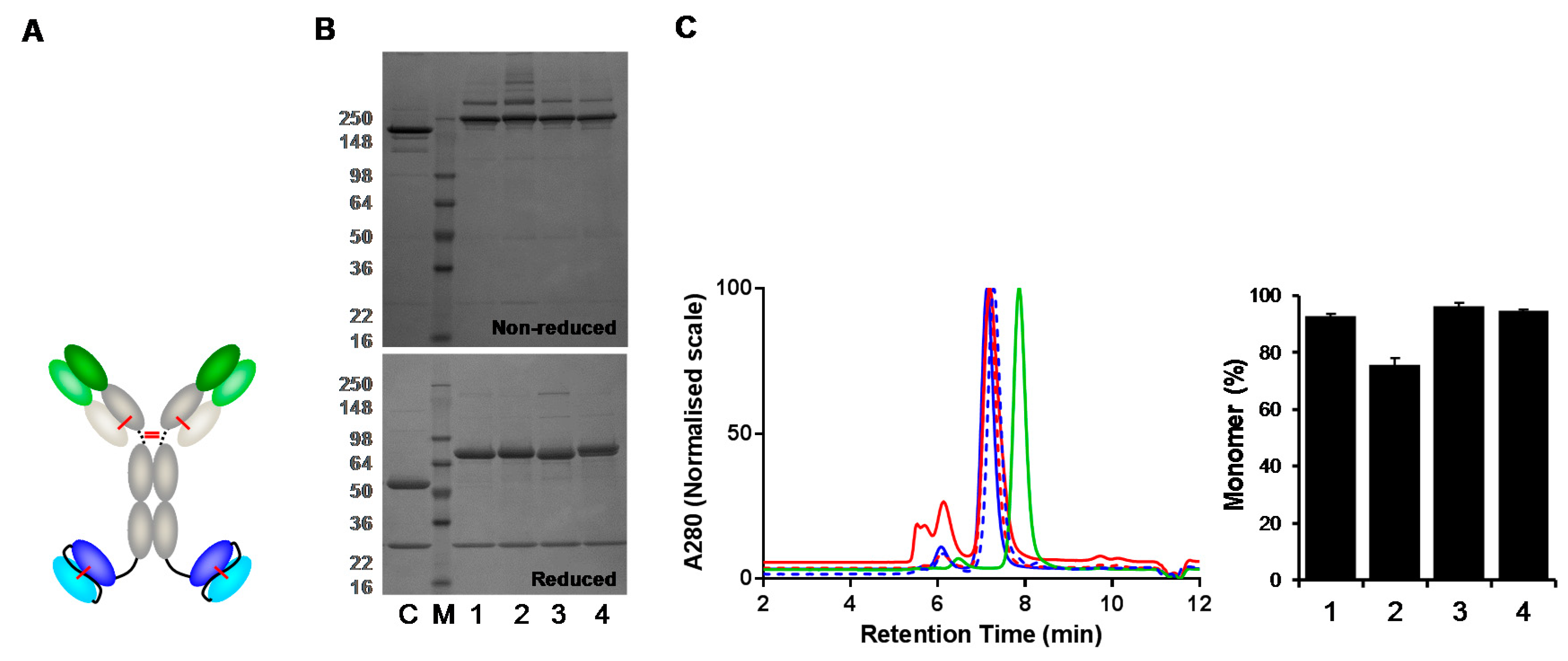
3.4. Analysis of IgG(H)-dsscFv with Framework/CDR-‘Swapped’ Fv
4. Discussion
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Spiess, C.; Zhai, Q.; Carter, P.J. Alternative molecular formats and therapeutic applications for bispecific antibodies. Mol. Immunol. 2015, 67, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Weatherill, E.E.; Cain, K.L.; Heywood, S.P.; Compson, J.E.; Heads, J.T.; Adams, R.; Humphreys, D.P. Towards a universal disulphide stabilised single chain Fv format: Importance of interchain disulphide bond location and vL-vH orientation. Protein Eng. Des. Sel. 2012, 25, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Adams, R.; Griffin, L.; Compson, J.E.; Jairaj, M.; Baker, T.; Ceska, T.; West, S.; Zaccheo, O.; Dave, E.; Lawson, A.D.; et al. Extending the half-life of a fab fragment through generation of a humanized anti-human serum albumin Fv domain: An investigation into the correlation between affinity and serum half-life. MAbs 2016, 8, 1336–1346. [Google Scholar] [CrossRef] [PubMed]
- Dave, E.; Adams, R.; Zaccheo, O.; Carrington, B.; Compson, J.E.; Dugdale, S.; Airey, M.; Malcolm, S.; Hailu, H.; Wild, G.; et al. Fab-dsFv: A bispecific antibody format with extended serum half-life through albumin binding. MAbs 2016, 8, 1319–1335. [Google Scholar] [CrossRef] [PubMed]
- Metz, S.; Panke, C.; Haas, A.K.; Schanzer, J.; Lau, W.; Croasdale, R.; Hoffmann, E.; Schneider, B.; Auer, J.; Gassner, C.; et al. Bispecific antibody derivatives with restricted binding functionalities that are activated by proteolytic processing. Protein Eng. Des. Sel. 2012, 25, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.G.; Bautista, C.; Pong, E.; Nguyen, D.H.; Jacinto, J.; Eivazi, A.; Muchhal, U.S.; Karki, S.; Chu, S.Y.; Lazar, G.A. A novel bispecific antibody format enables simultaneous bivalent and monovalent co-engagement of distinct target antigens. MAbs 2011, 3, 546–557. [Google Scholar] [CrossRef] [PubMed]
- Coloma, M.J.; Morrison, S.L. Design and production of novel tetravalent bispecific antibodies. Nat. Biotechnol. 1997, 15, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Croasdale, R.; Wartha, K.; Schanzer, J.M.; Kuenkele, K.P.; Ries, C.; Mayer, K.; Gassner, C.; Wagner, M.; Dimoudis, N.; Herter, S.; et al. Development of tetravalent IgG1 dual targeting IGF-1R-EGFR antibodies with potent tumor inhibition. Arch. Biochem. Biophys. 2012, 526, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Michaelson, S.J.; Demarest, S.J.; Miller, B.; Amatucci, A.; Snyder, W.B.; Wu, X.; Huang, F.; Phan, S.; Gao, S.; Doern, A.; et al. Anti-tumor activity of stability-engineered IgG-like bispecific antibodies targeting TRAIL-R2 and LTbetaR. MAbs 2009, 1, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Orcutt, D.K.; Ackerman, M.E.; Cieslewicz, M.; Quiroz, E.; Slusarczyk, A.L.; Frangioni, J.V.; Wittrup, K.D. A modular IgG-scFv bispecific antibody topology. Protein Eng. Des. Sel. 2010, 23, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Schanzer, J.; Jekle, A.; Nezu, J.; Lochner, A.; Croasdale, R.; Dioszegi, M.; Zhang, J.; Hoffmann, E.; Dormeyer, W.; Stracke, J.; et al. Development of tetravalent, bispecific CCR5 antibodies with antiviral activity against CCR5 monoclonal antibody-resistant HIV-1 strains. Antimicrob. Agents Chemother. 2011, 55, 2369–2378. [Google Scholar] [CrossRef] [PubMed]
- Yazaki, J.P.; Lee, B.; Channappa, D.; Cheung, C.W.; Crow, D.; Chea, J.; Poku, E.; Li, L.; Andersen, J.T.; Sandlie, I.; et al. A series of anti-CEA/anti-DOTA bispecific antibody formats evaluated for pre-targeting: Comparison of tumor uptake and blood clearance. Protein Eng. Des. Sel. 2013, 26, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Heads, T.J.; Adams, R.; D’Hooghe, L.E.; Page, M.J.; Humphreys, D.P.; Popplewell, A.G.; Lawson, A.D.; Henry, A.J. Relative stabilities of IgG1 and IgG4 Fab domains: Influence of the light-heavy interchain disulfide bond architecture. Protein Sci. 2012, 21, 1315–1322. [Google Scholar] [CrossRef] [PubMed]
- Desplancq, D.; King, D.J.; Lawson, A.D.; Mountain, A. Multimerization behaviour of single chain Fv variants for the tumour-binding antibody B72.3. Protein Eng. Des. Sel. 1994, 7, 1027–1033. [Google Scholar] [CrossRef]
- Reiter, Y.; Brinkmann, U.; Kreitman, R.J.; Jung, S.H.; Lee, B.; Pastan, I. Stabilization of the Fv fragments in recombinant immunotoxins by disulfide bonds engineered into conserved framework regions. Biochemistry 1994, 33, 5451–5459. [Google Scholar] [CrossRef] [PubMed]
- Rothlisberger, D.; Honegger, A.; Pluckthun, A. Domain interactions in the Fab fragment: A comparative evaluation of the single-chain Fv and Fab format engineered with variable domains of different stability. J. Mol. Biol. 2005, 347, 773–789. [Google Scholar] [CrossRef] [PubMed]
- Schlapschy, M.; Theobald, I.; Mack, H.; Schottelius, M.; Wester, H.J.; Skerra, A. Fusion of a recombinant antibody fragment with a homo-amino-acid polymer: Effects on biophysical properties and prolonged plasma half-life. Protein Eng. Des. Sel. 2007, 20, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Abhinandan, R.K.; Martin, A.C. Analysis and improvements to Kabat and structurally correct numbering of antibody variable domains. Mol. Immunol. 2008, 45, 3832–3839. [Google Scholar] [CrossRef] [PubMed]
- Egan, J.T.; Diem, D.; Weldon, R.; Neumann, T.; Meyer, S.; Urech, D.M. Novel multispecific heterodimeric antibody format allowing modular assembly of variable domain fragments. MAbs 2017, 9, 68–84. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.T.; Park, C.S.; Jang, J.; Kim, M.R.; Na, H.J.; Lee, K.; Kim, H.J.; Heo, K.; Yoo, B.C.; Kim, Y.M.; et al. Inhibition of VEGF-dependent angiogenesis and tumor angiogenesis by an optimized antibody targeting CLEC14a. Mol. Oncol. 2018, 12, 356–372. [Google Scholar] [CrossRef] [PubMed]
- Ewert, S.; Honegger, A.; Pluckthun, A. Structure-based improvement of the biophysical properties of immunoglobulin VH domains with a generalizable approach. Biochemistry 2003, 42, 1517–1528. [Google Scholar] [CrossRef] [PubMed]
- Ewert, S.; Honegger, A.; Pluckthun, A. Stability improvement of antibodies for extracellular and intracellular applications: CDR grafting to stable frameworks and structure-based framework engineering. Methods 2004, 34, 184–199. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.B.; Demarest, S.J.; Lugovskoy, A.; Huang, F.; Wu, X.; Snyder, W.B.; Croner, L.J.; Wang, N.; Amatucci, A.; Michaelson, J.S.; et al. Stability engineering of scFvs for the development of bispecific and multivalent antibodies. Protein Eng. Des. Sel. 2010, 23, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Fennell, J.B.; McDonnell, B.; Tam, A.S.; Chang, L.; Steven, J.; Broadbent, I.D.; Gao, H.; Kieras, E.; Alley, J.; Luxenberg, D.; et al. CDR-restricted engineering of native human scFvs creates highly stable and soluble bifunctional antibodies for subcutaneous delivery. MAbs 2013, 5, 882–895. [Google Scholar] [CrossRef] [PubMed]
| dsFv | dsscFv | |||||||
|---|---|---|---|---|---|---|---|---|
| Expression (μg/mL) | μg Final Product/mL Expression Culture | Monomer (%) | Tm (°C) | Expression (μg/mL) | μg Final Product/mL Expression Culture | Monomer (%) | Tm (°C) | |
| #1 | <LOD | 2.4 | 44.4 ± 5.8 # | 59.5 ± 0.9 * | 14.9 ± 1.3 | 6.4 | 97.0 ± 3.4 | 57.8 ± 0.7 |
| #2 | 29.4 ± 1.3 | 12.8 | 100.0 ± 0.0 | 75.2 ± 0.2 | 58.3 ± 5.7 | 27.0 | 100.0 ± 0.0 | 76.9 ± 0.4 |
| #3 | 37.6 ± 0.4 | 11.7 | 99.4 ± 1.1 | 66.8 ± 0.3 | 47.3 ± 8.7 | 18.1 | 99.5 ± 1.0 | 61.2 ± 0.6 |
| #4 | <LOD | 1.3 | 40.3 ± 5.5 # | 75.4 ± 0.2 * | 15.3 ± 1.4 | 4.1 | 93.4 ± 1.2 | 74.7 ± 0.0 |
| dsFv/dsscFv | Fab-dsFv | Fab-dsscFv | ||||||
|---|---|---|---|---|---|---|---|---|
| Expression (μg/mL) | Monomer (%) | Fab Tm (°C) | dsFv Tm (°C) | Expression (μg/mL) | Monomer (%) | Fab Tm (°C) | dsscFv Tm (°C) | |
| #1 | 23.4 ± 3.1 | 87.5 ± 0.5 | 78.7 ± 0.2 | 58.4 ± 0.1 | 20.0 ± 2.9 | 91.2 ± 0.2 | 78.8 ± 0.2 | 59.0 ± 1.2 |
| #2 | 28.1 ± 2.2 | 37.8 ± 1.0 | 78.8 ± 0.0 | 72.4 ± 0.9 | 21.7 ± 1.0 | 70.9 ± 1.6 | 78.9 ± 0.4 | 73.1 ± 0.9 |
| #3 | 24.5.0 ± 2.1 | 83.7 ± 1.7 | 78.8 ± 0.2 | 62.5 ± 0.5 | 15.8 ± 0.1 | 91.8 ± 0.5 | 78.8 ± 0.3 | 61.4 ± 0.9 |
| #4 | 15.1 ± 2.6 | 76.0 ± 0.4 | 78.8 ± 0.2 | 73.6 ± 0.3 | 21.8 ± 3.9 | 79.9 ± 0.8 | 79.0 ± 0.4 | 72.7 ± 0.3 |
| dsFv/dsscFv | Fab-dsFv | Fab-dsscFv | ||||||
|---|---|---|---|---|---|---|---|---|
| Expression (μg/mL) | Monomer (%) | Fab Tm (°C) | dsFv Tm (°C) | Expression (μg/m) | Monomer (%) | Fab Tm (°C) | dsscFv Tm (°C) | |
| #1 * | 23.4 ± 3.1 | 87.5 ± 0.5 | 78.7 ± 0.2 | 58.4 ± 0.1 | 20.0 ± 2.9 | 91.2 ± 0.2 | 78.8 ± 0.2 | 59.0 ± 1.2 |
| #2 * | 28.1 ± 2.2 | 37.8 ± 1.0 | 78.8 ± 0.0 | 72.4 ± 0.9 | 21.7 ± 1.0 | 70.9 ± 1.6 | 78.9 ± 0.4 | 73.1 ± 0.9 |
| FW#1/CDR#2 | 34.0 ± 2.1 | 47.1 ± 1.3 | 79.1 ± 0.4 | 68.4 ± 0.4 | 28.5 ± 1.4 | 90.1 ± 0.2 | 79.1 ± 0.3 | 72.4 ± 0.2 |
| FW#2/CDR#1 | 10.6 ± 0.3 | 91.6 ± 0.4 | 80.8 ± 0.5 | 53.7 ± 1.2 | 6.7 ± 0.6 | 91.9 ± 0.5 | 81.5 ± 0.1 | 53.6 ± 1.4 |
| dsFv/dsscFv | IgG(H)-dsscFv | |
|---|---|---|
| Expression (μg/mL) | Monomer (%) | |
| #1 | 22.0 ± 2.4 | 92.8 ± 0.6 |
| #2 | 24.9 ± 0.6 | 75.7 ± 2.3 |
| FW#1/CDR#2 | 39.1 ± 0.6 | 96.3 ± 1.0 |
| FW#2/CDR#1 | 3.7 ± 0.4 | 94.6 ± 0.6 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhatta, P.; Humphreys, D.P. Relative Contribution of Framework and CDR Regions in Antibody Variable Domains to Multimerisation of Fv- and scFv-Containing Bispecific Antibodies. Antibodies 2018, 7, 35. https://doi.org/10.3390/antib7030035
Bhatta P, Humphreys DP. Relative Contribution of Framework and CDR Regions in Antibody Variable Domains to Multimerisation of Fv- and scFv-Containing Bispecific Antibodies. Antibodies. 2018; 7(3):35. https://doi.org/10.3390/antib7030035
Chicago/Turabian StyleBhatta, Pallavi, and David P. Humphreys. 2018. "Relative Contribution of Framework and CDR Regions in Antibody Variable Domains to Multimerisation of Fv- and scFv-Containing Bispecific Antibodies" Antibodies 7, no. 3: 35. https://doi.org/10.3390/antib7030035
APA StyleBhatta, P., & Humphreys, D. P. (2018). Relative Contribution of Framework and CDR Regions in Antibody Variable Domains to Multimerisation of Fv- and scFv-Containing Bispecific Antibodies. Antibodies, 7(3), 35. https://doi.org/10.3390/antib7030035




