Genetic Fusion of an Anti-BclA Single-Domain Antibody with Beta Galactosidase
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Periplasmic and Cytoplasmic Production of sdAbs
2.3. Surface Plasmon Resonance
2.4. Determining Melting Temperature by Fluorescent Dye Melt Assay
2.5. Producing Fusion of sdAbs with β-gal
2.6. ELISA
3. Results
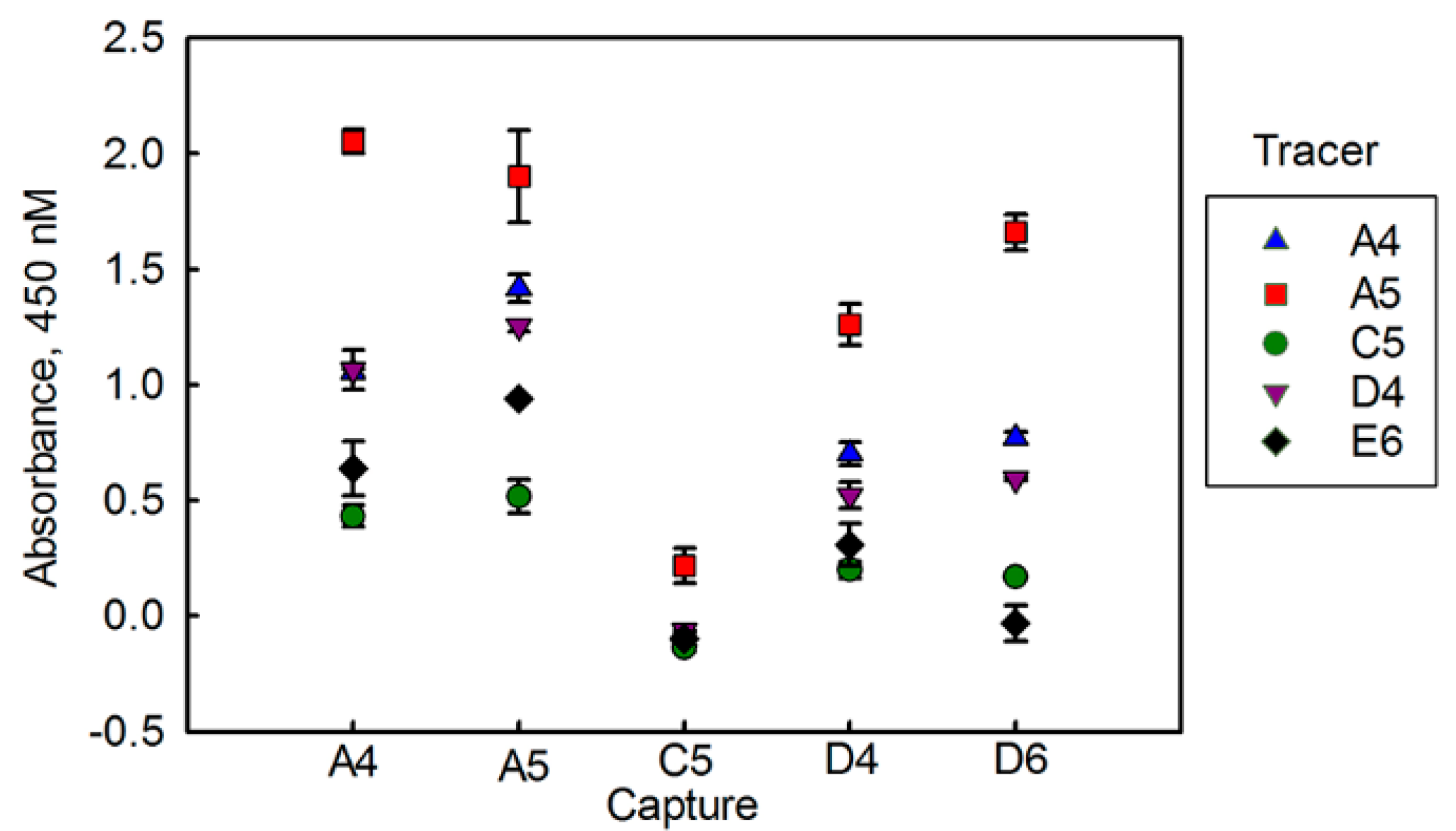
3.1. Evaluating BclA Binding sdAbs
3.2. Construction and Evaluation of β-gal Fusion
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
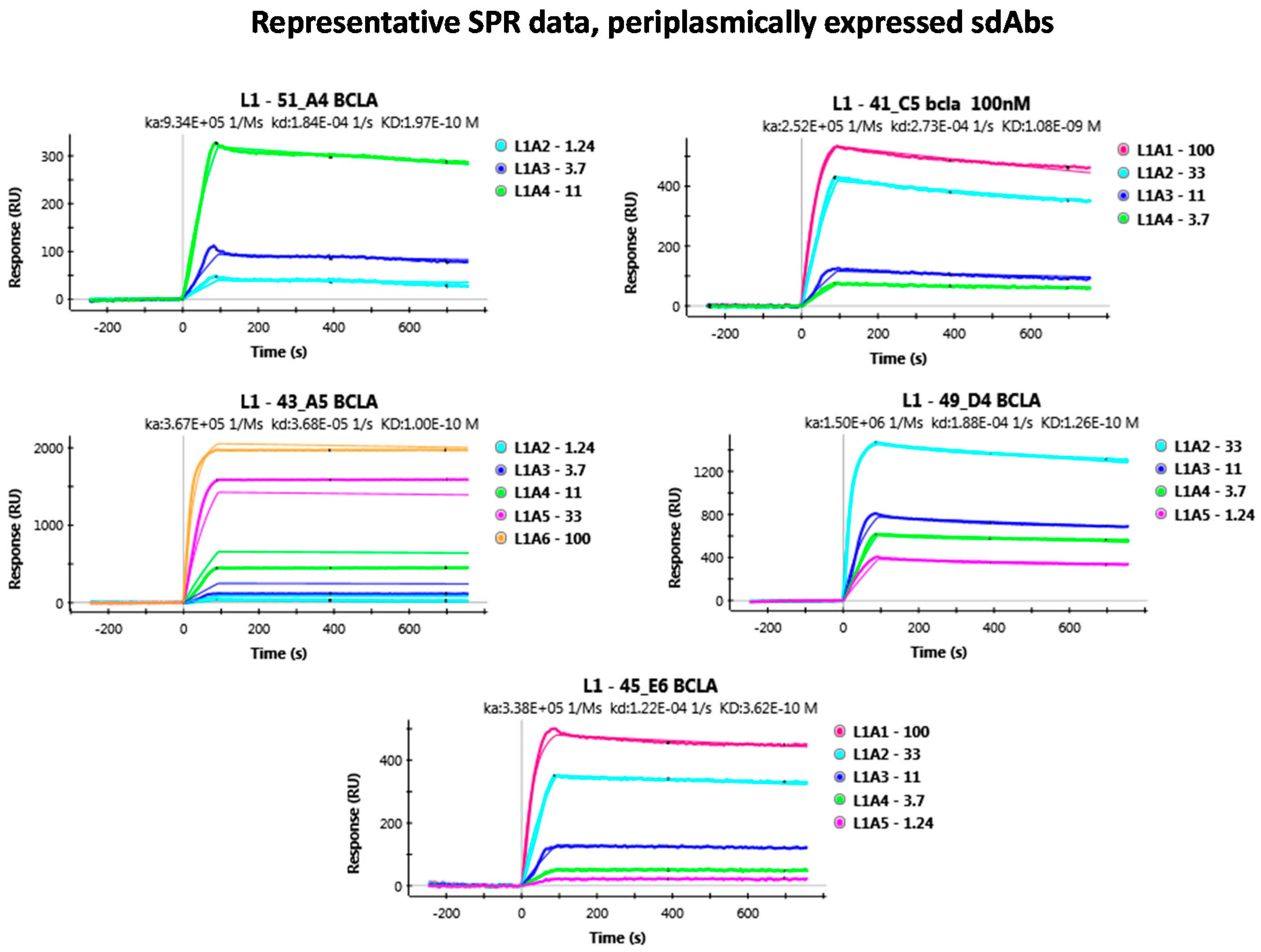
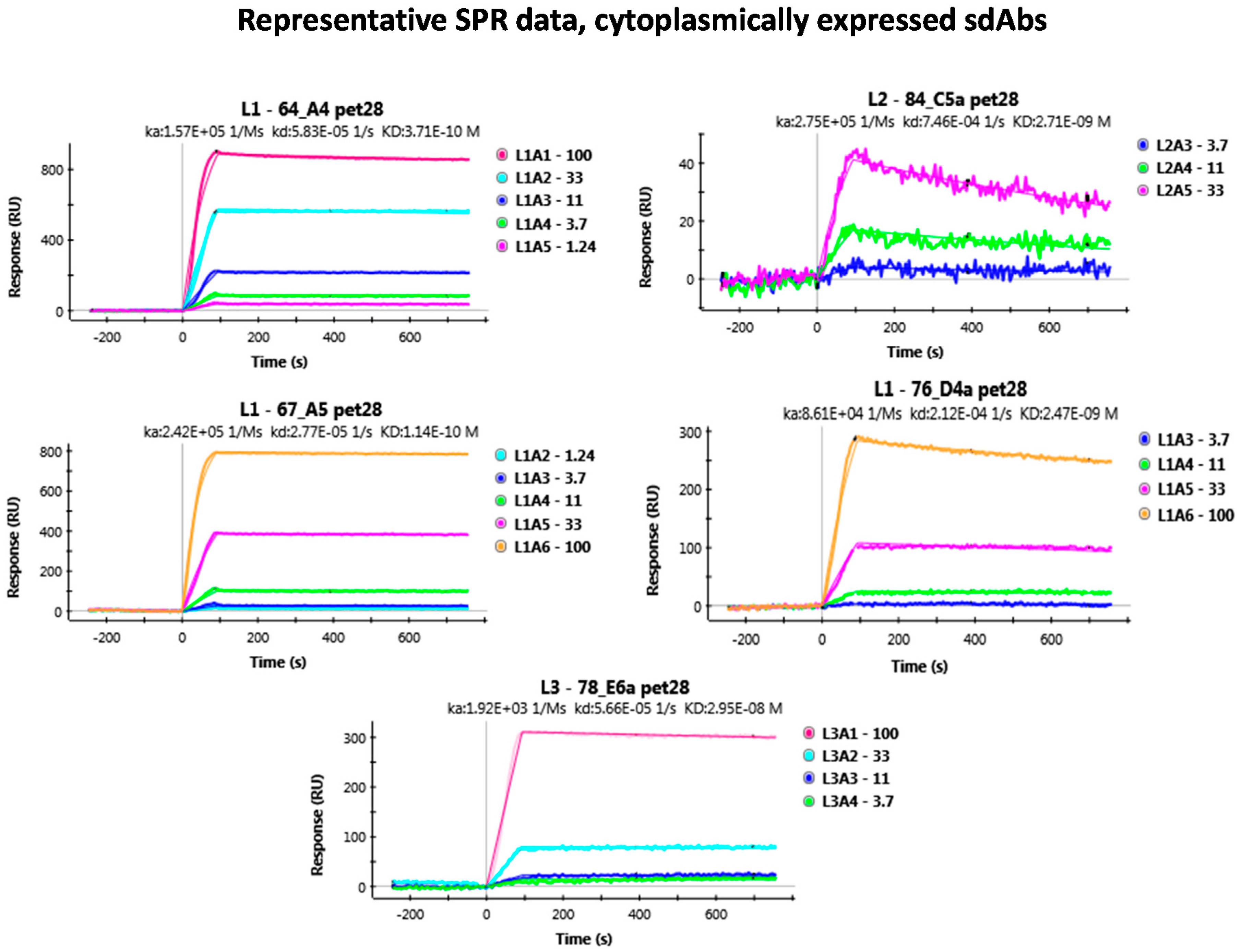
Appendix B
| SdAb | Production | ka (1/Ms) | kd (1/s) | KD (M) |
| A4 | periplasmic | 9.1 × 105/2.2 × 104 | 1.4 × 10−4/4.1 × 10−5 | 1.6 × 10−10/4.1 × 10−11 |
| cytoplasmic | 1.6 × 105/1.5 × 103 | 6.6 × 10−5/9.5 × 10−6 | 4.3 × 10−10/5.6 × 10−11 | |
| A5 | periplasmic | 3.6 × 105/9.5 × 103 | 3.7 × 10−5/7.0 × 10−7 | 1.0 × 10−10/5.0 × 10−12 |
| cytoplasmic | 2.2 × 105/2.2 × 104 | 2.4 × 10−5/3.7 × 10−6 | 1.1 × 10−10/6.0 × 10−12 | |
| C5 | periplasmic | 2.0 × 105/5.3 × 104 | 2.2 × 10−4/5.3 × 10−5 | 1.1 × 10−9/2.5 × 10−11 |
| cytoplasmic | 3.0 × 105/2.8 × 104 | 6.9 × 10−4/5.4 × 10−5 | 2.4 × 10−9/3.0 × 10−10 | |
| D4 | periplasmic | 1.5 × 106/3.0 × 104 | 2.0 × 10−4/1.3 × 10−5 | 1.4 × 10−10/1.1 × 10−11 |
| cytoplasmic | 9.5 × 104/9.0 × 103 | 2.3 × 10−4/2.1 × 10−5 | 2.5 × 10−9/1.0 × 10−11 | |
| E6 | periplasmic | 3.0 × 105/3.2 × 104 | 1.2 × 10−4/4.0 × 10−6 | 4.0 × 10−10/3.3 × 10−11 |
| cytoplasmic | 9.8 × 102/9.4 × 102 | 4.7 × 10−5/9.0 × 10−6 | 6.0 × 10−7/5.7 × 10−7 |
References
- Spencer, R.C. Bacillus anthracis. J. Clin. Pathol. 2003, 56, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Inglesby, T.V.; O’Toole, T.; Henderson, D.A.; Bartlett, J.G.; Ascher, M.S.; Eitzen, E.; Friedlander, A.M.; Gerberding, J.; Hauer, J.; Hughes, J.; et al. Anthrax as a biological weapon, 2002: Updated recommendations for management. JAMA 2002, 287, 2236–2252. [Google Scholar] [CrossRef] [PubMed]
- Buhr, T.L.; Young, A.A.; Barnette, H.K.; Minter, Z.A.; Kennihan, N.L.; Johnson, C.A.; Bohmke, M.D.; DePaola, M.; Cora-Laó, M.; Page, M.A. Test methods and response surface models for hot, humid air decontamination of materials contaminated with dirty spores of bacillus anthracis ∆sterne and bacillus thuringiensis al hakam. J. Appl. Microbiol. 2015, 119, 1263–1277. [Google Scholar] [CrossRef] [PubMed]
- Buhr, T.L.; Young, A.A.; Bensman, M.; Minter, Z.A.; Kennihan, N.L.; Johnson, C.A.; Bohmke, M.D.; Borgers-Klonkowski, E.; Osborn, E.B.; Avila, S.D.; et al. Hot, humid air decontamination of a c-130 aircraft contaminated with spores of two acrystalliferous bacillus thuringiensis strains, surrogates for bacillus anthracis. J. Appl. Microbiol. 2016, 120, 1074–1084. [Google Scholar] [CrossRef] [PubMed]
- Walper, S.A.; Anderson, G.P.; Brozozog Lee, P.A.; Glaven, R.H.; Liu, J.L.; Bernstein, R.D.; Zabetakis, D.; Johnson, L.; Czarnecki, J.M.; Goldman, E.R. Rugged single domain antibody detection elements for bacillus anthracis spores and vegetative cells. PLoS ONE 2012, 7, e32801. [Google Scholar] [CrossRef] [PubMed]
- Walper, S.A.; Lee, P.A.B.; Anderson, G.P.; Goldman, E.R. Selection and characterization of single domain antibodies specific for bacillus anthracis spore proteins. Antibodies 2013, 2, 152–167. [Google Scholar] [CrossRef]
- Morel, N.; Volland, H.; Dano, J.; Lamourette, P.; Sylvestre, P.; Mock, M.; Creminon, C. Fast and sensitive detection of bacillus anthracis spores by immunoassay. Appl. Environ. Microbiol. 2012, 78, 6491–6498. [Google Scholar] [CrossRef] [PubMed]
- Kuehn, A.; Kovác, P.; Saksena, R.; Bannert, N.; Klee, S.R.; Ranisch, H.; Grunow, R. Development of antibodies against anthrose tetrasaccharide for specific detection of bacillus anthracis spores. Clin. Vaccine Immunol. CVI 2009, 16, 1728–1737. [Google Scholar] [CrossRef] [PubMed]
- Redmond, C.; Baillie, L.W.; Hibbs, S.; Moir, A.J.; Moir, A. Identification of proteins in the exosporium of bacillus anthracis. Microbiology 2004, 150, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Steichen, C.; Chen, P.; Kearney, J.F.; Turnbough, C.L., Jr. Identification of the immunodominant protein and other proteins of the bacillus anthracis exosporium. J. Bacteriol. 2003, 185, 1903–1910. [Google Scholar] [CrossRef] [PubMed]
- Sylvestre, P.; Couture-Tosi, E.; Mock, M. A collagen-like surface glycoprotein is a structural component of the bacillus anthracis exosporium. Mol. Microbiol. 2002, 45, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Todd, S.J.; Moir, A.J.; Johnson, M.J.; Moir, A. Genes of bacillus cereus and bacillus anthracis encoding proteins of the exosporium. J. Bacteriol. 2003, 185, 3373–3378. [Google Scholar] [CrossRef] [PubMed]
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hamers, C.; Songa, E.B.; Bendahman, N.; Hamers, R. Naturally occurring antibodies devoid of light chains. Nature 1993, 363, 446–448. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, A.S.; Avila, D.; Hughes, M.; Hughes, A.; McKinney, E.C.; Flajnik, M.F. A new antigen receptor gene family that undergods rearrangement and extensive somatice diversificatino in sharks. Nature 1995, 374, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Ghahroudi, M.A.; Desmyter, A.; Wyns, L.; Hamers, R.; Muyldermans, S. Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS Lett. 1997, 414, 521–526. [Google Scholar] [CrossRef]
- Muyldermans, S. Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef] [PubMed]
- De Marco, A. Biotechnological applications of recombinant single-domain antibody fragments. Microb. Cell Fact. 2011, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- Wesolowski, J.; Alzogaray, V.; Reyelt, J.; Unger, M.; Juarez, K.; Urrutia, M.; Cauerhff, A.; Danquah, W.; Rissiek, B.; Scheuplein, F.; et al. Single domain antibodies: Promising experimental and therapeutic tools in infection and immunity. Med. Microbiol. Immunol. 2009, 198, 157–174. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, L.J.; Osborn, L.E.; Carrion, R.; Patterson, J.L.; Hayhurst, A. Rapid assembly of sensitive antigen-capture assays for marburg virus, using in vitro selection of llama single-domain antibodies, at biosafety level 4. J. Infect. Dis. 2007, 196, S213–S219. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.L.; Zabetakis, D.; Brozozog Lee, P.A.; Goldman, E.R.; Anderson, G.P. Single domain antibody alkaline phosphatase fusion proteins for antigen detection—Analysis of affinity and thermal stability of single domain antibody. J. Immunol. Methods 2013, 393, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Walper, S.A.; Battle, S.R.; Lee, P.A.B.; Zabetakis, D.; Turner, K.D.; Buckley, P.E.; Calm, A.M.; Welsh, H.S.; Warner, C.R.; Zacharko, M.A.; et al. Thermostable single domain antibody–maltose binding protein fusion for Bacillus anthracis spore protein bcla detection. Anal. Biochem. 2014, 447, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.L.; Zabetakis, D.; Walper, S.A.; Goldman, E.R.; Anderson, G.P. Bioconjugates of rhizavidin with single domain antibodies as bifunctional immunoreagents. J. Immunol. Methods 2014, 411, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Pleschberger, M.; Saerens, D.; Weigert, S.; Sleytr, U.B.; Muyldermans, S.; Sara, M.; Egelseer, E.M. An s-layer heavy chain camel antibody fusion protein for generation of a nanopatterned sensing layer to detect the prostate-specific antigen by surface plasmon resonance technology. Bioconjugate Chem. 2004, 15, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, L.J.; Hayhurst, A. Hapten mediated display and pairing of recombinant antibodies accelerates assay assembly for biothreat countermeasures. Sci. Rep. 2012, 2, 807. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xu, Y.; Wan, D.-B.; Xiong, Y.-H.; He, Z.-Y.; Wang, X.-X.; Gee, S.J.; Ryu, D.; Hammock, B.D. Development of a nanobody–alkaline phosphatase fusion protein and its application in a highly sensitive direct competitive fluorescence enzyme immunoassay for detection of ochratoxin a in cereal. Anal. Chem. 2015, 87, 1387–1394. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, L.J.; Hayhurst, A. Ebolavirus nucleoprotein C-termini potently attract single domain antibodies enabling monoclonal affinity reagent sandwich assay (MARSA) formulation. PLoS ONE 2013, 8, e61232. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gurlo, T.; von Grafenstein, H. Cell-elisa using β-galactosidase conjugated antibodies. J. Immunol. Methods 2000, 234, P153–P167. [Google Scholar] [CrossRef]
- Gaylord, S.T.; Dinh, T.L.; Goldman, E.R.; Anderson, G.P.; Ngan, K.C.; Walt, D.R. Ultrasensitive detection of ricin toxin in multiple sample matrixes using single-domain antibodies. Anal. Chem. 2015, 87, 6570–6577. [Google Scholar] [CrossRef] [PubMed]
- Alcalá, P.; Ferrer-Miralles, N.; Villaverde, A. Engineering of Escherichia coli β-galactosidase for solvent display of a functional scfv antibody fragment. FEBS Lett. 2003, 533, 115–118. [Google Scholar] [CrossRef]
- Snyder, W.B.; Silhavy, T.J. Beta-galactosidase is inactivated by intermolecular disulfide bonds and is toxic when secreted to the periplasm of Escherichia coli. J. Bacteriol. 1995, 177, 953–963. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, R.S.; Malinverni, J.C.; Boyd, D.; Beckwith, J.; Silhavy, T.J. Folding lacz in the periplasm of Escherichia coli. J. Bacteriol. 2014, 196, 3343–3350. [Google Scholar] [CrossRef] [PubMed]
- Corpet, F. Multiple sequence alignment with hierarchical-clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Walper, S.A.; Liu, J.L.; Zabetakis, D.; Anderson, G.P.; Goldman, E.R. Development and evaluation of single domain antibodies for vaccinia and the L1 antigen. PLoS ONE 2014, 9, e106263. [Google Scholar] [CrossRef] [PubMed]
- Shriver-Lake, L.C.; Zabetakis, D.; Goldman, E.R.; Anderson, G.P. Evaluation of anti-botulinum neurotoxin single domain antibodies with additional optimization for improved production and stability. Toxicon 2017, 135, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.L.; Zabetakis, D.; Goldman, E.R.; Anderson, G.P. Selection and evaluation of single domain antibodies toward ms2 phage and coat protein. Mol. Immunol. 2013, 53, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Brown Iii, C.W.; Oh, E.; Hastman, D.A.; Walper, S.A.; Susumu, K.; Stewart, M.H.; Deschamps, J.R.; Medintz, I.L. Kinetic enhancement of the diffusion-limited enzyme beta-galactosidase when displayed with quantum dots. RSC Adv. 2015, 5, 93089–93094. [Google Scholar] [CrossRef]
- Anderson, G.P.; Bernstein, R.D.; Swain, M.D.; Zabetakis, D.; Goldman, E.R. Binding kinetics of antiricin single domain antibodies and improved detection using a b chain specific binder. Anal. Chem. 2010, 82, 7202–7207. [Google Scholar] [CrossRef] [PubMed]
- Goldman, E.R.; Brozozog-Lee, P.A.; Zabetakis, D.; Turner, K.B.; Walper, S.A.; Liu, J.L.; Anderson, G.P. Negative tail fusions can improve ruggedness of single domain antibodies. Protein Expr. Purif. 2014, 95, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Lavinder, J.J.; Hari, S.B.; Sullivan, B.J.; Magliery, T.J. High-throughput thermal scanning: A general, rapid dye-binding thermal shift screen for protein engineering. J. Am. Chem. Soc. 2009, 131, 3794–3795. [Google Scholar] [CrossRef] [PubMed]
- McConnell, A.D.; Spasojevich, V.; Macomber, J.L.; Krapf, I.P.; Chen, A.; Sheffer, J.C.; Berkebile, A.; Horlick, R.A.; Neben, S.; King, D.J.; et al. An integrated approach to extreme thermostabilization and affinity maturation of an antibody. Protein Eng. Des. Sel. 2013, 26, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Hagihara, Y.; Mine, S.; Uegaki, K. Stabilization of an immunoglobulin fold domain by an engineered disulfide bond at the buried hydrophobic region. J. Biol. Chem. 2007, 282, 36489–36495. [Google Scholar] [CrossRef] [PubMed]
- Saerens, D.; Conrath, K.; Govaert, J.; Muyldermans, S. Disulfide bond introduction for general stabilization of immunoglobulin heavy-chain variable domains. J. Mol. Biol. 2008, 377, 478–488. [Google Scholar] [CrossRef] [PubMed]
- George, J.; Compton, J.R.; Leary, D.H.; Olson, M.A.; Legler, P.M. Structural and mutational analysis of a monomeric and dimeric form of a single domain antibody with implications for protein misfolding. Proteins Struct. Funct. Bioinform. 2014, 82, 3101–3116. [Google Scholar] [CrossRef] [PubMed]
- Saerens, D.; Pellis, M.; Loris, R.; Pardon, E.; Dumoulin, M.; Matagne, A.; Wyns, L.; Muyldermans, S.; Conrath, K. Identification of a universal vhh framework to graft non-canonical antigen-binding loops of camel single-domain antibodies. J. Mol. Biol. 2005, 352, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Veggiani, G.; de Marco, A. Improved quantitative and qualitative production of single-domain intrabodies mediated by the co-expression of erv1p sulfhydryl oxidase. Protein Expr. Purif. 2011, 79, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Zarschler, K.; Witecy, S.; Kapplusch, F.; Foerster, C.; Stephan, H. High-yield production of functional soluble single-domain antibodies in the cytoplasm of Escherichia coli. Microb. Cell Fact. 2013, 12, 97. [Google Scholar] [CrossRef] [PubMed]
- Djender, S.; Schneider, A.; Beugnet, A.; Crepin, R.; Desrumeaux, K.E.; Romani, C.; Moutel, S.; Perez, F.; de Marco, A. Bacterial cytoplasm as an effective cell compartment for producing functional vhh-based affinity reagents and camelidae igg-like recombinant antibodies. Microb. Cell Fact. 2014, 13, 140. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hayman, R.B.; Walt, D.R. Detection of single-molecule DNA hybridization using enzymatic amplification in an array of femtoliter-sized reaction vessels. J. Am. Chem. Soc. 2008, 130, 12622–12623. [Google Scholar] [CrossRef] [PubMed]
- Walt, D.R. Optical methods for single molecule detection and analysis. Anal. Chem. 2013, 85, 1258–1263. [Google Scholar] [CrossRef] [PubMed]
- Walt, D.R. Protein measurements in microwells. Lab Chip 2014, 14, 3195–3200. [Google Scholar] [CrossRef] [PubMed]
- Dinh, T.L.; Ngan, K.C.; Shoemaker, C.B.; Walt, D.R. Rapid and ultrasensitive detection of botulinum neurotoxin serotype A1 in human serum and urine using single-molecule array method. Forensic Toxicol. 2017, 35, 179–184. [Google Scholar] [CrossRef]

| SdAb | Production | Yield (mg/L) | ka (1/Ms) | kd (1/s) | KD (M) | Tm (°C) |
|---|---|---|---|---|---|---|
| A4 | periplasmic | 15 | 9.3 × 105 | 1.8 × 10−4 | 2.0 × 10−10 | 57 |
| cytoplasmic | 0.3 | 1.6 × 105 | 5.8 × 10−5 | 3.7 × 10−10 | 58 | |
| A5 | periplasmic | 7 | 3.7 × 105 | 3.7 × 10−5 | 1.0 × 10−10 | 67 |
| cytoplasmic | 6 | 2.4 × 105 | 2.8 × 10−5 | 1.1 × 10−10 | 56 | |
| C5 | periplasmic | 6 | 2.5 × 105 | 2.7 × 10−4 | 1.1 × 10−9 | 58 |
| cytoplasmic | 4 | 2.8 × 105 | 7.5 × 10−4 | 2.7 × 10−9 | 45 | |
| D4 | periplasmic | 11 | 1.5 × 106 | 1.9 × 10−4 | 1.3 × 10−10 | 67 |
| cytoplasmic | 3 | 8.6 × 104 | 2.1 × 10−4 | 2.5 × 10−9 | 50 | |
| E6 | periplasmic | 20 | 3.4 × 105 | 1.2 × 10−4 | 3.6 × 10−10 | 60 |
| cytoplasmic | 5 | 1.9 × 103 | 5.7 × 10−5 | 2.9 × 10−8 | 59 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anderson, G.P.; Shriver-Lake, L.C.; Walper, S.A.; Ashford, L.; Zabetakis, D.; Liu, J.L.; Breger, J.C.; Brozozog Lee, P.A.; Goldman, E.R. Genetic Fusion of an Anti-BclA Single-Domain Antibody with Beta Galactosidase. Antibodies 2018, 7, 36. https://doi.org/10.3390/antib7040036
Anderson GP, Shriver-Lake LC, Walper SA, Ashford L, Zabetakis D, Liu JL, Breger JC, Brozozog Lee PA, Goldman ER. Genetic Fusion of an Anti-BclA Single-Domain Antibody with Beta Galactosidase. Antibodies. 2018; 7(4):36. https://doi.org/10.3390/antib7040036
Chicago/Turabian StyleAnderson, George P., Lisa C. Shriver-Lake, Scott A. Walper, Lauryn Ashford, Dan Zabetakis, Jinny L. Liu, Joyce C. Breger, P. Audrey Brozozog Lee, and Ellen R. Goldman. 2018. "Genetic Fusion of an Anti-BclA Single-Domain Antibody with Beta Galactosidase" Antibodies 7, no. 4: 36. https://doi.org/10.3390/antib7040036
APA StyleAnderson, G. P., Shriver-Lake, L. C., Walper, S. A., Ashford, L., Zabetakis, D., Liu, J. L., Breger, J. C., Brozozog Lee, P. A., & Goldman, E. R. (2018). Genetic Fusion of an Anti-BclA Single-Domain Antibody with Beta Galactosidase. Antibodies, 7(4), 36. https://doi.org/10.3390/antib7040036






