Comparison between Neutralization Capacity of Antibodies Elicited by COVID-19 Natural Infection and Vaccination in Indonesia: A Prospective Cohort
Abstract
:1. Introduction
2. Materials and Methods
3. Results
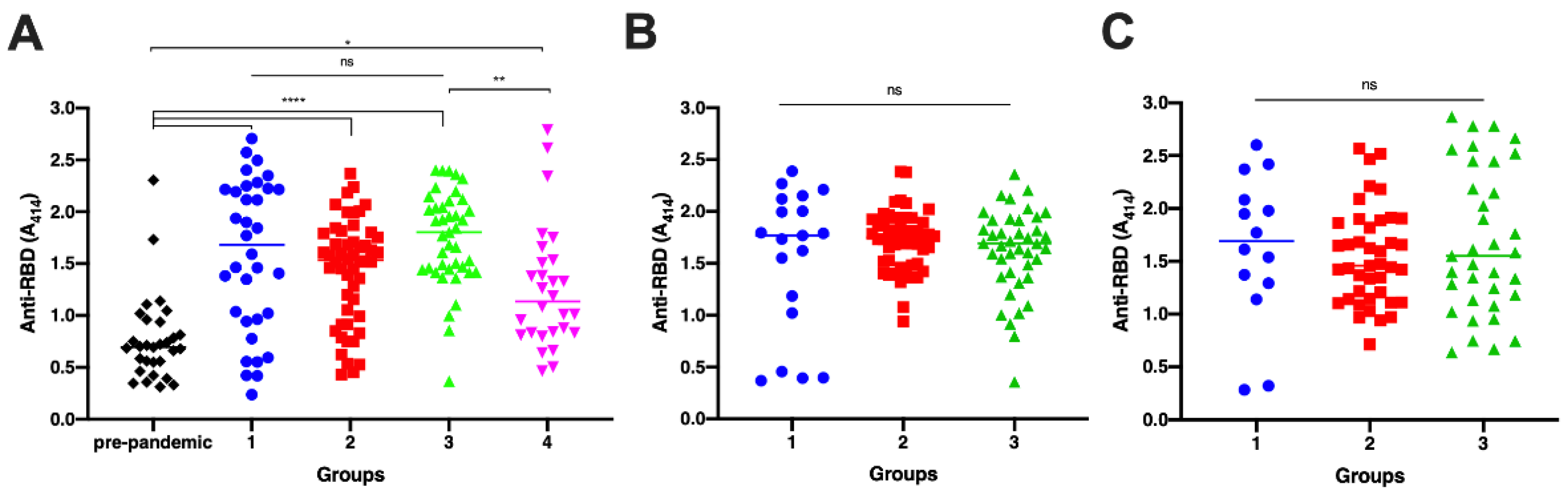
3.1. Antibody Titer Elicited by Vaccination and Natural Infection
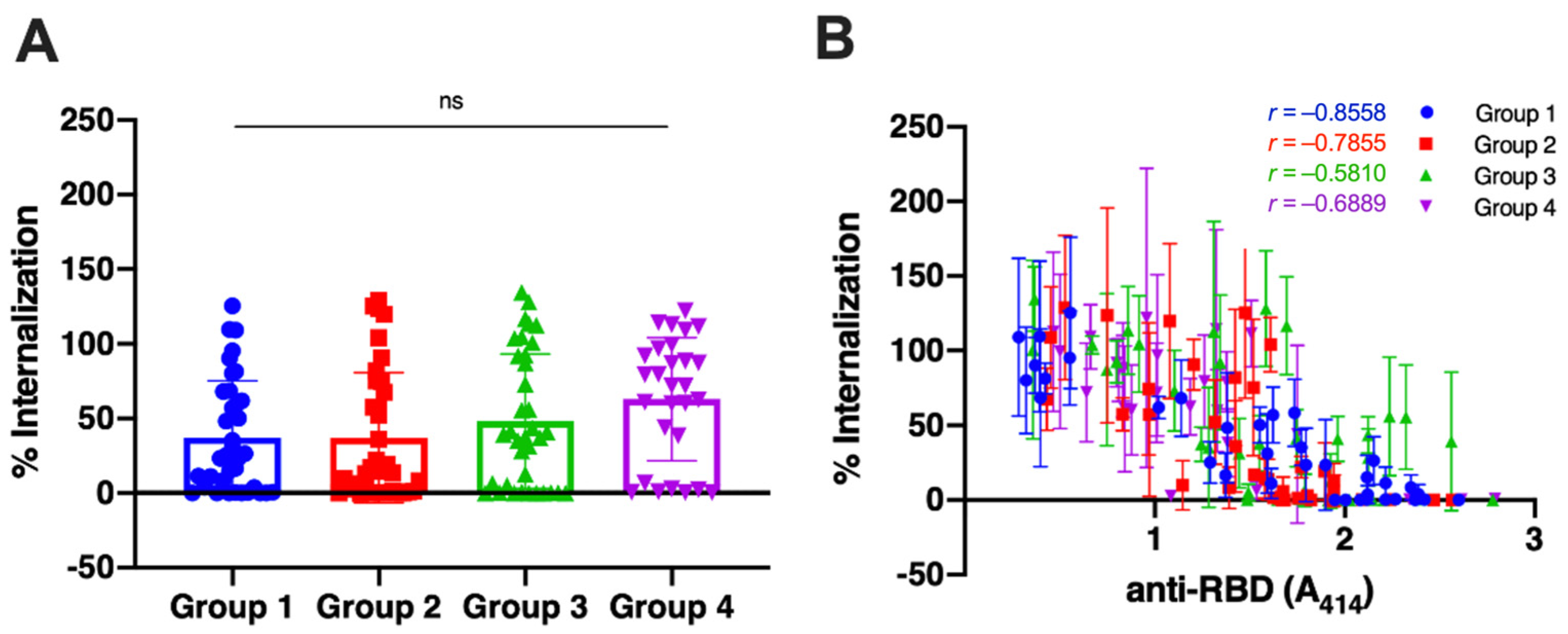
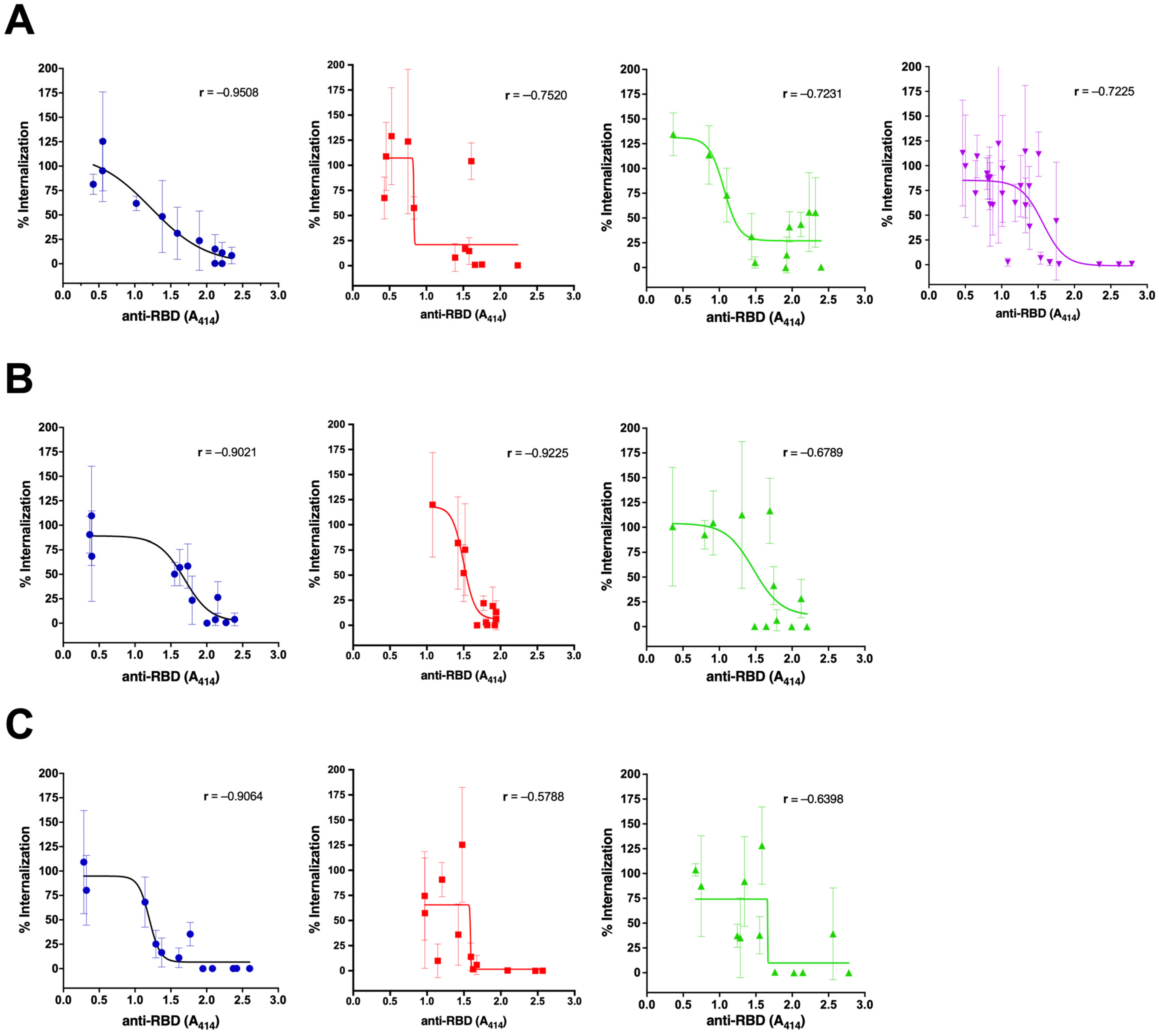
3.2. The Neutralization Capacity of the Antibody
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luo, J.; Brakel, A.; Krizsan, A.; Ludwig, T.; Mötzing, M.; Volke, D.; Lakowa, N.; Grünewald, T.; Lehmann, C.; Wolf, J.; et al. Sensitive and specific serological ELISA for the detection of SARS-CoV-2 infections. Virol. J. 2022, 19, 50. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Liu, L.; Wang, X.; Luo, N.; Li, L. Clinical Outcomes in 55 Patients With Severe Acute Respiratory Syndrome Coronavirus 2 Who Were Asymptomatic at Hospital Admission in Shenzhen, China. J. Infect. Dis. 2020, 221, 1770–1774. [Google Scholar] [CrossRef] [PubMed]
- Vabret, N.; Britton, G.J.; Gruber, C.; Hegde, S.; Kim, J.; Kuksin, M.; Levantovsky, R.; Malle, L.; Moreira, A.; Park, M.D.; et al. Immunology of COVID-19: Current State of the Science. Immunity 2020, 52, 910–941. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.M.; Heesterbeek, H.; Klinkenberg, D.; Hollingsworth, T.D. How will country-based mitigation measures influence the course of the COVID-19 epidemic? Lancet 2020, 395, 931–934. [Google Scholar] [CrossRef] [PubMed]
- Nicola, M.; Alsafi, Z.; Sohrabi, C.; Kerwan, A.; Al-Jabir, A.; Iosifidis, C.; Agha, M.; Agha, R. The socio-economic implications of the coronavirus pandemic (COVID-19): A review. Int. J. Surg. 2020, 78, 185–193. [Google Scholar] [CrossRef]
- Legido-Quigley, H.; Asgari, N.; Teo, Y.Y.; Leung, G.M.; Oshitani, H.; Fukuda, K.; Cook, A.R.; Hsu, L.Y.; Shibuya, K.; Heymann, D. Are high-performing health systems resilient against the COVID-19 epidemic? Lancet 2020, 395, 848–850. [Google Scholar] [CrossRef]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Raj, R. Analysis of non-structural proteins, NSPs of SARS-CoV-2 as targets for computational drug designing. Biochem. Biophys. Rep. 2021, 25, 100847. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Zeng, W.; Liu, G.; Ma, H.; Zhao, D.; Yang, Y.; Liu, M.; Mohammed, A.; Zhao, C.; Yang, Y.; Xie, J.; et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020, 527, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-K.; Sue, S.-C.; Yu, T.-H.; Hsieh, C.-M.; Tsai, C.-K.; Chiang, Y.-C.; Lee, S.-J.; Hsiao, H.-H.; Wu, W.-J.; Chang, W.-L.; et al. Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 2006, 13, 59–72. [Google Scholar] [CrossRef] [PubMed]
- McBride, R.; Van Zyl, M.; Fielding, B.C. The Coronavirus Nucleocapsid Is a Multifunctional Protein. Viruses 2014, 6, 2991–3018. [Google Scholar] [CrossRef] [PubMed]
- Hajissa, K.; Mussa, A.; Karobari, M.I.; Abbas, M.A.; Ibrahim, I.K.; Assiry, A.A.; Iqbal, A.; Alhumaid, S.; Mutair, A.A.; Rabaan, A.A.; et al. The SARS-CoV-2 Antibodies, Their Diagnostic Utility, and Their Potential for Vaccine Development. Vaccines 2022, 10, 1346. [Google Scholar] [CrossRef]
- Dhawan, M.; Sharma, A.; Priyanka; Thakur, N.; Rajkhowa, T.K.; Choudhary, O.P. Delta variant (B.1.617.2) of SARS-CoV-2: Mutations, impact, challenges and possible solutions. Hum. Vaccines Immunother. 2022, 18, 2068883. [Google Scholar] [CrossRef]
- Lau, E.H.Y.; Tsang, O.T.Y.; Hui, D.S.C.; Kwan, M.Y.W.; Chan, W.-H.; Chiu, S.S.; Ko, R.L.W.; Chan, K.H.; Cheng, S.M.S.; Perera, R.A.P.M.; et al. Neutralizing antibody titres in SARS-CoV-2 infections. Nat. Commun. 2021, 12, 63. [Google Scholar] [CrossRef]
- To, K.K.-W.; Tsang, O.T.-Y.; Leung, W.-S.; Tam, A.R.; Wu, T.-C.; Lung, D.C.; Yip, C.C.-Y.; Cai, J.-P.; Chan, J.M.-C.; Chik, T.S.-H.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef]
- Fafi-Kremer, S.; Bruel, T.; Madec, Y.; Grant, R.; Tondeur, L.; Grzelak, L.; Staropoli, I.; Anna, F.; Souque, P.; Fernandes-Pellerin, S.; et al. Serologic responses to SARS-CoV-2 infection among hospital staff with mild disease in eastern France. eBioMedicine 2020, 59, 102915. [Google Scholar] [CrossRef]
- Iyer, A.S.; Jones, F.K.; Nodoushani, A.; Kelly, M.; Becker, M.; Slater, D.; Mills, R.; Teng, E.; Kamruzzaman, M.; Garcia-Beltran, W.F.; et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci. Immunol. 2020, 5, eabe0367. [Google Scholar] [CrossRef]
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 584, 115–119. [Google Scholar] [CrossRef]
- L’Huillier, A.G.; Meyer, B.; Andrey, D.O.; Arm-Vernez, I.; Baggio, S.; Didierlaurent, A.; Eberhardt, C.S.; Eckerle, I.; Grasset-Salomon, C.; Huttner, A.; et al. Antibody persistence in the first 6 months following SARS-CoV-2 infection among hospital workers: A prospective longitudinal study. Clin. Microbiol. Infect. 2021, 27, e781–e784. [Google Scholar] [CrossRef] [PubMed]
- Addetia, A.; Crawford Katharine, H.D.; Dingens, A.; Zhu, H.; Roychoudhury, P.; Huang, M.-L.; Jerome Keith, R.; Bloom Jesse, D.; Greninger Alexander, L. Neutralizing Antibodies Correlate with Protection from SARS-CoV-2 in Humans during a Fishery Vessel Outbreak with a High Attack Rate. J. Clin. Microbiol. 2020, 58, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.T.; Garcia-Carreras, B.; Hitchings, M.D.T.; Yang, B.; Katzelnick, L.C.; Rattigan, S.M.; Borgert, B.A.; Moreno, C.A.; Solomon, B.D.; Trimmer-Smith, L.; et al. A systematic review of antibody mediated immunity to coronaviruses: Kinetics, correlates of protection, and association with severity. Nat. Commun. 2020, 11, 4704. [Google Scholar] [CrossRef]
- Lumley, S.F.; O’Donnell, D.; Stoesser, N.E.; Matthews, P.C.; Howarth, A.; Hatch, S.B.; Marsden, B.D.; Cox, S.; James, T.; Warren, F.; et al. Antibody Status and Incidence of SARS-CoV-2 Infection in Health Care Workers. N. Engl. J. Med. 2020, 384, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, P.B.; Montefiori, D.C.; McDermott, A.B.; Fong, Y.; Benkeser, D.; Deng, W.; Zhou, H.; Houchens, C.R.; Martins, K.; Jayashankar, L.; et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science 2022, 375, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Sette, A.; Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 861–880. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, M.; Peng, Y.; Liang, Y.; Wei, J.; Xing, L.; Guo, L.; Li, X.; Li, J.; Wang, J.; et al. Longitudinal analysis of antibody dynamics in COVID-19 convalescents reveals neutralizing responses up to 16 months after infection. Nat. Microbiol. 2022, 7, 423–433. [Google Scholar] [CrossRef]
- Corbett, K.S.; Flynn, B.; Foulds, K.E.; Francica, J.R.; Boyoglu-Barnum, S.; Werner, A.P.; Flach, B.; O’Connell, S.; Bock, K.W.; Minai, M.; et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020, 383, 1544–1555. [Google Scholar] [CrossRef]
- Crotty, S. Hybrid immunity. Science 2021, 372, 1392–1393. [Google Scholar] [CrossRef]
- Seaman, M.S.; Siedner, M.J.; Boucau, J.; Lavine, C.L.; Ghantous, F.; Liew, M.Y.; Mathews, J.I.; Singh, A.; Marino, C.; Regan, J.; et al. Vaccine breakthrough infection leads to distinct profiles of neutralizing antibody responses by SARS-CoV-2 variant. JCI Insight 2022, 7, e159944. [Google Scholar] [CrossRef]
- Krammer, F.; Srivastava, K.; Alshammary, H.; Amoako, A.A.; Awawda, M.H.; Beach, K.F.; Bermúdez-González, M.C.; Bielak, D.A.; Carreño, J.M.; Chernet, R.L.; et al. Antibody Responses in Seropositive Persons after a Single Dose of SARS-CoV-2 mRNA Vaccine. N. Engl. J. Med. 2021, 384, 1372–1374. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Wang, X.; Gong, W.; Zhong, J.; Leng, Z.; Ren, L.; Feng, L.; Guo, L.; Gao, L.; Liang, X.; et al. Humoral responses after inactivated COVID-19 vaccination in individuals with and without prior SARS-CoV-2 infection: A prospective cohort study. J Med. Virol. 2022, 94, 5746–5757. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.-L.; Shi, D.-W.; Li, Y.; Hong, W.; Lai, D.-Y.; Xue, J.-B.; Jiang, H.-W.; Zhang, H.-N.; Qi, H.; Meng, Q.-F.; et al. Systematic profiling of SARS-CoV-2-specific IgG responses elicited by an inactivated virus vaccine identifies peptides and proteins for predicting vaccination efficacy. Cell Discov. 2021, 7, 67. [Google Scholar] [CrossRef] [PubMed]
- Ebinger, J.E.; Fert-Bober, J.; Printsev, I.; Wu, M.; Sun, N.; Prostko, J.C.; Frias, E.C.; Stewart, J.L.; Van Eyk, J.E.; Braun, J.G.; et al. Antibody responses to the BNT162b2 mRNA vaccine in individuals previously infected with SARS-CoV-2. Nat. Med. 2021, 27, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Favresse, J.; Gillot, C.; Di Chiaro, L.; Eucher, C.; Elsen, M.; Van Eeckhoudt, S.; David, C.; Morimont, L.; Dogné, J.-M.; Douxfils, J. Neutralizing Antibodies in COVID-19 Patients and Vaccine Recipients after Two Doses of BNT162b2. Viruses 2021, 13, 1364. [Google Scholar] [CrossRef]
- Hall, V.; Foulkes, S.; Insalata, F.; Kirwan, P.; Saei, A.; Atti, A.; Wellington, E.; Khawam, J.; Munro, K.; Cole, M.; et al. Protection against SARS-CoV-2 after Covid-19 Vaccination and Previous Infection. N. Engl. J. Med. 2022, 386, 1207–1220. [Google Scholar] [CrossRef]
- Druedahl, L.C.; Minssen, T.; Price, W.N. Collaboration in times of crisis: A study on COVID-19 vaccine R&D partnerships. Vaccine 2021, 39, 6291–6295. [Google Scholar] [CrossRef]
- Perveen, S.; Akram, M.; Nasar, A.; Arshad-Ayaz, A.; Naseem, A. Vaccination-hesitancy and vaccination-inequality as challenges in Pakistan’s COVID-19 response. J. Community Psychol. 2022, 50, 666–683. [Google Scholar] [CrossRef]
- Jeyanathan, M.; Afkhami, S.; Smaill, F.; Miller, M.S.; Lichty, B.D.; Xing, Z. Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 2020, 20, 615–632. [Google Scholar] [CrossRef]
- Sparrow, R.; Dartanto, T.; Hartwig, R. Indonesia Under the New Normal: Challenges and the Way Ahead. Bull. Indones. Econ. Stud. 2020, 56, 269–299. [Google Scholar] [CrossRef]
- Haldane, V.; De Foo, C.; Abdalla, S.M.; Jung, A.-S.; Tan, M.; Wu, S.; Chua, A.; Verma, M.; Shrestha, P.; Singh, S.; et al. Health systems resilience in managing the COVID-19 pandemic: Lessons from 28 countries. Nat. Med. 2021, 27, 964–980. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, A.; Osawa, H.; Hashimoto, H.; Mizuno, T.; Hasyim, A.A.; Abe, Y.-I.; Okahashi, Y.; Ogawa, R.; Iyori, M.; Shida, H.; et al. A replication-competent smallpox vaccine LC16m8Δ-based COVID-19 vaccine. Emerg. Microbes Infect. 2022, 11, 2359–2370. [Google Scholar] [CrossRef] [PubMed]
- Widge, A.T.; Rouphael, N.G.; Jackson, L.A.; Anderson, E.J.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Durability of Responses after SARS-CoV-2 mRNA-1273 Vaccination. N. Engl. J. Med. 2020, 384, 80–82. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Esposito, D.; Kang, Z.; Lu, J.; Remaley, A.T.; De Giorgi, V.; Chen, L.N.; West, K.; Cao, L. mRNA vaccine-induced antibodies more effective than natural immunity in neutralizing SARS-CoV-2 and its high affinity variants. Sci. Rep. 2022, 12, 2628. [Google Scholar] [CrossRef] [PubMed]
- Uriu, K.; Kimura, I.; Shirakawa, K.; Takaori-Kondo, A.; Nakada, T.-A.; Kaneda, A.; Nakagawa, S.; Sato, K. Neutralization of the SARS-CoV-2 Mu Variant by Convalescent and Vaccine Serum. N. Engl. J. Med. 2021, 385, 2397–2399. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Dong, Y.; Dai, T.; Wang, B.; Zhang, L.; Zeng, L.-H.; Huang, J.; Yan, H.; Zhang, L.; Zhou, F. The way of SARS-CoV-2 vaccine development: Success and challenges. Signal Transduct. Target. Ther. 2021, 6, 387. [Google Scholar] [CrossRef]
- Chen, L.-L.; Chua, G.T.; Lu, L.; Chan, B.P.-C.; Wong, J.S.-C.; Chow, C.C.-K.; Yu, T.-C.; Leung, A.S.-Y.; Lam, S.-Y.; Wong, T.-W.; et al. Omicron variant susceptibility to neutralizing antibodies induced in children by natural SARS-CoV-2 infection or COVID-19 vaccine. Emerg. Microbes Infect. 2022, 11, 543–547. [Google Scholar] [CrossRef]
- Sherwani, S.; Khan, M.W.A.; Mallik, A.; Khan, M.; Saleem, M.; Raafat, M.; Shati, A.A.; Alam, N. Seroprevalence of Anti-S1-RBD Antibodies in Pre-pandemic and Pandemic Subjects From Hail Region, KSA. Front. Public Health 2022, 10, 874741. [Google Scholar] [CrossRef]
- Qi, H.; Liu, B.; Wang, X.; Zhang, L. The humoral response and antibodies against SARS-CoV-2 infection. Nat. Immunol. 2022, 23, 1008–1020. [Google Scholar] [CrossRef]
- Kodde, C.; Tafelski, S.; Balamitsa, E.; Nachtigall, I.; Bonsignore, M. Factors Influencing Antibody Response to SARS-CoV-2 Vaccination. Vaccines 2023, 11, 451. [Google Scholar] [CrossRef] [PubMed]
- Camacho Moll, M.E.; Mata Tijerina, V.L.; Silva Ramirez, B.; Penuelas Urquides, K.; Gonzalez Escalante, L.A.; Escobedo Guajardo, B.L.; Cruz Luna, J.E.; Corrales Perez, R.; Gomez Garcia, S.; Bermudez de Leon, M. Sex, Age, and Comorbidities Are Associated with SARS-CoV-2 Infection, COVID-19 Severity, and Fatal Outcome in a Mexican Population: A Retrospective Multi-Hospital Study. J. Clin. Med. 2023, 12, 2676. [Google Scholar] [CrossRef] [PubMed]
- Shim, H.W.; Shin, J.H.; Shin, S.C.; Lee, H.J.; So, K.S.; Lee, S.Y.; Jun, J.W.; Seo, J.K.; Lee, H.S.; Lee, S.Y.; et al. Analysis of Factors Affecting Neutralizing Antibody Production after COVID-19 Vaccination Using Newly Developed Rapid Point-of-Care Test. Diagnostics 2022, 12, 1924. [Google Scholar] [CrossRef] [PubMed]
- Kang, I.S.; Kong, K.A. Body mass index and severity/fatality from coronavirus disease 2019: A nationwide epidemiological study in Korea. PLoS ONE 2021, 16, e0253640. [Google Scholar] [CrossRef] [PubMed]
- Tanoto, R. COVID-19 Vaccination Post Introduction Evaluation (cPIE) in Indonesia. Available online: https://www.who.int/indonesia/news/detail/05-07-2023-covid-19-vaccination-post-introduction-evaluation-(cpie)-in-indonesia (accessed on 25 July 2023).
| Group | COVID-19 Infection | Vaccination | n |
|---|---|---|---|
| 1 | Yes | No | 35 |
| 2 | Yes | Yes (After infection) | 52 |
| 3 | No | Yes | 41 |
| 4 | Yes | Yes (Before infection) | 28 |
| Variable | Group 1 | Group 2 | Group 3 | Group4 |
|---|---|---|---|---|
| Age | ||||
| Mean | 46.7 | 36.6 | 34.4 | 40.1 |
| Median (Range) | 48 (19–72) | 36.0 (24–56) | 31.0 (18–70) | 41.0 (23–64) |
| Sex | ||||
| Female (%) | 19 (54.3) | 18 (34.6) | 19 (46.3) | 11 (39.3) |
| Male (%) | 16 (45.7) | 34 (65.4) | 22 (53.7) | 17 (60.7) |
| Severity | ||||
| Severe (%) | 17 (48.6) | 0 (0) | N/A | 1 (3.6) |
| Non-Severe (%) | 18 (51.4) | 52 (100) | N/A | 27 (96.4) |
| Body Mass Index (WHO) | ||||
| Underweight (%) | 11 (31.4) | 12 (23.1) | 23 (56.1) | 11 (39.3) |
| Normal (%) | 21 (60.0) | 26 (50.0) | 16 (39.0) | 8 (28.6) |
| Overweight (%) | 3 (8.6) | 13 (25.0) | 2 (4.9) | 9 (32.1) |
| Obese (%) | 0 (0) | 1 (1.9) | 0 (0) | 0 (0) |
| Health Workers | ||||
| Yes (%) | 1 (2.9) | 32 (61.5) | 0 (0) | 3 (10.7) |
| No (%) | 34 (97.1) | 20 (38.5) | 41 (100) | 25 (89.3) |
| Duration between infection and vaccine (months) | ||||
| Mean | N/A | 4.70 | N/A | 4.034 |
| Median (Range) | N/A | 4 (3–11) | N/A | 4 (2–6) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nurisyah, S.; Iyori, M.; Hasyim, A.A.; Sakamoto, A.; Hashimoto, H.; Yamagata, K.; Yamauchi, S.; Amru, K.; Zainal, K.H.; Idris, I.; et al. Comparison between Neutralization Capacity of Antibodies Elicited by COVID-19 Natural Infection and Vaccination in Indonesia: A Prospective Cohort. Antibodies 2023, 12, 60. https://doi.org/10.3390/antib12030060
Nurisyah S, Iyori M, Hasyim AA, Sakamoto A, Hashimoto H, Yamagata K, Yamauchi S, Amru K, Zainal KH, Idris I, et al. Comparison between Neutralization Capacity of Antibodies Elicited by COVID-19 Natural Infection and Vaccination in Indonesia: A Prospective Cohort. Antibodies. 2023; 12(3):60. https://doi.org/10.3390/antib12030060
Chicago/Turabian StyleNurisyah, Sitti, Mitsuhiro Iyori, Ammar Abdurrahman Hasyim, Akihiko Sakamoto, Hinata Hashimoto, Kyouhei Yamagata, Saya Yamauchi, Khaeriah Amru, Kartika Hardianti Zainal, Irfan Idris, and et al. 2023. "Comparison between Neutralization Capacity of Antibodies Elicited by COVID-19 Natural Infection and Vaccination in Indonesia: A Prospective Cohort" Antibodies 12, no. 3: 60. https://doi.org/10.3390/antib12030060
APA StyleNurisyah, S., Iyori, M., Hasyim, A. A., Sakamoto, A., Hashimoto, H., Yamagata, K., Yamauchi, S., Amru, K., Zainal, K. H., Idris, I., Yoshida, S., Djaharuddin, I., Syafruddin, D., Bukhari, A., Asih, P. B. S., & Yusuf, Y. (2023). Comparison between Neutralization Capacity of Antibodies Elicited by COVID-19 Natural Infection and Vaccination in Indonesia: A Prospective Cohort. Antibodies, 12(3), 60. https://doi.org/10.3390/antib12030060







