Complement Activation by an Anti-Dengue/Zika Antibody with Impaired Fcγ Receptor Binding Provides Strong Efficacy and Abrogates Risk of Antibody-Dependent Enhancement
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Proteins and Antibodies
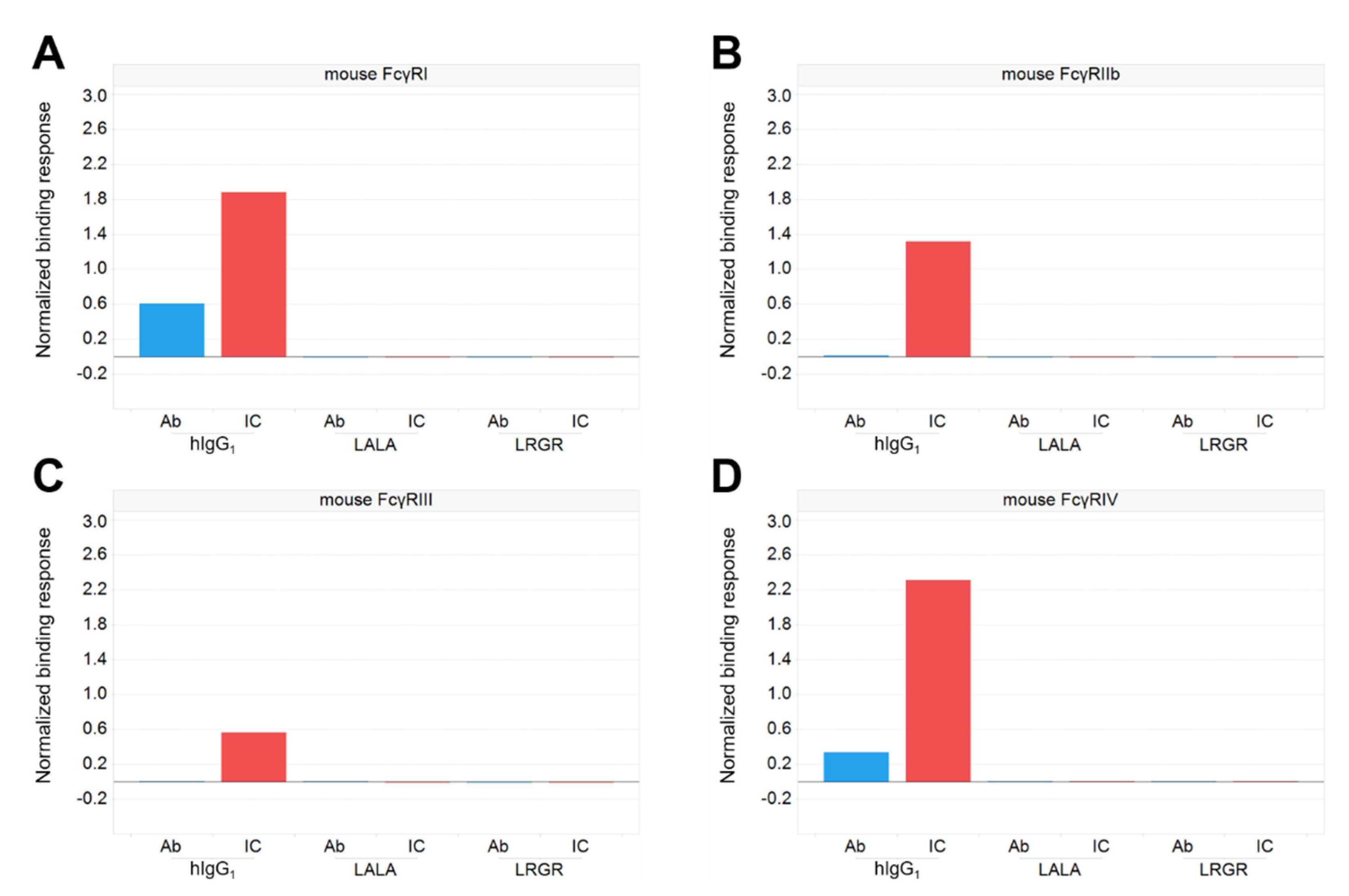
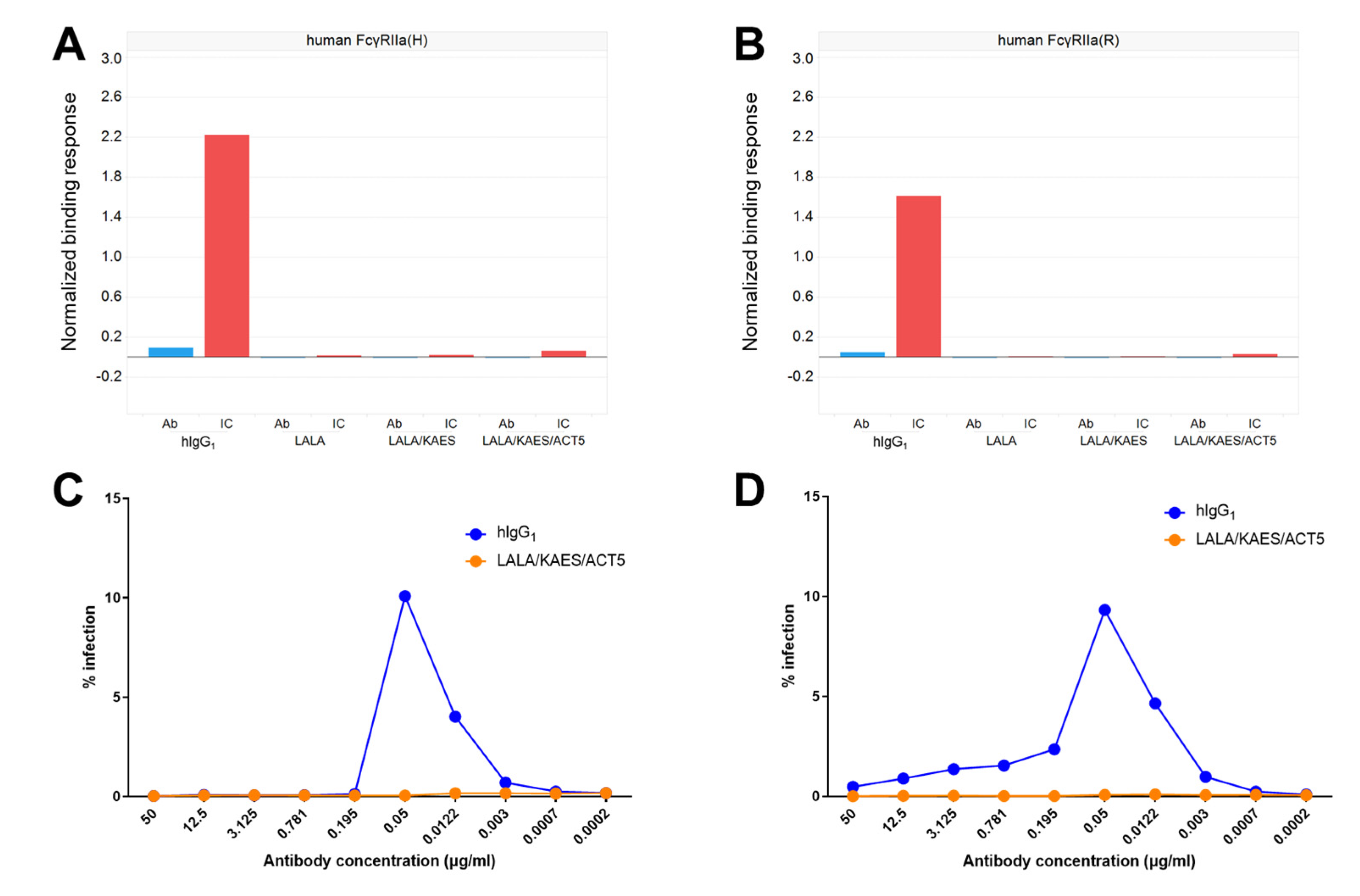
2.2. Evaluation of Binding Activity to FcγRs
2.3. Evaluation of Binding Activity to Human C1q
2.4. Evaluation of Binding Activity to Mouse C1q
2.5. In Vivo Efficacy Study
2.6. In Vitro Complement Activation Assay
2.7. In Vitro ADE Assay
3. Results
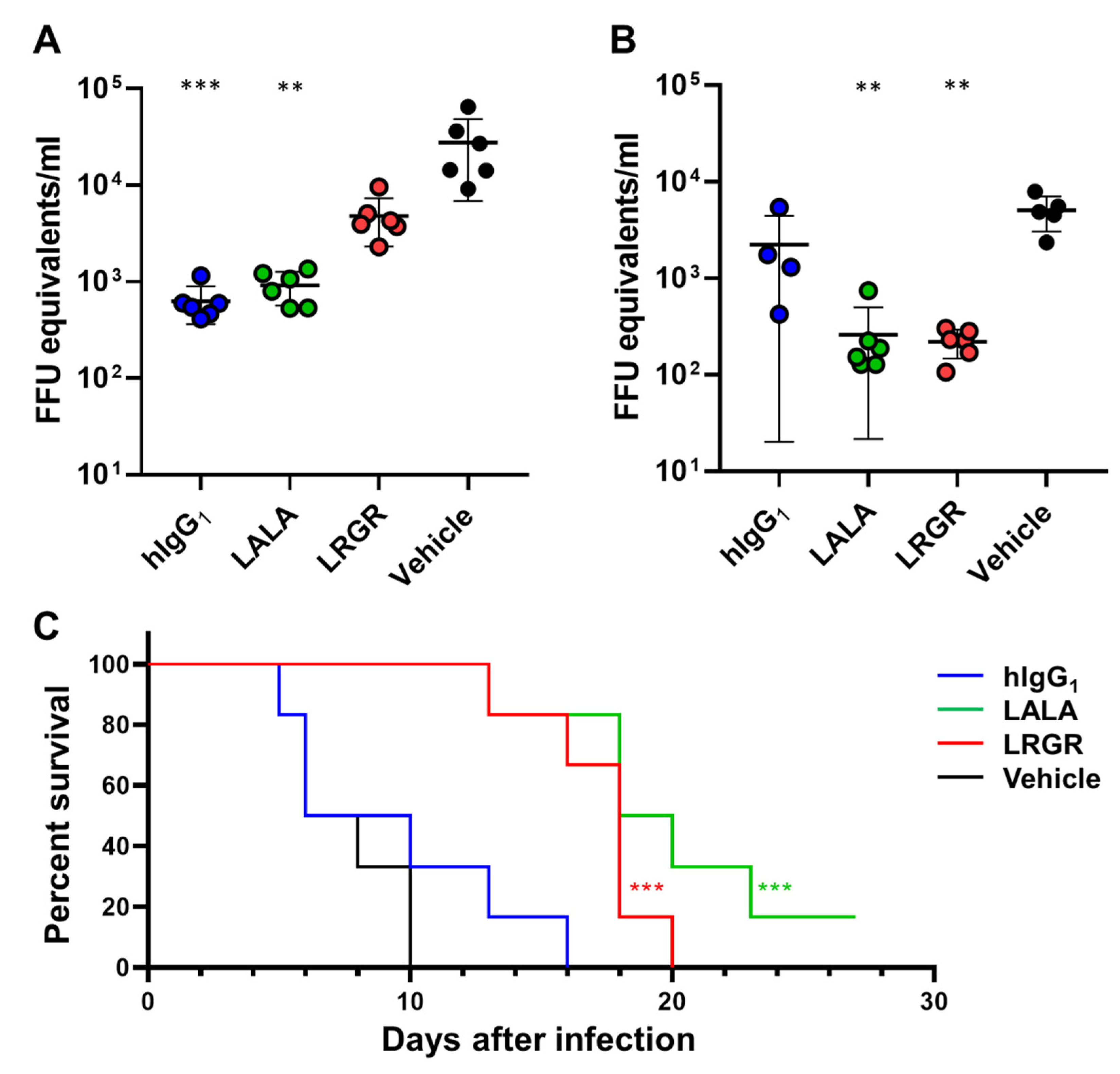
3.1. Therapeutic Efficacy in Mice Was Affected by Fc Engineering
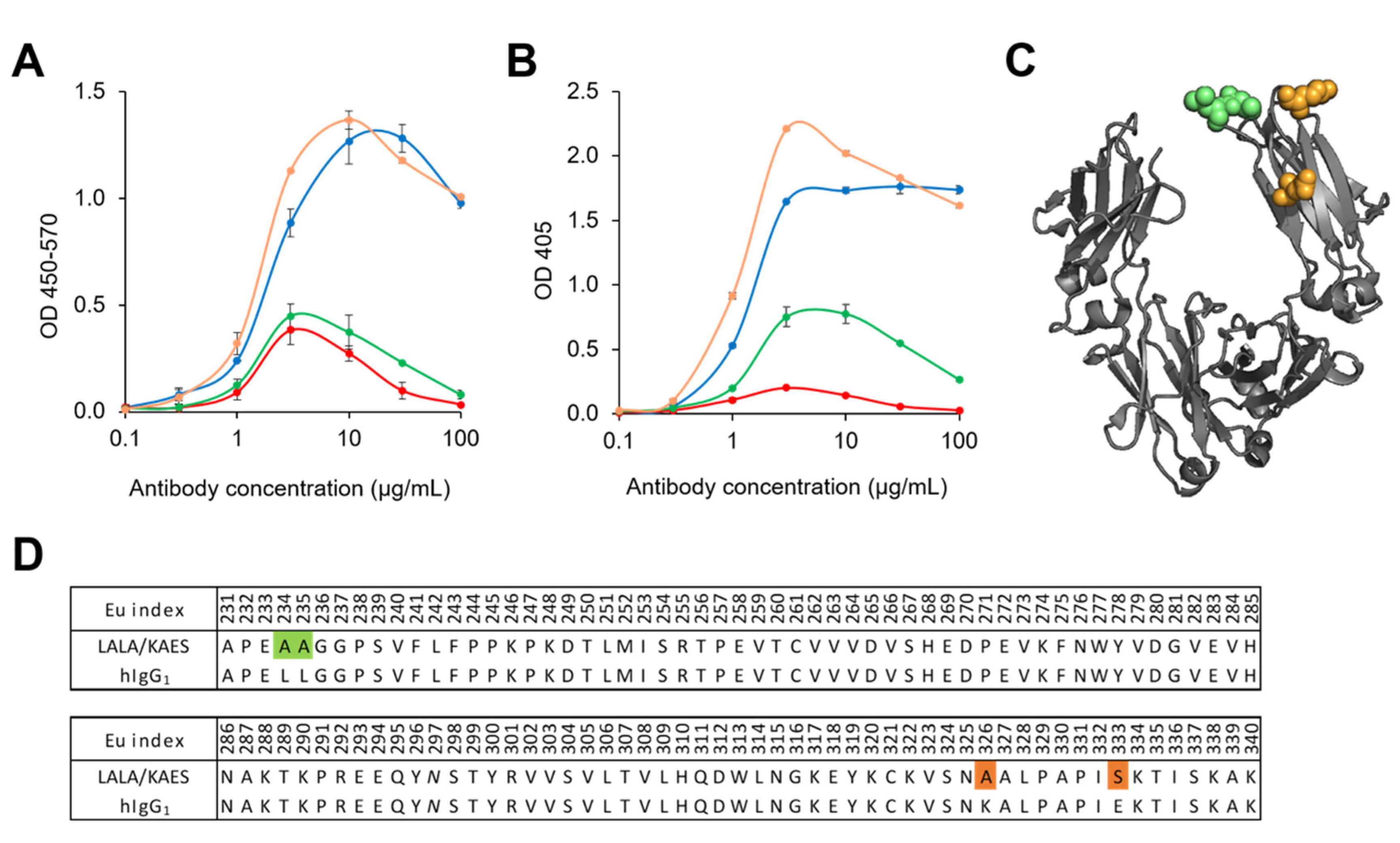
3.2. Antibody Engineering for Therapeutic Application
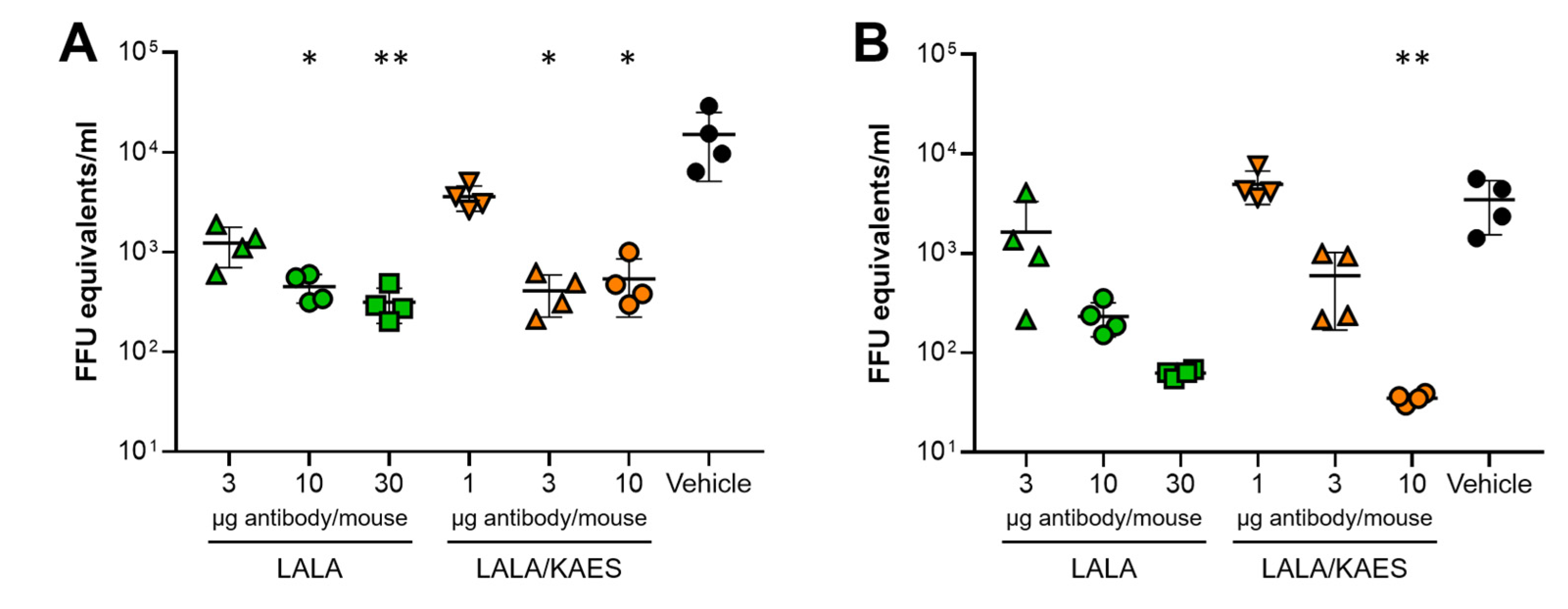
3.3. In Vivo Efficacy Was Improved by Fc Engineering to Enhance C1q Binding
3.4. No ADE Risk Was Observed by the Engineered Antibody AID351 In Vitro
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simmons, C.P.; Farrar, J.J.; van Vinh Chau, N.; Wills, B. Dengue. N. Engl. J. Med. 2012, 366, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Guzman, M.G.; Alvarez, M.; Halstead, S.B. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: An historical perspective and role of antibody-dependent enhancement of infection. Arch. Virol. 2013, 158, 1445–1459. [Google Scholar] [CrossRef] [PubMed]
- Bournazos, S.; Gupta, A.; Ravetch, J.V. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 2020, 20, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo, W.W.; Jin, X.; Blackley, S.D.; Rose, R.C.; Schlesinger, J.J. Differential enhancement of dengue virus immune complex infectivity mediated by signaling-competent and signaling-incompetent human FcγRIA (CD64) or FcγRIIA (CD32). J. Virol. 2006, 80, 10128–10138. [Google Scholar] [CrossRef]
- Taylor, A.; Foo, S.S.; Bruzzone, R.; Dinh, L.V.; King, N.J.; Mahalingam, S. Fc receptors in antibody-dependent enhancement of viral infections. Immunol. Rev. 2015, 268, 340–364. [Google Scholar] [CrossRef]
- Deng, S.Q.; Yang, X.; Wei, Y.; Chen, J.T.; Wang, X.J.; Peng, H.J. A review on dengue vaccine development. Vaccines 2020, 8, 63. [Google Scholar] [CrossRef]
- Idris, F.; Ting, D.H.R.; Alonso, S. An update on dengue vaccine development, challenges, and future perspectives. Expert Opin. Drug Discov. 2021, 16, 47–58. [Google Scholar] [CrossRef]
- Katzelnick, L.C.; Narvaez, C.; Arguello, S.; Lopez Mercado, B.; Collado, D.; Ampie, O.; Elizondo, D.; Miranda, T.; Bustos Carillo, F.; Mercado, J.C.; et al. Zika virus infection enhances future risk of severe dengue disease. Science 2020, 369, 1123–1128. [Google Scholar] [CrossRef]
- Ferguson, N.M.; Rodriguez-Barraquer, I.; Dorigatti, I.; Mier, Y.T.-R.L.; Laydon, D.J.; Cummings, D.A. Benefits and risks of the Sanofi-Pasteur dengue vaccine: Modeling optimal deployment. Science 2016, 353, 1033–1036. [Google Scholar] [CrossRef]
- Chan, K.R.; Ong, E.Z.; Ooi, E.E. Therapeutic antibodies as a treatment option for dengue fever. Expert Rev. Anti-Infect. Ther. 2013, 11, 1147–1157. [Google Scholar] [CrossRef]
- Xu, M.; Zuest, R.; Velumani, S.; Tukijan, F.; Toh, Y.X.; Appanna, R.; Tan, E.Y.; Cerny, D.; MacAry, P.; Wang, C.I.; et al. A potent neutralizing antibody with therapeutic potential against all four serotypes of dengue virus. NPJ Vaccines 2017, 2, 2. [Google Scholar] [CrossRef]
- Kam, Y.W.; Lee, C.Y.; Teo, T.H.; Howland, S.W.; Amrun, S.N.; Lum, F.M.; See, P.; Kng, N.Q.; Huber, R.G.; Xu, M.H.; et al. Cross-reactive dengue human monoclonal antibody prevents severe pathologies and death from Zika virus infections. JCI Insight 2017, 2, e92428. [Google Scholar] [CrossRef]
- Barba-Spaeth, G.; Dejnirattisai, W.; Rouvinski, A.; Vaney, M.C.; Medits, I.; Sharma, A.; Simon-Loriere, E.; Sakuntabhai, A.; Cao-Lormeau, V.M.; Haouz, A.; et al. Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature 2016, 536, 48–53. [Google Scholar] [CrossRef]
- Zhang, S.; Loy, T.; Ng, T.S.; Lim, X.N.; Chew, S.V.; Tan, T.Y.; Xu, M.; Kostyuchenko, V.A.; Tukijan, F.; Shi, J.; et al. A human antibody neutralizes different flaviviruses by using different mechanisms. Cell Rep. 2020, 31, 107584. [Google Scholar] [CrossRef]
- Hezareh, M.; Hessell, A.J.; Jensen, R.C.; van de Winkel, J.G.; Parren, P.W. Effector function activities of a panel of mutants of a broadly neutralizing antibody against human immunodeficiency virus type 1. J. Virol. 2001, 75, 12161–12168. [Google Scholar] [CrossRef]
- Winkler, E.S.; Gilchuk, P.; Yu, J.; Bailey, A.L.; Chen, R.E.; Chong, Z.; Zost, S.J.; Jang, H.; Huang, Y.; Allen, J.D.; et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell 2021, 184, 1804–1820.e16. [Google Scholar] [CrossRef]
- Schafer, A.; Muecksch, F.; Lorenzi, J.C.C.; Leist, S.R.; Cipolla, M.; Bournazos, S.; Schmidt, F.; Maison, R.M.; Gazumyan, A.; Martinez, D.R.; et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 2021, 218, e20201993. [Google Scholar] [CrossRef]
- Tan, G.K.; Ng, J.K.; Trasti, S.L.; Schul, W.; Yip, G.; Alonso, S. A non mouse-adapted dengue virus strain as a new model of severe dengue infection in AG129 mice. PLoS Negl. Trop. Dis. 2010, 4, e672. [Google Scholar] [CrossRef]
- Zust, R.; Toh, Y.X.; Valdes, I.; Cerny, D.; Heinrich, J.; Hermida, L.; Marcos, E.; Guillen, G.; Kalinke, U.; Shi, P.Y.; et al. Type I interferon signals in macrophages and dendritic cells control dengue virus infection: Implications for a new mouse model to test dengue vaccines. J. Virol. 2014, 88, 7276–7285. [Google Scholar] [CrossRef]
- Sampei, Z.; Haraya, K.; Tachibana, T.; Fukuzawa, T.; Shida-Kawazoe, M.; Gan, S.W.; Shimizu, Y.; Ruike, Y.; Feng, S.; Kuramochi, T.; et al. Antibody engineering to generate SKY59, a long-acting anti-C5 recycling antibody. PLoS ONE 2018, 13, e0209509. [Google Scholar] [CrossRef] [PubMed]
- Conde, J.N.; Silva, E.M.; Barbosa, A.S.; Mohana-Borges, R. The complement system in flavivirus infections. Front. Microbiol. 2017, 8, 213. [Google Scholar] [CrossRef] [PubMed]
- Maeda, A.; Iwayanagi, Y.; Haraya, K.; Tachibana, T.; Nakamura, G.; Nambu, T.; Esaki, K.; Hattori, K.; Igawa, T. Identification of human IgG1 variant with enhanced FcRn binding and without increased binding to rheumatoid factor autoantibody. mAbs 2017, 9, 844–853. [Google Scholar] [CrossRef] [PubMed]
- Boonnak, K.; Slike, B.M.; Donofrio, G.C.; Marovich, M.A. Human FcγRII cytoplasmic domains differentially influence antibody-mediated dengue virus infection. J. Immunol. 2013, 190, 5659–5665. [Google Scholar] [CrossRef]
- Salazar, G.; Zhang, N.; Fu, T.M.; An, Z. Antibody therapies for the prevention and treatment of viral infections. NPJ Vaccines 2017, 2, 19. [Google Scholar] [CrossRef]
- Ali, M.G.; Zhang, Z.; Gao, Q.; Pan, M.; Rowan, E.G.; Zhang, J. Recent advances in therapeutic applications of neutralizing antibodies for virus infections: An overview. Immunol. Res. 2020, 68, 325–339. [Google Scholar] [CrossRef]
- Taylor, P.C.; Adams, A.C.; Hufford, M.M.; de la Torre, I.; Winthrop, K.; Gottlieb, R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021, 21, 382–393. [Google Scholar] [CrossRef]
- Weinreich, D.M.; Sivapalasingam, S.; Norton, T.; Ali, S.; Gao, H.; Bhore, R.; Musser, B.J.; Soo, Y.; Rofail, D.; Im, J.; et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with COVID-19. N. Engl. J. Med. 2021, 384, 238–251. [Google Scholar] [CrossRef]
- Weinreich, D.M.; Sivapalasingam, S.; Norton, T.; Ali, S.; Gao, H.; Bhore, R.; Xiao, J.; Hooper, A.T.; Hamilton, J.D.; Musser, B.J.; et al. REGEN-COV antibody combination and outcomes in outpatients with COVID-19. N. Engl. J. Med. 2021, 385, e81. [Google Scholar] [CrossRef]
- Dougan, M.; Nirula, A.; Azizad, M.; Mocherla, B.; Gottlieb, R.L.; Chen, P.; Hebert, C.; Perry, R.; Boscia, J.; Heller, B.; et al. Bamlanivimab plus etesevimab in mild or moderate COVID-19. N. Engl. J. Med. 2021, 385, 1382–1392. [Google Scholar] [CrossRef]
- Igawa, T.; Tsunoda, H.; Kuramochi, T.; Sampei, Z.; Ishii, S.; Hattori, K. Engineering the variable region of therapeutic IgG antibodies. mAbs 2011, 3, 243–252. [Google Scholar] [CrossRef]
- Wang, X.; Mathieu, M.; Brezski, R.J. IgG Fc engineering to modulate antibody effector functions. Protein Cell 2018, 9, 63–73. [Google Scholar] [CrossRef]
- Mimoto, F.; Katada, H.; Kadono, S.; Igawa, T.; Kuramochi, T.; Muraoka, M.; Wada, Y.; Haraya, K.; Miyazaki, T.; Hattori, K. Engineered antibody Fc variant with selectively enhanced FcγRIIb binding over both FcγRIIa(R131) and FcγRIIa(H131). Protein Eng. Des. Sel. PEDS 2013, 26, 589–598. [Google Scholar] [CrossRef]
- Mimoto, F.; Igawa, T.; Kuramochi, T.; Katada, H.; Kadono, S.; Kamikawa, T.; Shida-Kawazoe, M.; Hattori, K. Novel asymmetrically engineered antibody Fc variant with superior FcγR binding affinity and specificity compared with afucosylated Fc variant. mAbs 2013, 5, 229–236. [Google Scholar] [CrossRef]
- Mimoto, F.; Kadono, S.; Katada, H.; Igawa, T.; Kamikawa, T.; Hattori, K. Crystal structure of a novel asymmetrically engineered Fc variant with improved affinity for FcγRs. Mol. Immunol. 2014, 58, 132–138. [Google Scholar] [CrossRef]
- Byrne, A.B.; Talarico, L.B. Role of the complement system in antibody-dependent enhancement of flavivirus infections. Int. J. Infect. Dis. 2021, 103, 404–411. [Google Scholar] [CrossRef]
- Mehlhop, E.; Nelson, S.; Jost, C.A.; Gorlatov, S.; Johnson, S.; Fremont, D.H.; Diamond, M.S.; Pierson, T.C. Complement protein C1q reduces the stoichiometric threshold for antibody-mediated neutralization of West Nile virus. Cell Host Microbe 2009, 6, 381–391. [Google Scholar] [CrossRef]
- Vogt, M.R.; Dowd, K.A.; Engle, M.; Tesh, R.B.; Johnson, S.; Pierson, T.C.; Diamond, M.S. Poorly neutralizing cross-reactive antibodies against the fusion loop of West Nile virus envelope protein protect in vivo via Fcγ receptor and complement-dependent effector mechanisms. J. Virol. 2011, 85, 11567–11580. [Google Scholar] [CrossRef]
- Idusogie, E.E.; Presta, L.G.; Gazzano-Santoro, H.; Totpal, K.; Wong, P.Y.; Ultsch, M.; Meng, Y.G.; Mulkerrin, M.G. Mapping of the C1q binding site on rituxan, a chimeric antibody with a human IgG1 Fc. J. Immunol. 2000, 164, 4178–4184. [Google Scholar] [CrossRef]
- Idusogie, E.E.; Wong, P.Y.; Presta, L.G.; Gazzano-Santoro, H.; Totpal, K.; Ultsch, M.; Mulkerrin, M.G. Engineered antibodies with increased activity to recruit complement. J. Immunol. 2001, 166, 2571–2575. [Google Scholar] [CrossRef]
- Moore, G.L.; Chen, H.; Karki, S.; Lazar, G.A. Engineered Fc variant antibodies with enhanced ability to recruit complement and mediate effector functions. mAbs 2010, 2, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Natsume, A.; In, M.; Takamura, H.; Nakagawa, T.; Shimizu, Y.; Kitajima, K.; Wakitani, M.; Ohta, S.; Satoh, M.; Shitara, K.; et al. Engineered antibodies of IgG1/IgG3 mixed isotype with enhanced cytotoxic activities. Cancer Res. 2008, 68, 3863–3872. [Google Scholar] [CrossRef] [PubMed]
- Diebolder, C.A.; Beurskens, F.J.; de Jong, R.N.; Koning, R.I.; Strumane, K.; Lindorfer, M.A.; Voorhorst, M.; Ugurlar, D.; Rosati, S.; Heck, A.J.; et al. Complement is activated by IgG hexamers assembled at the cell surface. Science 2014, 343, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Romain, G.; Yan, W.; Watanabe, M.; Charab, W.; Todorova, B.; Lee, J.; Triplett, K.; Donkor, M.; Lungu, O.I.; et al. IgG Fc domains that bind C1q but not effector Fcγ receptors delineate the importance of complement-mediated effector functions. Nat. Immunol. 2017, 18, 889–898. [Google Scholar] [CrossRef]
- Kayentao, K.; Ongoiba, A.; Preston, A.C.; Healy, S.A.; Doumbo, S.; Doumtabe, D.; Traore, A.; Traore, H.; Djiguiba, A.; Li, S.; et al. Safety and efficacy of a monoclonal antibody against malaria in Mali. N. Engl. J. Med. 2022, 387, 1833–1842. [Google Scholar] [CrossRef]
- Wu, R.L.; Idris, A.H.; Berkowitz, N.M.; Happe, M.; Gaudinski, M.R.; Buettner, C.; Strom, L.; Awan, S.F.; Holman, L.A.; Mendoza, F.; et al. Low-dose subcutaneous or intravenous monoclonal antibody to prevent malaria. N. Engl. J. Med. 2022, 387, 397–407. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sampei, Z.; Koo, C.X.; Teo, F.J.; Toh, Y.X.; Fukuzawa, T.; Gan, S.W.; Nambu, T.; Ho, A.; Honda, K.; Igawa, T.; et al. Complement Activation by an Anti-Dengue/Zika Antibody with Impaired Fcγ Receptor Binding Provides Strong Efficacy and Abrogates Risk of Antibody-Dependent Enhancement. Antibodies 2023, 12, 36. https://doi.org/10.3390/antib12020036
Sampei Z, Koo CX, Teo FJ, Toh YX, Fukuzawa T, Gan SW, Nambu T, Ho A, Honda K, Igawa T, et al. Complement Activation by an Anti-Dengue/Zika Antibody with Impaired Fcγ Receptor Binding Provides Strong Efficacy and Abrogates Risk of Antibody-Dependent Enhancement. Antibodies. 2023; 12(2):36. https://doi.org/10.3390/antib12020036
Chicago/Turabian StyleSampei, Zenjiro, Christine Xing’er Koo, Frannie Jiuyi Teo, Ying Xiu Toh, Taku Fukuzawa, Siok Wan Gan, Takeru Nambu, Adrian Ho, Kiyofumi Honda, Tomoyuki Igawa, and et al. 2023. "Complement Activation by an Anti-Dengue/Zika Antibody with Impaired Fcγ Receptor Binding Provides Strong Efficacy and Abrogates Risk of Antibody-Dependent Enhancement" Antibodies 12, no. 2: 36. https://doi.org/10.3390/antib12020036
APA StyleSampei, Z., Koo, C. X., Teo, F. J., Toh, Y. X., Fukuzawa, T., Gan, S. W., Nambu, T., Ho, A., Honda, K., Igawa, T., Ahmed, F., Wang, C.-I., Fink, K., & Nezu, J. (2023). Complement Activation by an Anti-Dengue/Zika Antibody with Impaired Fcγ Receptor Binding Provides Strong Efficacy and Abrogates Risk of Antibody-Dependent Enhancement. Antibodies, 12(2), 36. https://doi.org/10.3390/antib12020036





