Scalable Shared Scripting for Spatial Structure of Regionalized Ratings
Abstract
:1. Introduction
2. Methodology of Implicit Topology
2.1. Compiling and Collecting Content
2.2. Proximal Propensity and Raised Regions
2.3. Compound Connectivity and Hyper-Hills of Intensified Indicators
2.4. Impaneled Indicators and Rating Relations
3. Representative Results
3.1. Spatial Structures: Virtual Vertices, Singletons, Sectors, Sheaths, and Sites
3.2. Spatial Structures: Truncated Trends of Topology
[1] 0 20 10 16 31 37 52 63 67 72 74 87 93 95 100 105 125 144 149 [20] 169 172 175
[1] 95 111 0 105 123 0 149 148 167 133 150 166 0 16 15 33 0 37 0 [20] 67 0 87 0 144 162 143 0 52 0 10 11 27 25 26 8 24 28 0 [39] 63 0 72 0 74 57 0 93 0 125 0 31 0 169 0 100 0 175 174 [58] 0 172 0
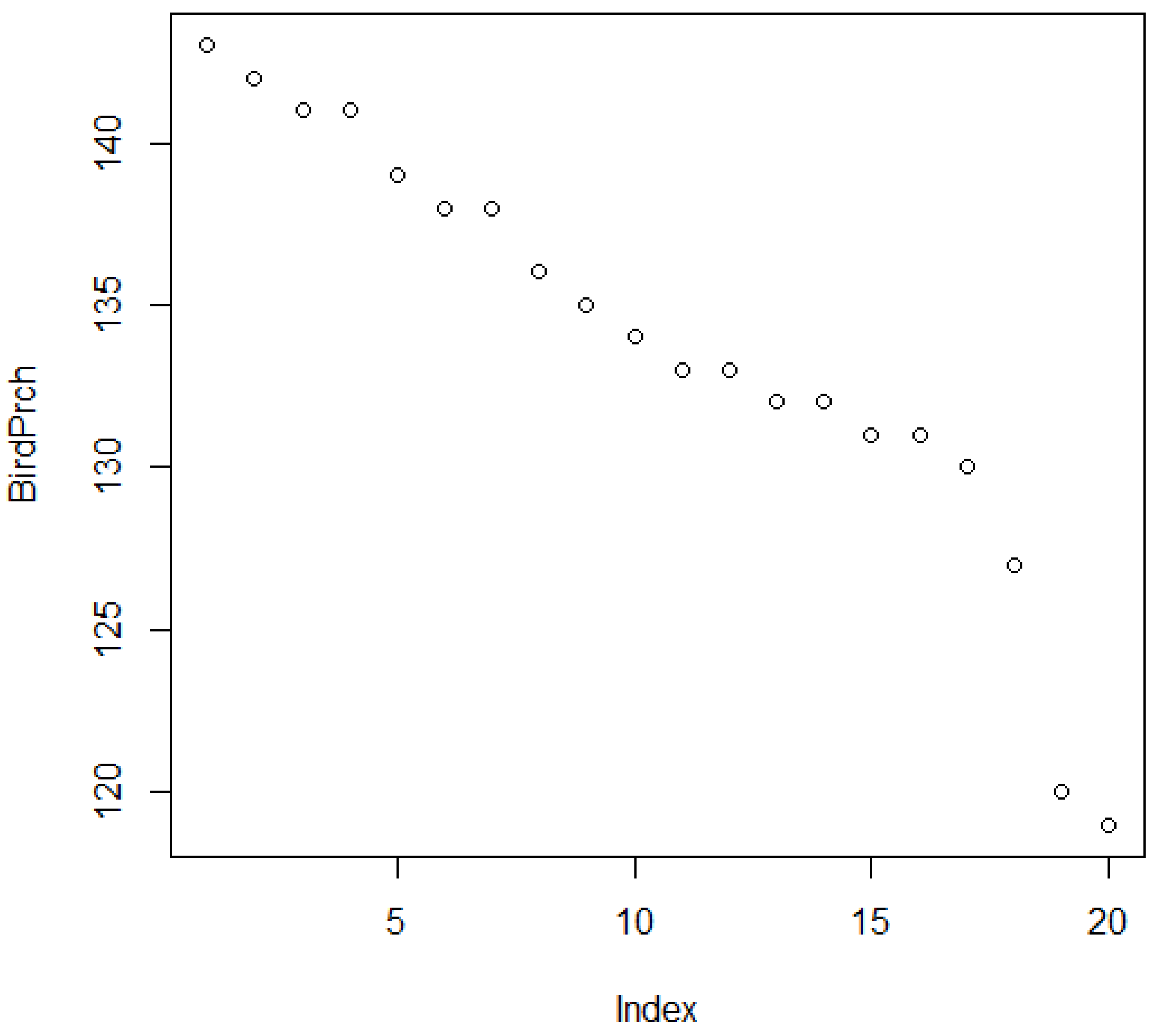
3.3. Spatial Structure: Rippled Rings of Relative Ratings
3.4. Regionalized Rating Relations: Impaneling Interrelated Indicators as Detector Networks
3.5. Regionalized Rating Relations: Data Domains
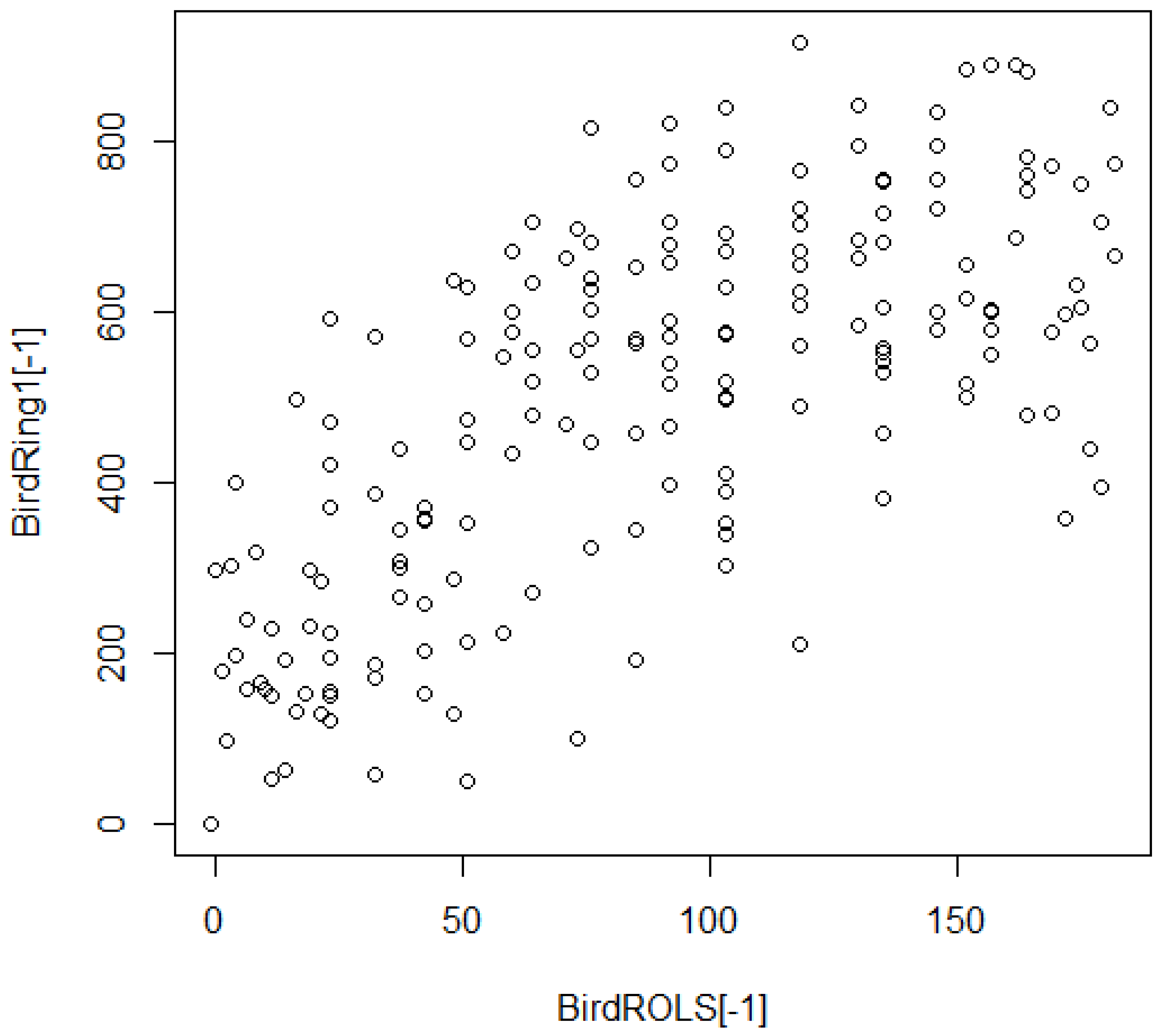
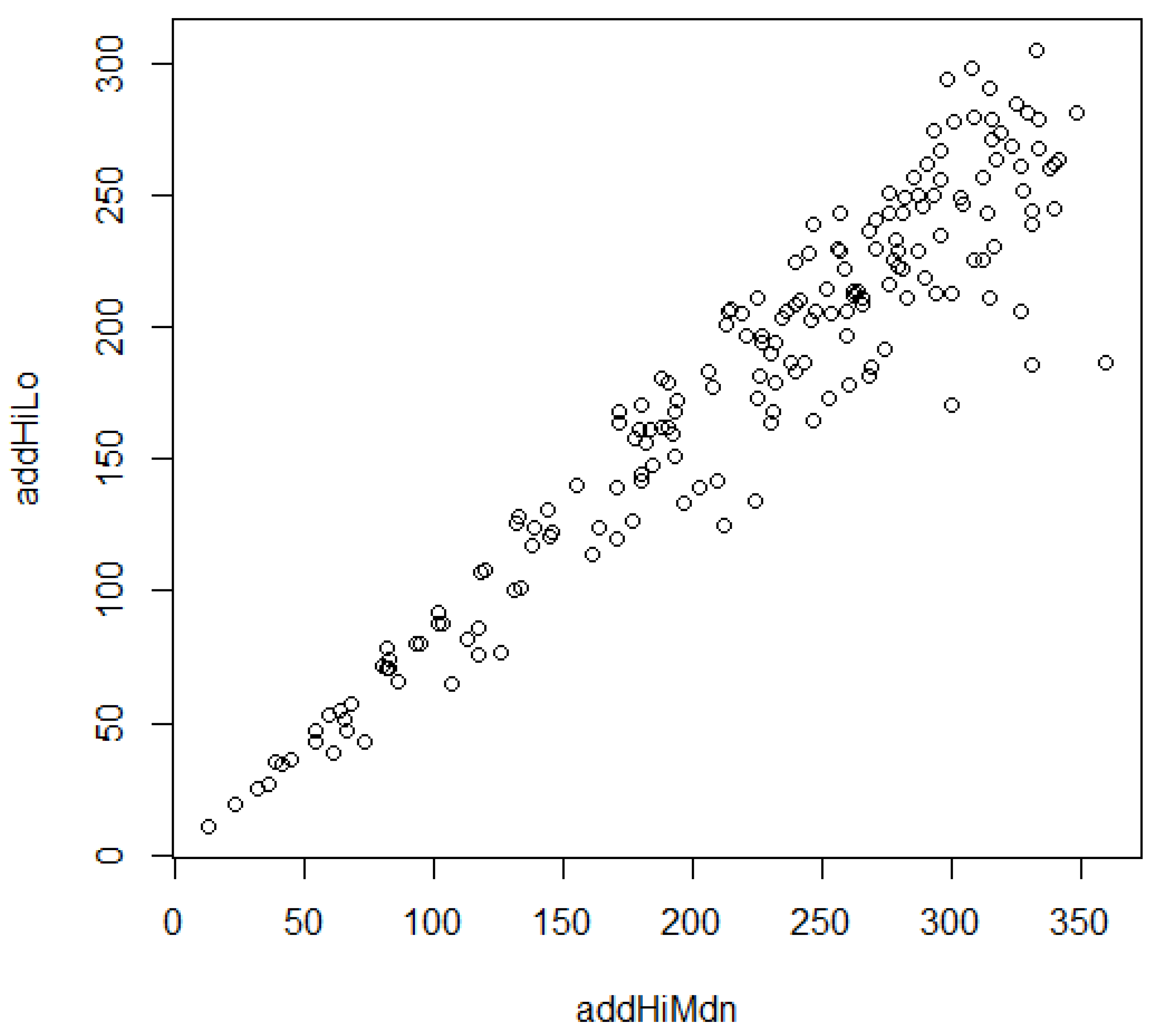
3.6. Regionalized Rating Relations: Cross-Coupled Combinations of ROLS Range Ratings
3.7. Regionalized Rating Relations: Contextual Collectives
3.8. Regionalized Rating Relations: Panel Paired Primacy
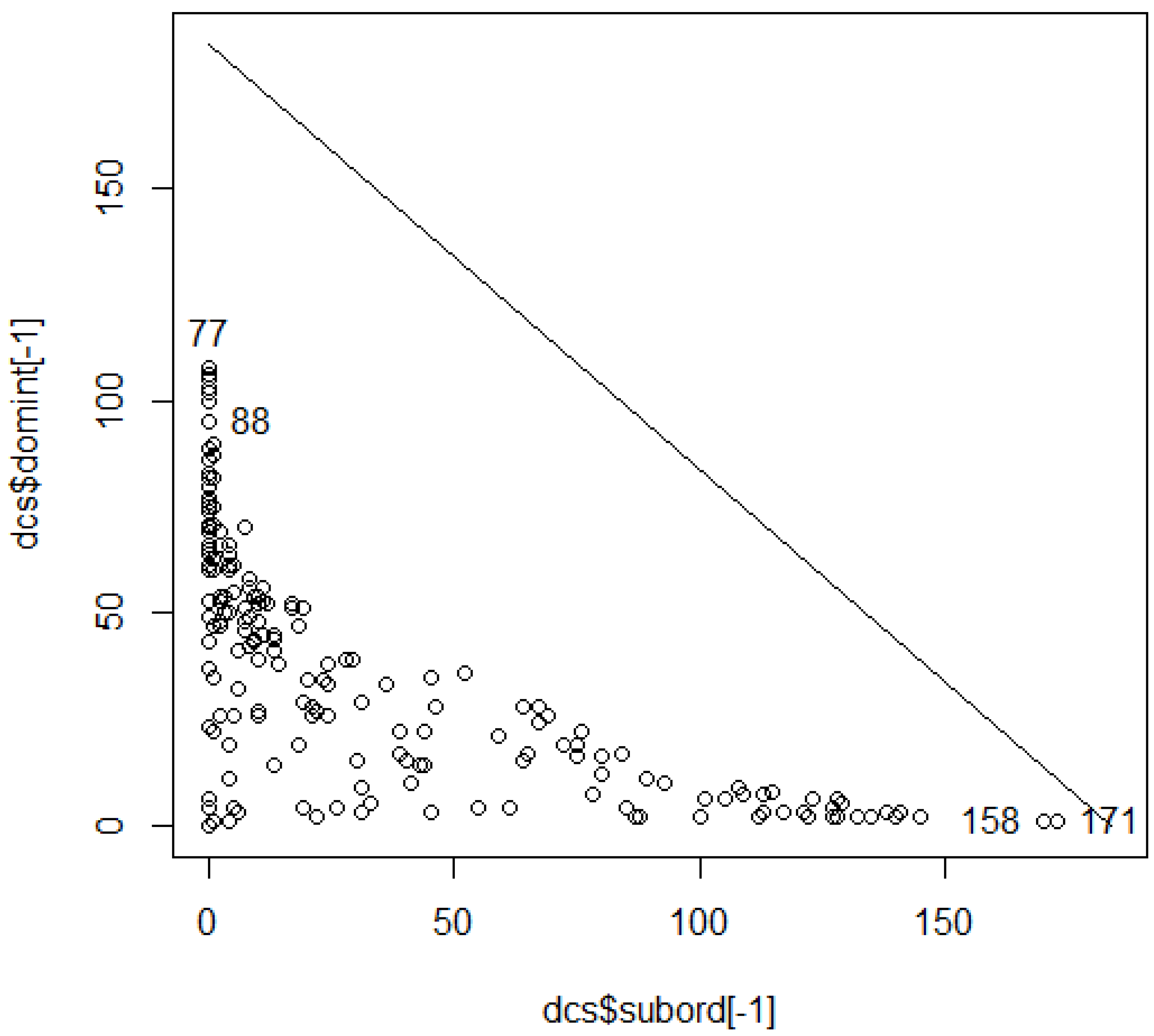
@ hxIDtabl[172,] seg col pin Birds Mamls ElevSD PctFC 1829 1 11 171 96 34 17 25.3 @ hxIDtabl[159,] seg col pin Birds Mamls ElevSD PctFC 2294 5 10 158 103 39 15 11.9
@ hxIDtabl[dcs$domint > 95,]
seg col pin Birds Mamls ElevSD PctFC
2647 4 3 38 133 48 135 95.7
2527 4 4 55 135 49 120 88.6
2648 5 4 56 129 52 126 98.5
2171 1 5 69 132 50 104 97.5
3143 9 5 77 132 50 118 91.0
3527 14 8 133 134 52 142 84.6
3.9. Regionalized Rating Relations: Detecting Discrepant Combinatorial Components

4. Discussion
5. Conclusions: Shared Scripting with Hybrid HEXIZON Resolves Regionalization
| “““Columnar Hexagonal Implicit Positioning: implicit network geometry |
| _hip (hexagonal implicit pattern) file |
| “““ |
| from tkinter import * |
| import tkinter.messagebox as msg |
| windOh = Tk() |
| windOh.title(‘HIDNhip’) |
| def hipspecs(): |
| # Request _hip file info. |
| # regn = int(input(‘Region reference number: ‘)) |
| regn = int(region.get()) |
| # ulccX = float(input(‘Upper-left containment corner X: ‘)) |
| ulccX = float(uplefccX.get()) |
| # ulccY = float(input(‘Upper-left containment corner Y: ‘)) |
| ulccY = float(uplefccY.get()) |
| # lrccX = float(input(‘Lower-right containment corner X: ‘)) |
| lrccX = float(loritccX.get()) |
| # lrccY = float(input(‘Lower-right containment corner Y: ‘)) |
| lrccY = float(loritccY.get()) |
| # starsiz = float(input(‘Hexagon star span between opposing points: ‘)) |
| starsiz = float(stars.get()) |
| # shhs = int(input(‘First whole hex as hump (1) or slump (-1): ‘)) |
| shhs = int(rippl.get()) |
| # hyprhalvs = int(input(‘Hyper-halves on major margins (1) or not (-1): ‘)) |
| hyprhalvs = −1 |
| hxht = 0.866 * starsiz |
| segspan = ulccY − lrccY − (hxht/2.0) |
| segs = int(segspan/hxht) |
| colspan = lrccX − ulccX − starsiz |
| spoke = starsiz/2.0 |
| colstep = 1.5 * spoke |
| cols = int(colspan/colstep) + 1 |
| wedgArea = (starsiz * hxht)/8.0 |
| doc = -1 #description of context |
| # print(‘ulccX = ‘,ulccX,’\tulccY = ‘,ulccY) |
| # print(‘lrccX = ‘,lrccX,’\tlrccY = ‘,lrccY) |
| # print(‘star-span = ‘,starsiz) |
| # print(‘hump/slump = ‘,shhs) |
| # print(‘hyper-halves = ‘,hyprhalvs) |
| # print(‘hxht = ‘,hxht) |
| # print(‘segs = ‘,segs) |
| # print(‘cols = ‘,cols) |
| if hyprhalvs > 0: segs * = −1 |
| colz = cols |
| if shhs < 0: cols * = −1 |
| # Generate _hip file. |
| # fnam = input(‘Base name for files: ‘) |
| fnam = filnam.get() |
| hipnam = fnam + ‘_hip.txt’ |
| hipfil = open(hipnam,’w’) |
| hipfil.write(‘regn_cols_segs_doc ccX_Star_Cstp ccY_Hxht_WdgA\n’) |
| hipfil.write(str(regn)+’ ‘+str(ulccX)+’ ‘+str(ulccY)+’\n’) |
| hipfil.write(str(cols)+’ ‘+str(lrccX)+’ ‘+str(lrccY)+’\n’) |
| hipfil.write(str(segs)+’ ‘+str(starsiz)+’ ‘+str(hxht)+’\n’) |
| hipfil.write(str(doc)+’ ‘+str(colstep)+’ ‘+str(wedgArea)+’\n’) |
| hipfil.close() |
| #generate _tap file. |
| tapnam = fnam + ‘_tap.txt’ |
| tapfil = open(tapnam,’w’) |
| tapfil.write(‘col tapX tapY\n’) |
| topY = ulccY |
| botmY = topY − (segs * hxht) − (0.5 * hxht) |
| col = 1 |
| colX = ulccX + spoke |
| hilo = 1 |
| if shhs < 0: hilo = -1 * hilo |
| while col < = colz: |
| if hilo > 0: |
| tapfil.write(str(col)+’ ‘+str(colX)+’ ‘+str(botmY)+’\n’) |
| if hilo < 0: |
| tapfil.write(str(col)+’ ‘+str(colX)+’ ‘+str(topY)+’\n’) |
| hilo * = −1 |
| colX + = colstep |
| col + = 1 |
| tapfil.close() |
| # print(‘hip file generated.’) |
| msg.showinfo(‘filing’,’files finished’) |
| # byebye = input(‘Press ENTER to exit:’) |
| labl1 = Label(windOh,text = ‘Enter region number (integer) : ‘) |
| region = Entry(windOh,width = 20) |
| labl2 = Label(windOh,text = ‘Enter up-left ccX : ‘) |
| uplefccX = Entry(windOh,width = 20) |
| labl3 = Label(windOh,text = ‘Enter up-left ccY : ‘) |
| uplefccY = Entry(windOh,width = 20) |
| labl4 = Label(windOh,text = ‘Enter lo-right ccX : ‘) |
| loritccX = Entry(windOh,width = 20) |
| labl5 = Label(windOh,text = ‘Enter lo-right ccY : ‘) |
| loritccY = Entry(windOh,width = 20) |
| labl6 = Label(windOh,text = ‘Enter inscribed star-span : ‘) |
| stars = Entry(windOh,width = 20) |
| labl7 = Label(windOh,text = ‘Enter 1 for hump/slump or −1 :’) |
| rippl = Entry(windOh,width = 20) |
| labl8 = Label(windOh,text = ‘Enter base name of files : ‘) |
| filnam = Entry(windOh,width = 40) |
| btn_DoTell = Button(windOh,text = ‘ReadyRun’,command = hipspecs) |
| labl1.grid(row = 0,column = 0) |
| region.grid(row = 0,column = 1,padx = 5) |
| labl2.grid(row = 1,column = 0) |
| uplefccX.grid(row = 1,column = 1) |
| labl3.grid(row = 2,column = 0) |
| uplefccY.grid(row = 2,column = 1) |
| labl4.grid(row = 3,column = 0) |
| loritccX.grid(row = 3,column = 1) |
| labl5.grid(row = 4,column = 0) |
| loritccY.grid(row = 4,column = 1) |
| labl6.grid(row = 5,column = 0) |
| stars.grid(row = 5,column = 1) |
| labl7.grid(row = 6,column = 0) |
| rippl.grid(row = 6,column = 1) |
| labl8.grid(row = 7,column = 0) |
| btn_DoTell.grid(row = 7,column = 1,pady = 5) |
| filnam.grid(row = 8,column = 0,pady = 10,padx = 5) |
| windOh.mainloop() |
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Patil, G.; Myers, W.; Luo, Z.; Johnson, G.; Taillie, C. Multiscale assessment of landscapes and watersheds with synoptic multivariate spatial data in environmental and ecological statistics. Math. Comput. Model. 2000, 32, 257–272. [Google Scholar] [CrossRef]
- Hong, H.-J.; Kim, C.-K.; Lee, H.-W.; Lee, W.-K. Conservation, Restoration, and Sustainable Use of Biodiversity Based on Habitat Quality Monitoring: A Case Study on Jeju Island, South Korea (1989–2019). Land 2021, 10, 774. [Google Scholar] [CrossRef]
- Valeri, S.; Zavattero, L.; Capotorti, G. Ecological Connectivity in Agricultural Green Infrastructure: Suggested Criteria for Fine Scale Assessment and Planning. Land 2021, 10, 807. [Google Scholar] [CrossRef]
- Myers, W.L.; Patil, G.P. Statistical Geoinformatics for Human Environment Interface; Informa UK Limited: London, UK, 2012; ISBN 978-1-4200-8287-6. [Google Scholar]
- Brüggemann, R.; Patil, G.P. Ranking and Prioritization for Multi-Indicator Systems; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Myers, W.; Patil, G.P. Biodiversity in the Age of Ecological Indicators. Acta Biotheor. 2006, 54, 119–123. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- McGrath, M. R for Data Analysis; Easy Steps Ltd.: Warwickshire, UK, 2018. [Google Scholar]
- McGrath, M. Python in Easy Steps; Easy Steps Ltd.: Warwickshire, UK, 2015. [Google Scholar]
- Webster, R.; Oliver, M.A. Geostatistics for Environmental Scientists, 2nd ed.; John Wiley & Sons Ltd.: Chichester, UK, 2007; ISBN 978-0-470-02858-2. [Google Scholar]
- Myers, W.; Patil, G.P.; Joly, K. Echelon approach to areas of concern in synoptic regional monitoring. Environ. Ecol. Stat. 1997, 4, 131–152. [Google Scholar] [CrossRef]
- Myers, W.L.; McKenney-Easterling, M.; Hychka, K.; Griscom, B.; Bishop, J.A.; Bayard, A.; Rocco, G.L.; Brooks, R.P.; Constantz, G.; Patil, G.P.; et al. Contextual clustering for configuring collaborative conservation of watersheds in the Mid-Atlantic Highlands. Environ. Ecol. Stat. 2006, 13, 391–407. [Google Scholar] [CrossRef]
- Myers, W.; Patil, G.P. Multivariate Methods of Representing Relations in R for Prioritization Purposes: Selective Scaling, Comparative Clustering, Collective Criteria and Sequenced Sets; Springer: New York, NY, USA, 2012; ISBN 978-1-4616-3121-3. [Google Scholar]
- Myers, W.; Bishop, J.; Brooks, R.; O’Connell, T.; Argent, D.; Storm, G.; Stauffer, J., Jr. The Pennsylvania GAP Analysis Final Report; The Pennsylvania State University: University Park, PA, USA, 2000. [Google Scholar]
- Myers, W.L.; Kurihara, K.; Patil, G.P.; Vraney, R. Finding upper-level sets in cellular surface data using echelons and saTScan. Environ. Ecol. Stat. 2006, 13, 379–390. [Google Scholar] [CrossRef]
- Smits, P.; Myers, W. Echelon approach to characterize and understand spatial structures of change in multitemporal remote sensing imagery. IEEE Trans. Geosci. Remote Sens. 2000, 38, 2299–2309. [Google Scholar] [CrossRef]
- Myers, W.; Patil, G. Preliminary prioritization based on partial order theory and R software for compositional complexes in landscape ecology, with applications to restoration, remediation, and enhancement. Environ. Ecol. Stat. 2010, 17, 411–436. [Google Scholar] [CrossRef]
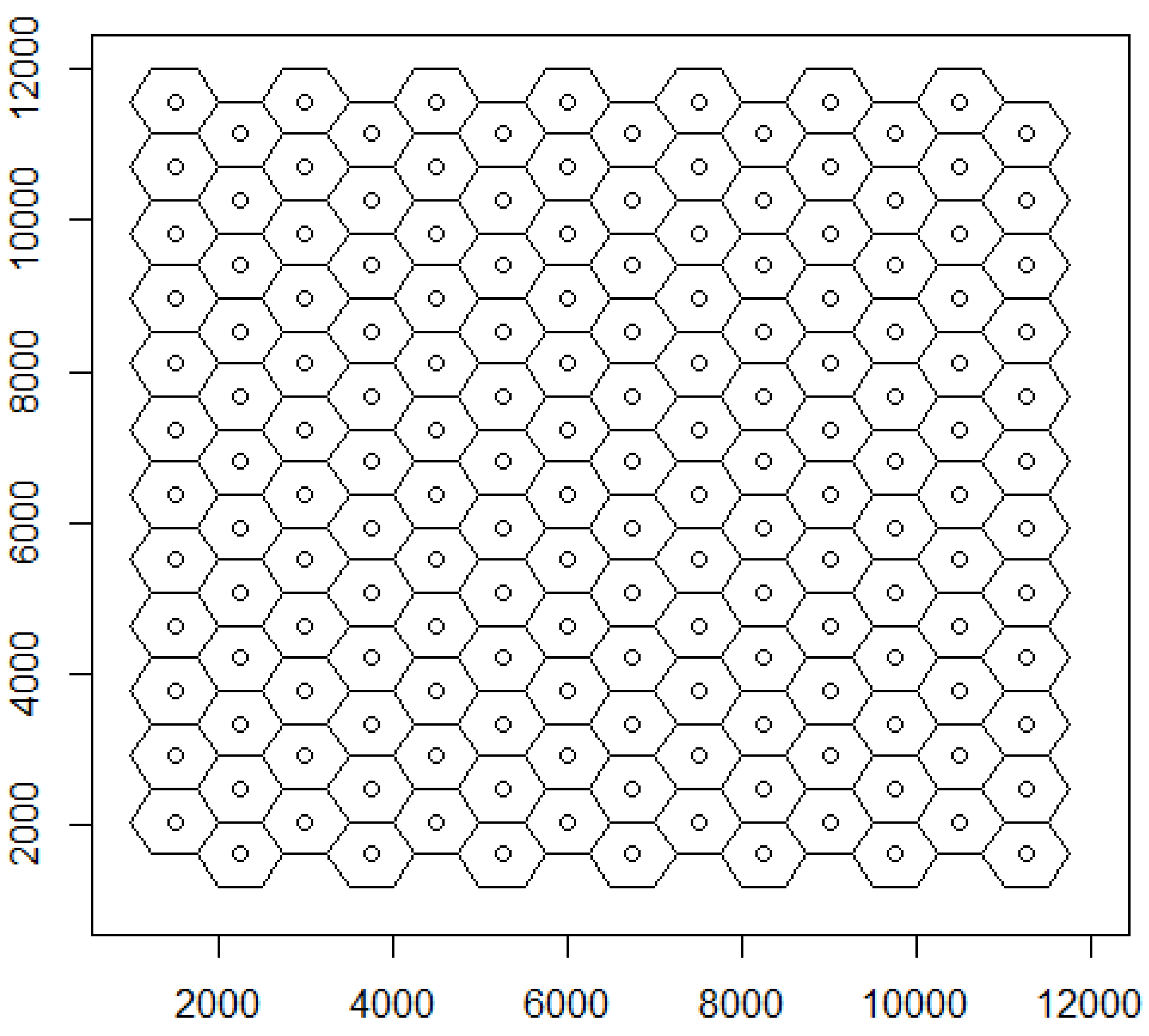











| regn_cols_segs_doc | ccX_Star_Cstp | ccY_Hxht_WdgA |
|---|---|---|
| 184 | 100.0 | 500.0 |
| −11 | 295.0 | 160.0 |
| 17 | 22.11 | 19.14726 |
| −1 | 16.5825 | 52.9182398249999 |
| seg | col | pin | Birds | Mamls | ElvSD | PctFC | PctFP | PctOP | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 17 | 10 | −187 | 1 | 1 | 1.0 | 1.0 | 1.0 | |
| 2404 | 1 | 1 | 1 | 104 | 48 | 181 | 99.5 | 99.5 | 0.2 |
| 2524 | 2 | 1 | 2 | 103 | 48 | 187 | 94.1 | 77.8 | 2.1 |
| 2645 | 3 | 1 | 3 | 115 | 48 | 205 | 91.9 | 91 | 2.6 |
| 2767 | 4 | 1 | 4 | 114 | 48 | 72 | 94.9 | 94.8 | 1.2 |
| 2890 | 5 | 1 | 5 | 106 | 47 | 87 | 81.2 | 78.4 | 8.7 |
| Birdubt | ||||
|---|---|---|---|---|
| site | upr | btm | tls | |
| 1 | 1 | 145 | 136 | 2 |
| 2 | 2 | 145 | 141 | 2 |
| 3 | 3 | 144 | 132 | 6 |
| 4 | 4 | 143 | 133 | 3 |
| 5 | 5 | 143 | 143 | 1 |
| 6 | 6 | 142 | 142 | 1 |
| 7 | 7 | 141 | 141 | 1 |
| 8 | 8 | 140 | 132 | 3 |
| 9 | 9 | 139 | 139 | 1 |
| 10 | 10 | 138 | 131 | 8 |
| 11 | 11 | 138 | 138 | 1 |
| 12 | 12 | 138 | 138 | 1 |
| 13 | 13 | 135 | 131 | 2 |
| 14 | 14 | 135 | 135 | 1 |
| 15 | 15 | 134 | 134 | 1 |
| 16 | 16 | 133 | 133 | 1 |
| 17 | 17 | 130 | 130 | 1 |
| 18 | 18 | 127 | 127 | 1 |
| 19 | 19 | 125 | 119 | 2 |
| 20 | 20 | 120 | 120 | 1 |
| Birds | Mamls | ElevSD | PctFC | PctFP | |
|---|---|---|---|---|---|
| 2404 | 6 | 107 | 181 | 179 | 179 |
| 2524 | 4 | 107 | 182 | 153 | 110 |
| 2645 | 23 | 107 | 183 | 150 | 150 |
| 2767 | 21 | 107 | 90 | 156 | 157 |
| 2890 | 8 | 86 | 117 | 114 | 112 |
| 3014 | 103 | 86 | 90 | 140 | 142 |
| Domain | Column | Connect | |
|---|---|---|---|
| [1,] | 1 | 4 | 0.984626 |
| [2,] | 1 | 5 | 0.984626 |
| [3,] | 1 | 6 | 0.943667 |
| [4,] | 2 | 2 | 0.562876 |
| [5,] | 2 | 3 | 0.562876 |
| [6,] | 2 | 1 | 0.532953 |
| fivROLScls9 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| fivROLScls12 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| 1 | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 0 | 0 | 20 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 0 | 0 | 0 | 22 | 0 | 0 | 0 | 0 | 0 |
| 5 | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7 | 0 | 0 | 0 | 0 | 25 | 0 | 0 | 0 | 0 |
| 8 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 0 | 0 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 18 | 0 | 0 |
| 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 |
| 11 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Myers, W.L. Scalable Shared Scripting for Spatial Structure of Regionalized Ratings. Land 2021, 10, 859. https://doi.org/10.3390/land10080859
Myers WL. Scalable Shared Scripting for Spatial Structure of Regionalized Ratings. Land. 2021; 10(8):859. https://doi.org/10.3390/land10080859
Chicago/Turabian StyleMyers, Wayne L. 2021. "Scalable Shared Scripting for Spatial Structure of Regionalized Ratings" Land 10, no. 8: 859. https://doi.org/10.3390/land10080859
APA StyleMyers, W. L. (2021). Scalable Shared Scripting for Spatial Structure of Regionalized Ratings. Land, 10(8), 859. https://doi.org/10.3390/land10080859





