Supporting Adaptive Connectivity in Dynamic Landscapes
Abstract
:1. Introduction
2. Materials and Methods
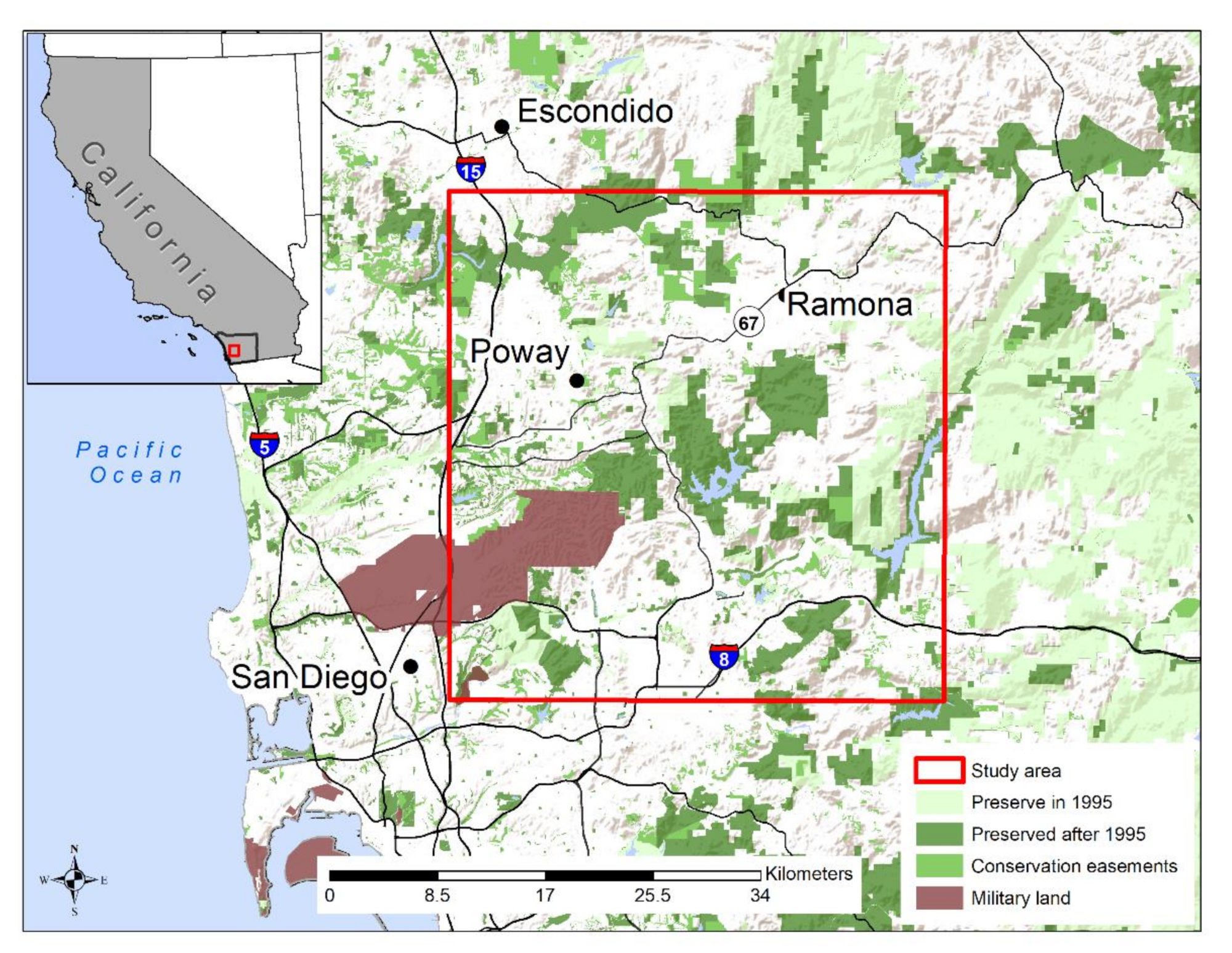
2.1. Study Area
2.2. Focal Species and Environmental Variables
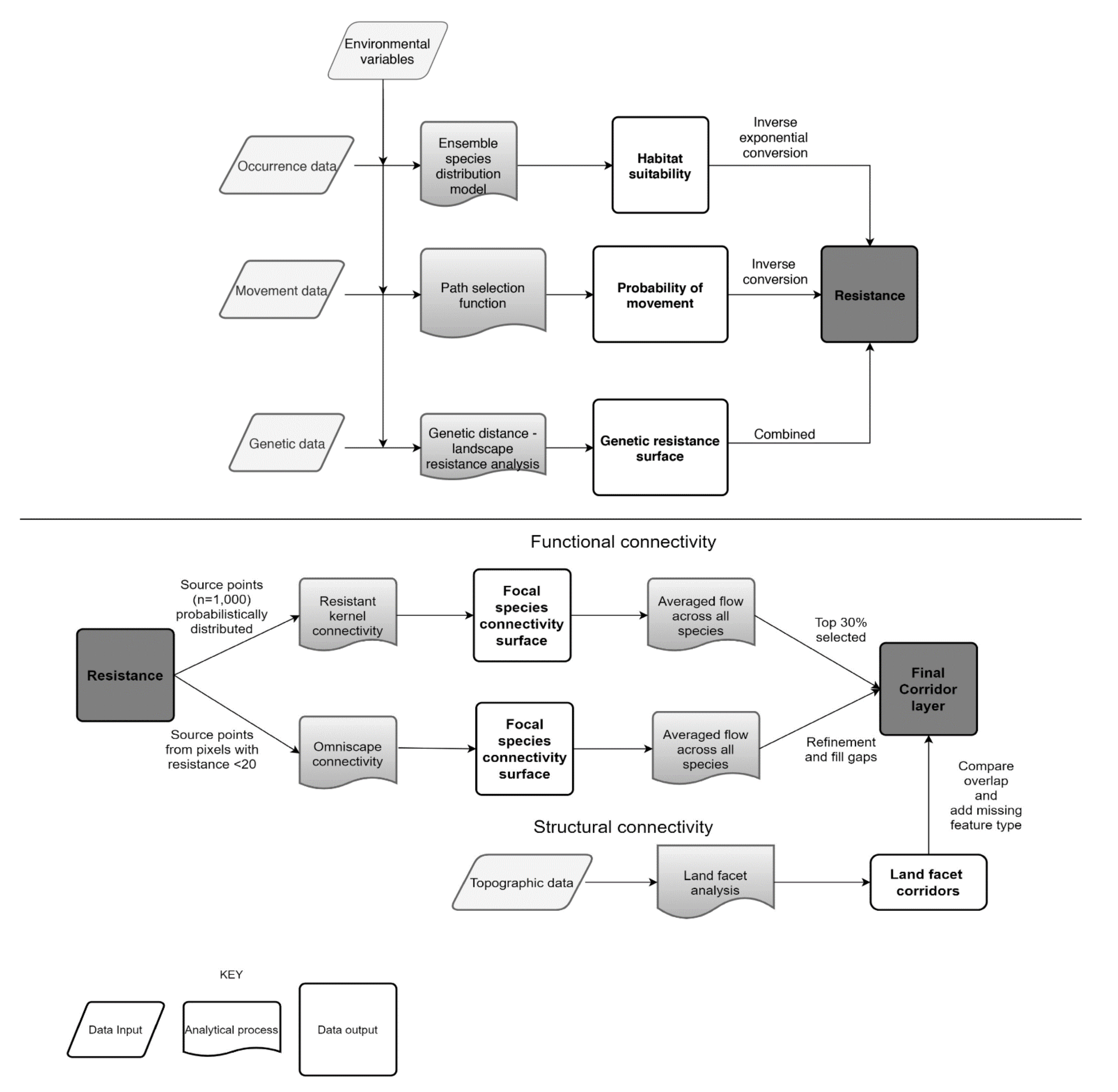
2.3. Modeling Approach Overview
2.4. Habitat Use, Movement, and Resistance Modeling
2.5. Connectivity Assessment
3. Results
3.1. Habitat Use, Movement, Resistance, and Corridor Modeling
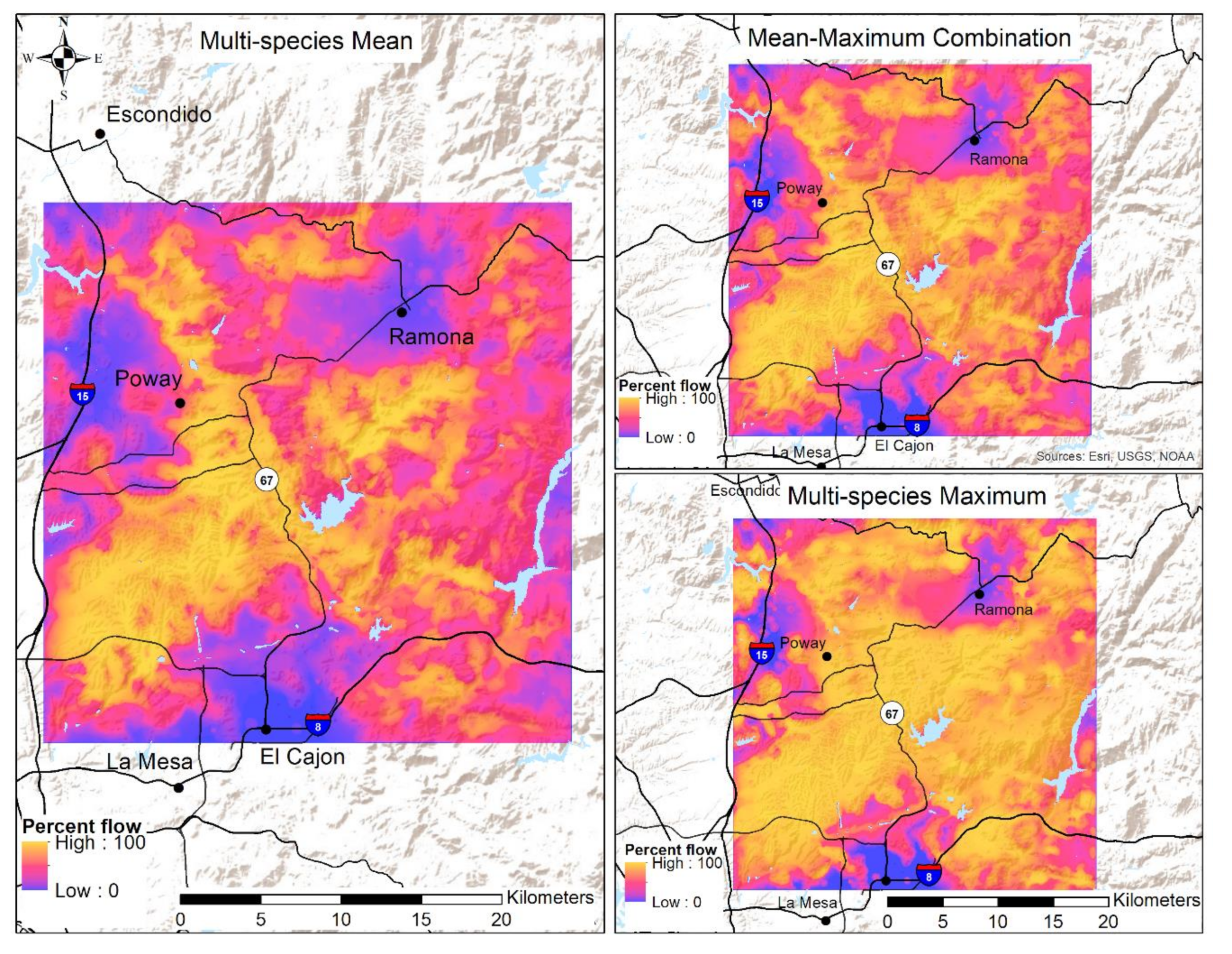
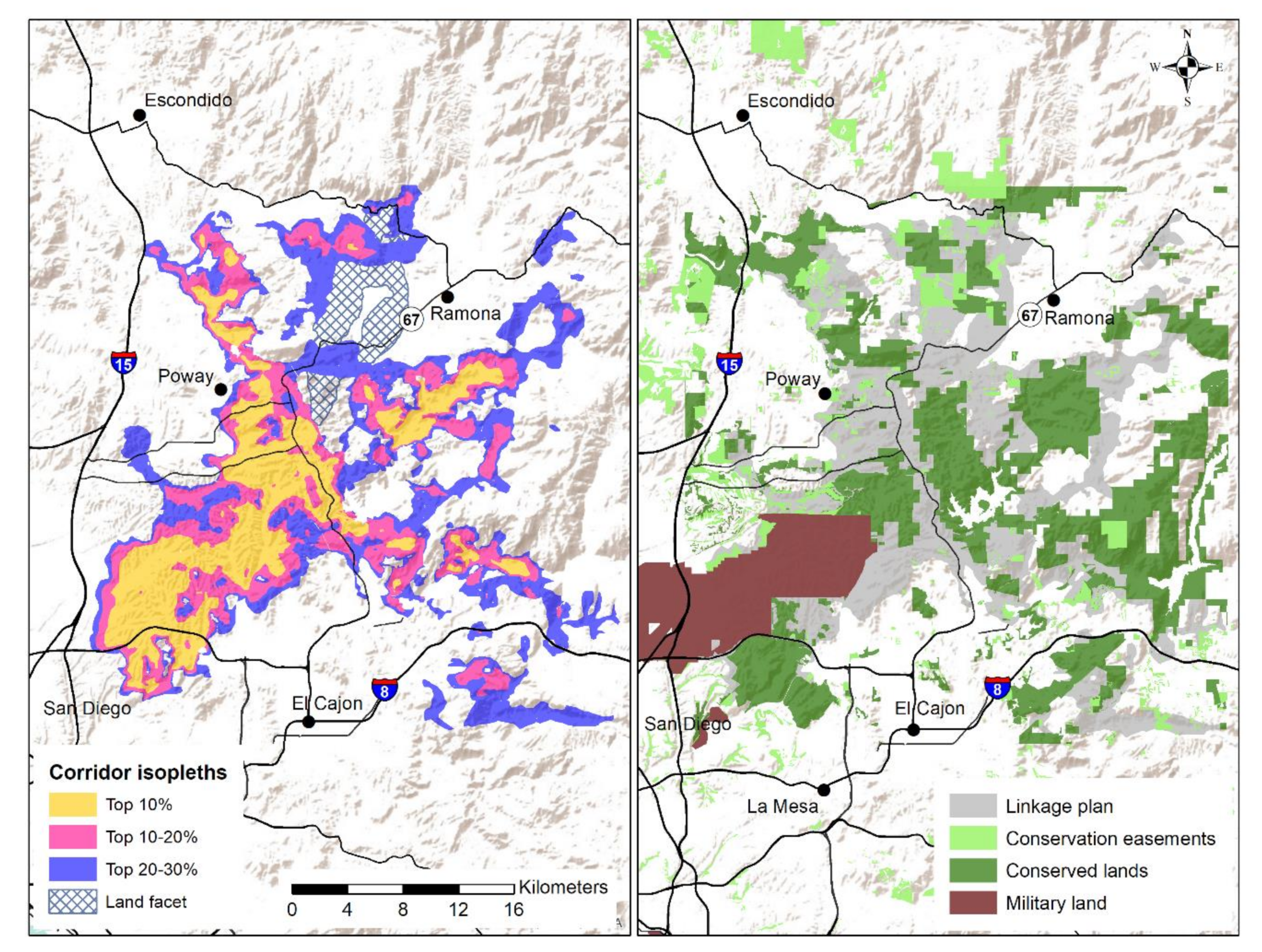
3.2. Connectivity Assessment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
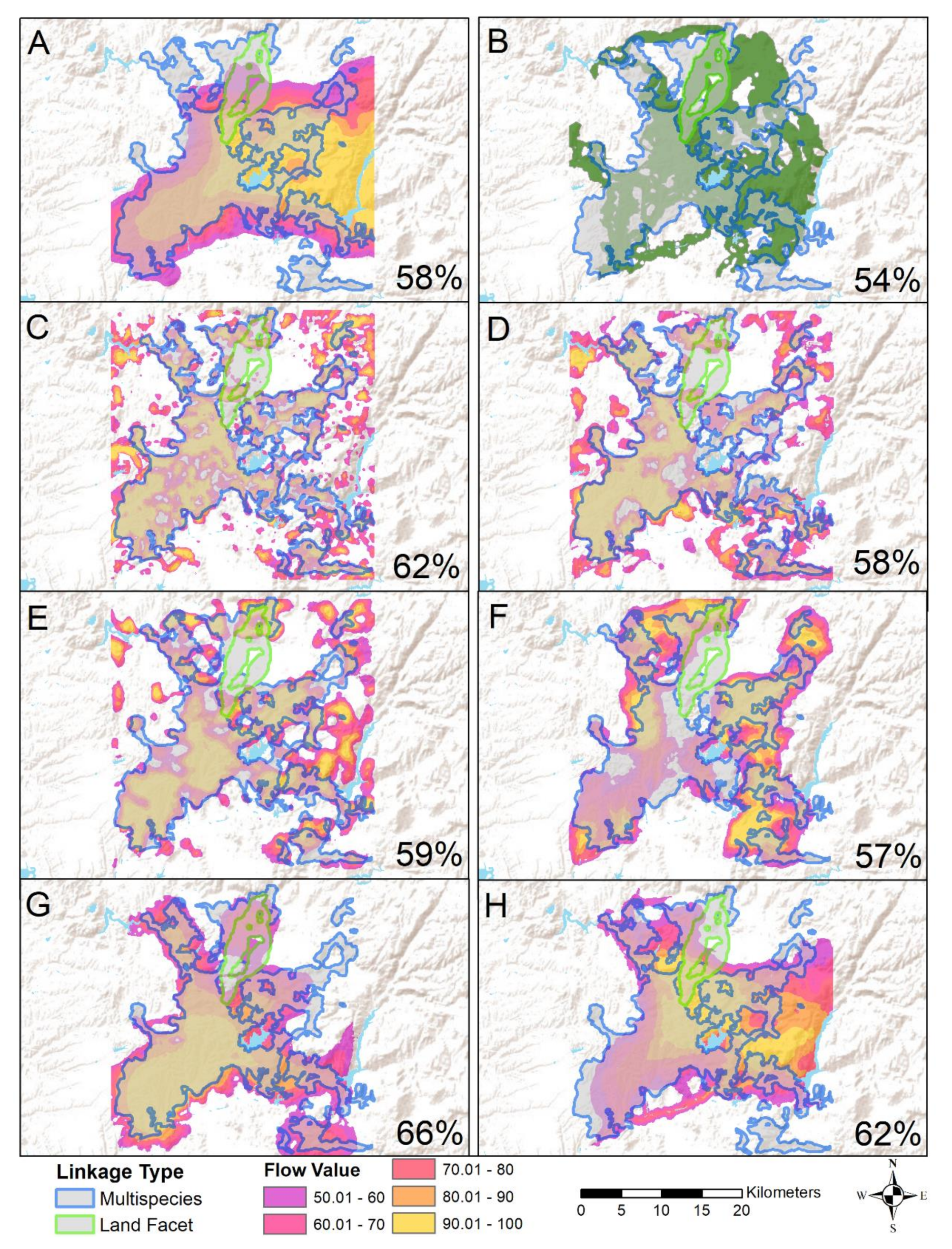
Appendix A.1. Conservation Framework of San Diego’s Multiple Species Conservation Plan
Appendix A.2. Multi-Species Flow Value Figure

References
- Boitani, L.; Falcucci, A.; Maiorano, L.; Rondinini, C. Ecological networks as conceptual frameworks or operational tools in conservation. Conserv. Biol. 2007, 21, 1414–1422. [Google Scholar] [CrossRef]
- Crooks, K.R.; Sanjayan, M. Connectivity Conservation Management; Cambridge University Press: Cambridge, UK, 2006; ISBN 9781849774727. [Google Scholar]
- Gilbert-Norton, L.; Wilson, R.; Stevens, J.R.; Beard, K.H. A Meta-Analytic Review of Corridor Effectiveness. Conserv. Biol. 2010, 24, 660–668. [Google Scholar] [CrossRef]
- Brown, J.H.; Kodric-Brown, A. Turnover Rates in Insular Biogeography: Effect of Immigration on Extinction. Ecology 1977, 58, 445–449. [Google Scholar] [CrossRef] [Green Version]
- Noss, R.F. Corridors in Real Landscapes: A Reply to Simberloff and Cox. Conserv. Biol. 1987, 1, 159–164. [Google Scholar] [CrossRef]
- Beier, P.; Noss, R.F. Do habitat corridors provide connectivity? Conserv. Biol. 1998, 12, 1241–1252. [Google Scholar] [CrossRef]
- Taylor, P.D.; Fahrig, L.; Henein, K.; Merriam, G. Connectivity Is a Vital Element of Landscape Structure. Oikos 1993, 68, 571. [Google Scholar] [CrossRef] [Green Version]
- Tischendorf, L.; Fahrig, L. On the usage and measurement of landscape connectivity. Oikos 2000, 90, 7–19. [Google Scholar] [CrossRef] [Green Version]
- Carter, S.K.; Januchowski-Hartley, S.R.; Pohlman, J.D.; Bergeson, T.L.; Pidgeon, A.M.; Radeloff, V.C. An evaluation of environmental, institutional and socio-economic factors explaining successful conservation plan implementation in the north-central United States. Biol. Conserv. 2015, 192, 135–144. [Google Scholar] [CrossRef]
- Jongman, R.H.G. Nature conservation planning in Europe: Developing ecological networks. Landsc. Urban Plan. 1995, 32, 169–183. [Google Scholar] [CrossRef]
- Convention on Biological Diversity. Decision UNEP/CBD/COP/DEC/X/2 Adopted by the Conference of the Parties to the Convention on Biological Diversity at its Tenth Meeting. 2011. Available online: https://www.cbd.int/decision/cop/?id=12268 (accessed on 16 July 2020).
- Saura, S.; Bertzky, B.; Bastin, L.; Battistella, L.; Mandrici, A.; Dubois, G. Global trends in protected area connectivity from 2010 to 2018. Biol. Conserv. 2019, 238, 108183. [Google Scholar] [CrossRef]
- Costello, C.; Polasky, S. Dynamic reserve site selection. Resour. Energy Econ. 2004, 26, 157–174. [Google Scholar] [CrossRef] [Green Version]
- Pressey, R.L.; Cabeza, M.; Watts, M.E.; Cowling, R.M.; Wilson, K.A. Conservation planning in a changing world. Trends Ecol. Evol. 2007, 22, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Van Teeffelen, A.J.A.; Vos, C.C.; Opdam, P. Species in a dynamic world: Consequences of habitat network dynamics on conservation planning. Biol. Conserv. 2012, 153, 239–253. [Google Scholar] [CrossRef] [Green Version]
- Opdam, P.; Steingröver, E.; van Rooij, S. Ecological networks: A spatial concept for multi-actor planning of sustainable landscapes. Landsc. Urban Plan. 2006, 75, 322–332. [Google Scholar] [CrossRef]
- Fagan, W.F.; Calabrese, J.M. Quantifying connectivity: Balancing metric performance with data requirements. In Connectivity Conservation; Crooks, K.R., Sanjayan, M., Eds.; Cambridge University Press: Cambridge, UK, 2006; pp. 297–317. ISBN 9780521857062. [Google Scholar]
- Kool, J.T.; Moilanen, A.; Treml, E.A. Population connectivity: Recent advances and new perspectives. Landsc. Ecol. 2013, 28, 165–185. [Google Scholar] [CrossRef]
- Epps, C.W.; Mutayoba, B.M.; Gwin, L.; Brashares, J.S. An empirical evaluation of the African elephant as a focal species for connectivity planning in East Africa. Divers. Distrib. 2011, 17, 603–612. [Google Scholar] [CrossRef]
- McClure, M.L.; Hansen, A.J.; Inman, R.M. Connecting models to movements: Testing connectivity model predictions against empirical migration and dispersal data. Landsc. Ecol. 2016, 31, 1419–1432. [Google Scholar] [CrossRef]
- Zeller, K.A.; Jennings, M.K.; Vickers, T.W.; Ernest, H.B.; Cushman, S.A.; Boyce, W.M. Are all data types and connectivity models created equal? Validating common connectivity approaches with dispersal data. Divers. Distrib. 2018, 24, 868–879. [Google Scholar] [CrossRef] [Green Version]
- Naidoo, R.; Kilian, J.W.; Du Preez, P.; Beytell, P.; Aschenborn, O.; Taylor, R.D.; Stuart-Hill, G. Evaluating the effectiveness of local- and regional-scale wildlife corridors using quantitative metrics of functional connectivity. Biol. Conserv. 2018, 217, 96–103. [Google Scholar] [CrossRef]
- Sanderson, E.W.; Redford, K.H.; Vedder, A.; Coppolillo, P.B.; Ward, S.E. A conceptual model for conservation planning based on landscape species requirements. Landsc. Urban Plan. 2002, 58, 41–56. [Google Scholar] [CrossRef]
- Lambeck, R.J. Focal species: A multi-species umbrella for nature conservation. Conserv. Biol. 1997, 11, 849–856. [Google Scholar] [CrossRef] [Green Version]
- Dilkina, B.; Houtman, R.; Gomes, C.P.; Montgomery, C.A.; McKelvey, K.S.; Kendall, K.; Graves, T.A.; Bernstein, R.; Schwartz, M.K. Trade-offs and efficiencies in optimal budget-constrained multispecies corridor networks. Conserv. Biol. 2017, 31, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Gippoliti, S.; Battisti, C. More cool than tool: Equivoques, conceptual traps and weaknesses of ecological networks in environmental planning and conservation. Land Use Policy 2017, 68, 686–691. [Google Scholar] [CrossRef]
- Albert, C.H.; Rayfield, B.; Dumitru, M.; Gonzalez, A. Applying network theory to prioritize multispecies habitat networks that are robust to climate and land-use change. Conserv. Biol. 2017, 31, 1383–1396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brost, B.M.; Beier, P. Comparing linkage designs based on land facets to linkage designs based on focal species. PLoS ONE 2012, 7, e48965. [Google Scholar] [CrossRef] [Green Version]
- Krosby, M.; Breckheimer, I.; Pierce, D.J.; Singleton, P.H.; Hall, S.A.; Halupka, K.C.; Gaines, W.L.; Long, R.A.; McRae, B.H.; Cosentino, B.L.; et al. Focal species and landscape “naturalness” corridor models offer complementary approaches for connectivity conservation planning. Landsc. Ecol. 2015, 30, 2121–2132. [Google Scholar] [CrossRef]
- Anderson, M.G.; Comer, P.J.; Beier, P.; Lawler, J.J.; Schloss, C.A.; Buttrick, S.; Albano, C.M.; Faith, D.P. Case studies of conservation plans that incorporate geodiversity. Conserv. Biol. 2015, 29, 680–691. [Google Scholar] [CrossRef]
- Beier, P.; Brost, B. Use of land facets to plan for climate change: Conserving the arenas, not the actors. Conserv. Biol. 2010, 24, 701–710. [Google Scholar] [CrossRef]
- Theobald, D.M.; Reed, S.E.; Fields, K.; Soulé, M. Connecting natural landscapes using a landscape permeability model to prioritize conservation activities in the United States. Conserv. Lett. 2012, 5, 123–133. [Google Scholar] [CrossRef]
- Greer, K.A. Habitat Conservation Planning in San Diego County, California: Lessons Learned After Five Years of Implementation. Environ. Pract. 2004, 6, 230–239. [Google Scholar] [CrossRef]
- Beier, P.; Penrod, K.; Luke, C.; Spencer, W.; Cabañero, C. South Coast missing linkages: Restoring connectivity to wildlands in the largest metropolitan area in the United States. In Connectivity Conservation; Crooks, K.R., Sanjayan, M.A., Eds.; Cambridge University Press: Cambridge, UK, 2006; pp. 555–586. [Google Scholar]
- South Coast Wildlands. South Coast Missing Linkages: A Wildland Network for the South Coast Ecoregion. 2008. Available online: http://www.scwildlands.org/reports/SCMLRegionalReport.pdf (accessed on 16 July 2020).
- Spencer, W.D.; Beier, P.; Penrod, K.; Paulman, K.; Rustigian-Romsos, H.; Strittholt, J.; Parisi, M.; Pettler, A. California Essential Habitat Connectivity Project: A Strategy for Conservation a Connected California; 2010. Available online: https://nrm.dfg.ca.gov/FileHandler.ashx?DocumentID=18366 (accessed on 16 July 2020).
- Crooks, K.R. Relative sensitivities of mammalian carnivores to habitat fragmentation. Conserv. Biol. 2002, 16, 488–502. [Google Scholar] [CrossRef]
- Mitelberg, A.; Vandergast, A.G. Non-Invasive Genetic Sampling of Southern Mule Deer (Odocoileus hemionus fuliginatus) Reveals Limited Movement Across California State Route 67 in San Diego County. West. Wildl. 2016, 3, 8–18. [Google Scholar]
- Baker, M.; Nur, N.; Geupel, G.R. Correcting biased estimates of dispersal and survival due to limited study area: Theory and an application using wrentits. Condor 1995, 97, 663–674. [Google Scholar] [CrossRef]
- Mendelsohn, M.B.; Brehme, C.S.; Rochester, C.J.; Stokes, D.C.; Hathaway, S.A.; Fisher, R.N. Responses in bird communities to wildland fires in southern California. Fire Ecol. Spec. Issue 2008, 4, 63–82. [Google Scholar] [CrossRef]
- Delaney, K.S.; Riley, S.P.D.; Fisher, R.N. A rapid, strong, and convergent genetic response to urban habitat fragmentation in four divergent and widespread vertebrates. PLoS ONE 2010, 5, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Tremor, S.; Stokes, D.; Spencer, W.; Diffendorfer, J.; Thomas, H.; Chivers, S.; Unitt, P. San Diego County Mammal Atlas. In San Diego Society of Natural History, 46th ed.; San Diego Natural History Museum: San Diego, CA, USA, 2017; ISBN 9780692895399. [Google Scholar]
- County of San Diego SanBIOS Biological Database. Available online: http://rdw.sandag.org/Account/GetFSFile.aspx?dir=Ecology&Name=SanBIOS.zip (accessed on 21 October 2016).
- Ebird Ebird: An Online Database of Bird Distribution and Abundance [Web Application]. Available online: http://www.ebird.org (accessed on 28 December 2016).
- Jennings, M.K.; Lewison, R.L. Planning for Connectivity under Climate Change: Using Bobcat Movement to Assess Landscape Connectivity across San Diego County’s Open Spaces; San Diego State University: San Diego, CA, USA, 2013. [Google Scholar]
- San Diego Management and Monitoring Program Master Occurrence Matrix Database. Available online: https://www.sciencebase.gov/catalog/item/53e27963e4b0fe532be3bddf (accessed on 28 December 2016).
- Ernest, H.B.; Vickers, T.W.; Morrison, S.A.; Buchalski, M.R.; Boyce, W.M. Fractured Genetic Connectivity Threatens a Southern California Puma (Puma concolor) Population. PLoS ONE 2014, 9, e107985. [Google Scholar] [CrossRef] [Green Version]
- Zeller, K.A.; McGarigal, K.; Cushman, S.A.; Beier, P.; Vickers, T.W.; Boyce, W.M. Using step and path selection functions for estimating resistance to movement: Pumas as a case study. Landsc. Ecol. 2016, 31, 1319–1335. [Google Scholar] [CrossRef]
- Johnson, C.J.; Seip, D.R.; Boyce, M.S. A quantitative approach to conservation planning: Using resource selection functions to map the distribution of mountain caribou at multiple spatial scales. J. Appl. Ecol. 2004, 41, 238–251. [Google Scholar] [CrossRef]
- McGarigal, K.; Wan, H.Y.; Zeller, K.A.; Timm, B.C.; Cushman, S.A. Multi-scale habitat selection modeling: A review and outlook. Landsc. Ecol. 2016, 31, 1161–1175. [Google Scholar] [CrossRef]
- Adriaensen, F.; Chardon, J.P.; De Blust, G.; Swinnen, E.; Villalba, S.; Gulinck, H.; Matthysen, E. The application of ‘least-cost’ modelling as a functional landscape model. Landsc. Urban Plan. 2003, 64, 233–247. [Google Scholar] [CrossRef]
- Zeller, K.A.; Vickers, T.W.; Ernest, H.B.; Boyce, W.M. Multi-level, multi-scale resource selection functions and resistance surfaces for conservation planning: Pumas as a case study. PLoS ONE 2017, 12, e0179570. [Google Scholar] [CrossRef] [PubMed]
- Keeley, A.T.H.; Beier, P.; Gagnon, J.W. Estimating landscape resistance from habitat suitability: Effects of data source and nonlinearities. Landsc. Ecol. 2016, 31, 2151–2162. [Google Scholar] [CrossRef]
- Trainor, A.M.; Walters, J.R.; Morris, W.F.; Sexton, J.; Moody, A. Empirical estimation of dispersal resistance surfaces: A case study with red-cockaded woodpeckers. Landsc. Ecol. 2013, 28, 755–767. [Google Scholar] [CrossRef]
- Mateo-Sánchez, M.C.; Balkenhol, N.; Cushman, S.; Pérez, T.; Domínguez, A.; Saura, S. A comparative framework to infer landscape effects on population genetic structure: Are habitat suitability models effective in explaining gene flow? Landsc. Ecol. 2015, 30, 1405–1420. [Google Scholar] [CrossRef]
- Compton, B.W.; McGarigal, K.; Cushman, S.A.; Gamble, L.R. A resistant-kernel model of connectivity for amphibians that breed in vernal pools. Conserv. Biol. 2007, 21, 788–799. [Google Scholar] [CrossRef]
- McRae, B.H.; Popper, K.; Jones, A.; Schindel, M.; Buttrick, S.; Hall, K.; Unnasch, R.S.; Platt, J. Conserving Nature’s Stage: Mapping Omnidirectional Connectivity for Resilient Terrestrial Landscapes in the Pacific Northwest; The Nature Conservancy: Portland, OR, USA, 2016. [Google Scholar]
- Landguth, E.L.; Hand, B.K.; Glassy, J.; Cushman, S.A.; Sawaya, M.A. UNICOR: A species connectivity and corridor network simulator. Ecography 2012, 35, 9–14. [Google Scholar] [CrossRef]
- Ribble, D.O. Dispersal in a monogamous rodent, Peromyscus californicus. Ecology 1992, 73, 859–866. [Google Scholar] [CrossRef] [Green Version]
- Beier, P. Dispersal of juvenile cougars in fragmented habitat. J. Wildl. Manag. 1995, 59, 228. [Google Scholar] [CrossRef]
- Smith, M.H. Dispersal capacity of the dusky-footed woodrat, Neotoma fuscipes. Am. Midl. Nat. 1965, 74, 457. [Google Scholar] [CrossRef]
- Beier, P.; Majka, D.R.; Spencer, W.D. Forks in the road: Choices in procedures for designing wildland linkages. Conserv. Biol. 2008, 22, 836–851. [Google Scholar] [CrossRef]
- Hesselbarth, M.H.K.; Sciaini, M.; With, K.A.; Wiegand, K.; Nowosad, J. Landscapemetrics: An open-source R tool to calculate landscape metrics. Ecography 2019, 42, 1648–1657. [Google Scholar] [CrossRef] [Green Version]
- Jin, S.; Yang, L.; Danielson, P.; Homer, C.; Fry, J.; Xian, G. A comprehensive change detection method for updating the National Land Cover Database to circa 2011. Remote Sens. Environ. 2013, 132, 159–175. [Google Scholar] [CrossRef] [Green Version]
- Buttrick, S.; Popper, K.; McRae, B.; Unnasch, B.; Schindel, M.; Jones, A.; Platt, J. Conserving Nature’s Stage: Identifying Resilient Terrestrial Landscapes in the Pacific Northwest; Portland, OR, USA. 2015. Available online: http://nature.ly/resilienceNW (accessed on 18 October 2019).
- Valerio, F.; Carvalho, F.; Barbosa, A.M.; Mira, A.; Santos, S.M. Accounting for Connectivity Uncertainties in Predicting Roadkills: A Comparative Approach between Path Selection Functions and Habitat Suitability Models. Environ. Manag. 2019, 64, 329–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keeley, A.T.H.; Beier, P.; Keeley, B.W.; Fagan, M.E. Habitat suitability is a poor proxy for landscape connectivity during dispersal and mating movements. Landsc. Urban Plan. 2017, 161, 90–102. [Google Scholar] [CrossRef]
- Abrahms, B.; Sawyer, S.C.; Jordan, N.R.; McNutt, J.W.; Wilson, A.M.; Brashares, J.S. Does wildlife resource selection accurately inform corridor conservation? J. Appl. Ecol. 2017, 54, 412–422. [Google Scholar] [CrossRef] [Green Version]
- Santini, L.; Saura, S.; Rondinini, C. Connectivity of the global network of protected areas. Divers. Distrib. 2016, 22, 199–211. [Google Scholar] [CrossRef] [Green Version]
- Saura, S.; Bastin, L.; Battistella, L.; Mandrici, A.; Dubois, G. Protected areas in the world’s ecoregions: How well connected are they? Ecol. Indic. 2017, 76, 144–158. [Google Scholar] [CrossRef]
| Focal Species (Scientific Name) | Data Type(s) | Data Source(s) | Analytical Method(s) |
|---|---|---|---|
| California mouse (Peromyscus californicus) | Occurrence points | SDNHM Mammal Atlas 1, SanBIOS 2 | Species Distribution Model |
| Big-eared woodrat (Neotoma macrotis) | Occurrence points | SDNHM Mammal Atlas 1, SanBIOS 2 | Species Distribution Model |
| Wrentit (Chamaea fasciata) | Occurrence points | eBIRD 3 | Species Distribution Model |
| Mule deer (Odocoileus hemionus californicus) | Occurrence points and genetic data | SDNHM Mammal Atlas 1, SanBIOS 2, SDSU 4, SDMMP MOM 5, USGS 6 | Species Distribution Model and Landscape genetics analysis |
| Bobcat (Lynx rufus) | GPS telemetry and genetic data | SDSU 4 | Resource and Movement Selection Functions and Landscape genetics analysis |
| Puma (Puma concolor) | GPS telemetry and genetic data | University of California, Davis 7 | Resource and Movement Selection Functions and Landscape genetics analysis |
| Area (km2) | Mean Corridor Length (km) | Mean Corridor Width (km) | Aggregation(%) | Cohesion(%) | |
|---|---|---|---|---|---|
| Preserve in 1995 | 142.5 | 96.2 | 98.8 | ||
| Preserve in 2017 | 284.9 | 96.6 | 99.0 | ||
| Combined multi-species linkage | 269.5 | 12.7 | 2.1 | 98.5 | 99.9 |
| Average multi-species linkage | 249.9 | 12.8 | 2.1 | 98.4 | 99.9 |
| Mean–max multi-species linkage | 339.7 | 16.2 | 2 | 98.7 | 99.9 |
| Maximum multi-species linkage | 525.0 | 12.7 | 2.2 | 99.3 | 100.0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jennings, M.K.; Zeller, K.A.; Lewison, R.L. Supporting Adaptive Connectivity in Dynamic Landscapes. Land 2020, 9, 295. https://doi.org/10.3390/land9090295
Jennings MK, Zeller KA, Lewison RL. Supporting Adaptive Connectivity in Dynamic Landscapes. Land. 2020; 9(9):295. https://doi.org/10.3390/land9090295
Chicago/Turabian StyleJennings, Megan K., Katherine A. Zeller, and Rebecca L. Lewison. 2020. "Supporting Adaptive Connectivity in Dynamic Landscapes" Land 9, no. 9: 295. https://doi.org/10.3390/land9090295
APA StyleJennings, M. K., Zeller, K. A., & Lewison, R. L. (2020). Supporting Adaptive Connectivity in Dynamic Landscapes. Land, 9(9), 295. https://doi.org/10.3390/land9090295







