1. Introduction
The genus
Legionella consists of facultative intracellular Gram-negative bacteria commonly found in freshwater and human-made water systems. Approximately 60 different species have been identified, over 20 of which are classified as human pathogens. The species of greatest relevance in healthcare is
Legionella pneumophila, which is further divided into 15 serogroups with varying pathogenic characteristics. The most dangerous serogroup for humans is serogroup 1 (SG1), which is responsible for more than 90% of confirmed cases of Legionnaires’ disease (LD) and 10–15% of the associated mortality [
1].
Infections caused by
Legionella occur exclusively through the inhalation of aerosolized water particles that carry the bacterium into the lungs. The clinical manifestation depends on the host’s immune status: the most severe form is LD, a potentially fatal pneumonia, while milder exposure may result in Pontiac fever, a flu-like illness that typically resolves without hospitalisation [
2].
According to a report published by the European Centre for Disease Prevention and Control (ECDC) in 2023, over 10,700 cases of LD were recorded in Europe in 2021 [
3]. This figure reflects a steady increase aligned with epidemiological projections. The majority of cases were reported in France, Spain, Italy, and Germany—together accounting for 75% of all cases—with Italy reporting over 2700 cases (4.6 per 100,000 inhabitants) [
3,
4].
Legionella is a ubiquitous environmental microorganism, commonly found in natural aquatic habitats such as springs, thermal waters, and surface water bodies. However, it can also colonise a wide variety of engineered water systems, including water distribution networks, storage tanks, cooling towers, and other infrastructure in hospitals, hotels, and large public buildings. Although
Legionella contamination is frequently reported in drinking water systems, there are few studies of
Legionella contamination in non-potable water systems such as wastewater treatment plants (WWTPs). In fact, treated and untreated wastewater systems, reclaimed water distribution networks, and reuse application, can serve as potential reservoirs or transmission routes for
Legionella. These aquatic environments often provide favourable conditions for
Legionella colonisation and proliferation, creating significant public health risks [
5].
In recent years, the area of wastewater reuse has gained increasing interest in order to counteract climate change. Rising global temperatures, the increasing frequency and intensity of heatwaves, prolonged droughts, and altered precipitation patterns are placing unprecedented pressure on both the quantity and quality of available freshwater resources. In this scenario, the reuse of treated wastewater is becoming not only a strategic response to water scarcity but also a necessity for agricultural and environmental resilience, especially in Mediterranean and arid regions [
6].
The principle of wastewater reuse is based on the use of effluent from wastewater treatment plants that have been previously treated to remove hazardous substances and pathogenic microorganisms, making it suitable for various applications. The reclaimed water can mainly be used for agricultural irrigation, but it is also used for industrial cooling systems, ornamental purposes (such as fountains, artificial ponds, and urban landscaping), and street cleaning. Wastewater reuse may also contribute to a reduction in freshwater consumption, allowing potable water to be preserved for drinking purposes. Moreover, recycled water offers lower environmental, economic, and ecological impacts [
6,
7].
An important factor to consider when evaluating the reuse of treated wastewater in agriculture is the potential risk to human health posed by the presence of pathogenic microorganisms that may not be completely removed during the treatment process [
8]. In this context,
Legionella spp. contamination may be a health risk associated with the potential inhalation of aerosolized water when effluents are reused for irrigation purposes [
9].
For this reason, current European and national legislation on wastewater reuse establishes the assessment of the chemical–physical and microbiological characteristics of the water intended for reuse as a compulsory requirement, as well as risk management when wastewater is reused for irrigation purposes [
10,
11,
12]. This regulation includes, for the first time, the requirement to monitor for the presence of
Legionella spp. However, these regulations currently rely solely on culture-based methods, which may fail to detect viable but non-culturable cells, leading to an underestimation of real health risks.
The importance of monitoring
Legionella spp. in treated wastewater thus arises from the convergence of three major factors: increasing water reuse to combat water scarcity, the aerosolization risk in agricultural applications, and the growing influence of climate change on pathogen ecology. In fact, climate change also alters the ecological dynamics of microbial pathogens in water systems. Elevated ambient temperatures can increase water temperatures in storage and treatment infrastructures, fostering the growth of thermophilic bacteria such as
Legionella [
13]. Additionally, warmer temperatures can accelerate biofilm formation in pipelines and tanks, which act as protective reservoirs for
Legionella spp., shielding them from disinfection processes.
Some studies have shown that, in wastewater treatment and specifically in biological oxidation tanks, microclimatic conditions may favour the growth and development of
Legionella [
9,
14,
15,
16,
17], and certain strains have demonstrated resistance to chlorine-based disinfection products [
18]. Moreover, some studies confirm that direct aerosol emissions from wastewater treatment plants have been implicated in Legionnaires’ disease outbreaks, with documented dispersion distances reaching up to 300 m [
19] and approximately 1.6 kilometres [
20].
Given this complex and evolving context, the aim of the present study is to assess the presence of Legionella spp. and L. pneumophila at the influent, throughout various treatment stages, and in the final effluent of a wastewater treatment plant (WWTP) designed for wastewater reuse. The study compares results obtained from traditional culture-based methods with those from molecular techniques (qPCR), in order to better understand Legionella persistence, proliferation dynamics, and the implications for public health risk in effluent reuse, particularly considering climate change.
2. Materials and Methods
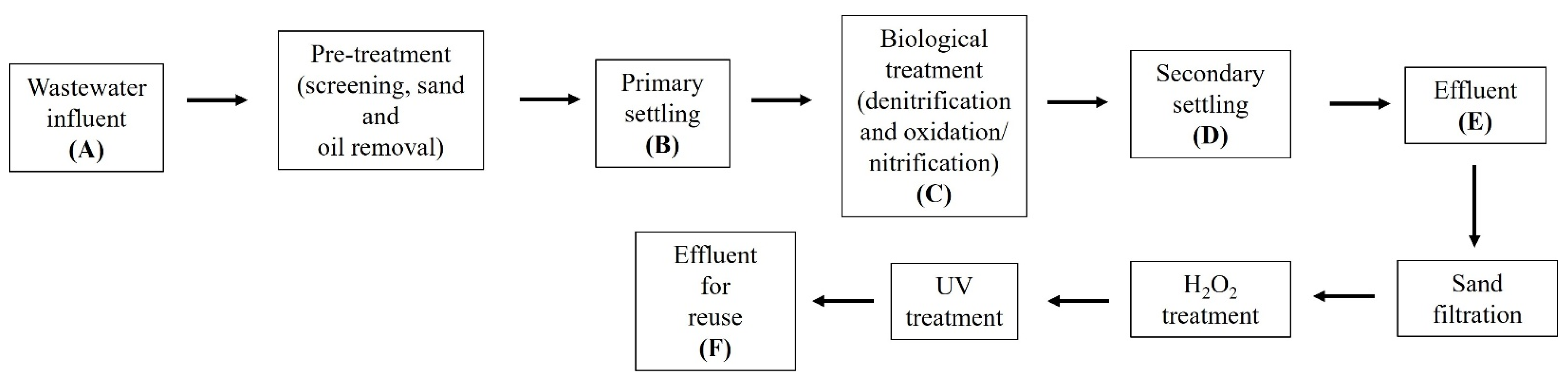
2.1. Wastewater Treatment Plant Characteristics and Sample Collection
The wastewater treatment plant (WWTP) investigated serves 280,000 population equivalent (p.e.) and processes 19 million m
3 per year, with 5.5 million m
3 annually reused for agricultural irrigation. The influent consists of mixed wastewater, with industrial effluent contributing 10% of the total. Upon entry, the wastewater is split into three treatment lines, with the third line being the largest, handling about 40% of the flow, until secondary treatment. Afterward, the three lines are merged into one, producing the WWTP effluent, which undergoes further treatments (sand filtration, hydrogen peroxide -H
2O
2, and UV treatment) to make it suitable for reuse. The wastewater treatment plant investigated was described in detail in a previous study [
10] and is represented in
Figure 1.
In this study, six distinct 24 h composite samples were collected, according to UNI EN ISO 5667-1:2022 [
21], at the following stages: A (WWTP influent); B (primary treatment inlet); C (biological treatment inlet); D (secondary treatment effluent); E (WWTP effluent); and F (effluent for agricultural reuse). The B, C, and D samples correspond to the third treatment line. The samples were collected during six monthly sessions (January–June) (
n = 36 samples). A 2 L volume of each sample was taken from each point, transported under refrigeration to the laboratory, and processed within 24 h.
Within the wastewater treatment plant, typical sewage temperatures were between 14.7 and 23.9 °C according to the season, pH values were between 7.3 and 8.1, and the Chemical Oxygen Demand (COD) was between 16 and 329 mg/L.
2.2. Culture-Dependent Method
For the determination of
Legionella spp. and
Legionella pneumophila, the culture method described in the ISO 11731:2017 standard “Water quality—Enumeration of
Legionella” was used [
22].
Briefly, the influent samples (A, B and C) were serially diluted while the effluent samples D, E and F were concentrated via centrifugation at 3000× g for 30 s. Each sample was then divided into two aliquots, one untreated and one heat-treated (50 °C for 30 min). The heat treatment was performed to reduce the presence of microorganisms that could interfere with the growth of Legionella. Each aliquot was then used for serial dilutions.
For each dilution, 500 µL of the sample was inoculated onto GVPC Agar (Glycine Vancomycin Polymyxin Cycloheximide). The plates were incubated at 36 ± 2 °C for 10 days. Legionella appears as circular colonies that are grey or white, although colour variations may occur, such as blue, purple, yellow, green, or pink. Colonies with the typical morphology were isolated by inoculating the same colony onto Buffered Charcoal Yeast Extract (BCYE) Agar plates without L-cysteine and BCYE agar plates with L-cysteine. Colonies that grew only on plates with L-cysteine were confirmed biochemically using a latex agglutination test (Legionella Latex Test, Oxoid, Basingstoke, UK). The results are expressed as CFU/L.
2.3. Culture-Independent Method
For molecular analysis, different volumes of the sample (ranging from 20 to 200 mL) were filtered onto polycarbonate filters with a porosity of 0.22 μm. The filters were stored at −20 °C until DNA extraction, which was performed using the DNeasy PowerWater Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
For the detection of
Legionella spp. and
Legionella pneumophila DNA, a Real-Time PCR (Quantitative PCR or qPCR) was performed using fluorescent hybridisation probes. The target genes used were 16S rRNA for
Legionella spp. and the
mip gene for
L. pneumophila, which is involved in the virulence, infection, and life cycle of the bacterium [
23].
For Real-Time PCR, the iQ-Check® Quanti Legionella spp. Real-Time PCR Quantification Kit and iQ-Check® Quanti L. pneumophila Real-Time PCR Quantification Kit (BioRad, Hercules, CA, USA) were used. A mix containing the amplification solution (primers, dNTP, Taq polymerase, MgCl2, PCR buffer, and UDG), fluorescent probes, and an internal plasmid DNA control was prepared. A total of 45 μL of this mix was distributed into the wells of the plate, and 5 μL of the previously extracted and diluted DNA or standards (Qs1, Qs2, Qs3, and Qs4) containing serial dilutions of the target DNA genomic units was added, along with a blank (sterile water).
The amplification reaction was performed using the CFX 96 C1000 instrument (Biorad), and the results were interpreted using the quantification curve based on the four standards. The results are expressed in genomic units (G.U.) per litre of sample (G.U./L).
2.4. Statistical Analysis
Statistical analysis was performed using the SPSS software (version 28.0.0). Data distribution was assessed using the Shapiro–Wilk normality test. To evaluate potential differences in contamination levels observed at different stages of the treatment process, a one-way ANOVA test followed by the Tukey post-hoc test for normally distributed data and the Kruskal–Wallis test for non-normally distributed data were used. p was considered significant when <0.05.
3. Results and Discussion
The culture-dependent method for
Legionella detection revealed the presence of
Legionella pneumophila SG 2-14 only in one influent (A) sample (5.7 log CFU/L). No other colonies attributed to
Legionella spp. or
L. pneumophila were detected in any of the other analysed samples. Considering that the current regulations [
11] require the maximum allowable concentration of
Legionella spp. in treated wastewater intended for reuse of 1000 CFU/L (3 log CFU/L), the results obtained indicated the suitability of the treated effluent for irrigation uses, including spraying.
Low percentages of
Legionella positivity have also been reported in other studies using the culture method in wastewater treatment plants. In a study conducted to assess the abatement efficiency of three Italian WWTPs, the culture-dependent method monitored
Legionella spp. in just one influent sample [
8]. Similar results were found in another study where the authors did not detect
Legionella in any of the effluent samples analysed [
24]. Likewise, in another study that evaluated the presence of
Legionella in the secondary treatment tanks of various WWTPs, no positivity for
Legionella spp. or
L. pneumophila was found in any of the samples analysed [
25]. Moreover, even in samples from Norwegian WWTPs, a low positivity rate for
Legionella (3%) was observed using the culture-dependent method [
26]. In addition, in another case, the authors found only one positive result in a WWTP effluent
(L. pneumophila SG 6) [
17].
In contrast, the culture-independent analysis (molecular analysis performed with qPCR) detected the presence of Legionella DNA at various steps in the wastewater treatment.
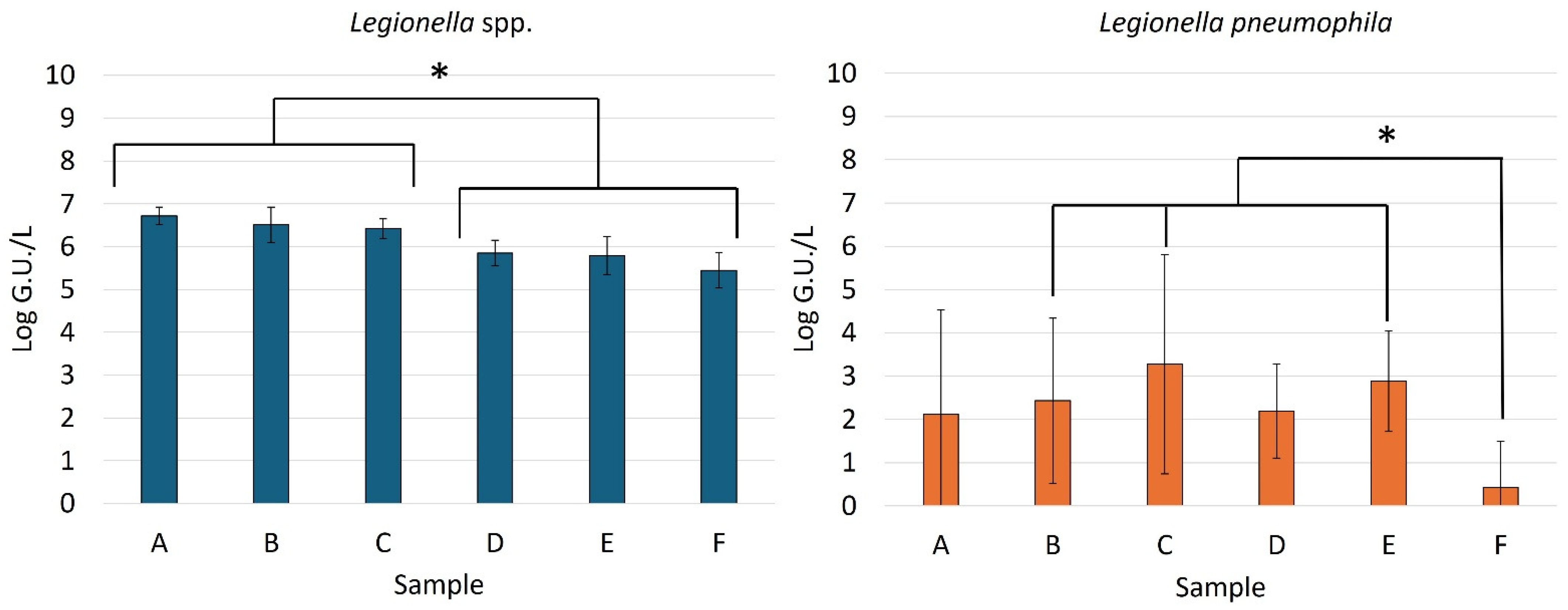
Figure 2 shows the average concentrations obtained using the molecular method for
Legionella spp. and
L. pneumophila in the influent wastewater samples, the effluent samples, and at different stages of the treatment process.
Specifically, all analysed samples tested positive for
Legionella spp., and 66% of the samples were also positive for
L. pneumophila (50% of the A samples, 67% of the B samples, 83% of the C samples, 83% of the D samples, 100% of the E samples, and 17% of the F samples) (
Figure 3).
The positivity rates found using the molecular method in this study are in agreement with those observed in other studies. In Bonetta et al., the molecular method identified
Legionella spp. in 100% of pre- and post-treatment samples from all monitored plants, while
L. pneumophila was found in 57% to 100% of pre-treatment samples and in 0% to 50% of post-treatment samples [
8]. Medema and colleagues detected the presence of
Legionella spp. in 100% of the analysed samples [
24] and Caicedo et al. identified
Legionella spp. in 20% to 80% of the samples from the WWTPs investigated [
25]. Lund and colleagues also found
Legionella spp. and
L. pneumophila in 98% and 30% of the analysed samples, respectively [
26]. In another study [
17], the authors found viable cells of
Legionella spp. and
L. pneumophila in 100% and 91.7% of the samples from a WWTP effluent, respectively.
As observed from the results, the positivity rates obtained using the culture-independent method were significantly different from those obtained using the culture-dependent method. As highlighted in other studies, the culture-dependent method can underestimate
Legionella presence in wastewater, while, conversely, the culture-independent method can overestimate it. Several factors can contribute to the underestimation of
Legionella detection using the cultural method, including the presence of abundant or competitive bacterial flora that inhibit microorganism growth and the inability of the method to detect viable but non-culturable (VBNC) forms. The ability of
Legionella to survive stressful conditions by entering a VBNC state, during which it exhibits low metabolic activity and cannot reproduce, is well-documented in the literature. A factor contributing to the transition to the VBNC form is exposure to high temperatures (50 °C), which are used in treatments defined by ISO 11731:2017 (the reference standard for
Legionella quantification in water) in the preparation of samples, necessary to reduce interfering flora [
22]. Additionally, the cultural method faces challenges in obtaining reproducible results [
9].
Culture-independent techniques (qPCR), on the other hand, can overestimate health risks because they detect the genetic material of dead or damaged microorganisms, which are not capable of causing disease. These differences between culture-dependent and culture-independent methods have been highlighted in several studies. In the Netherlands, all raw effluent samples analysed via qPCR (
n = 7) resulted in contamination by
Legionella spp., whereas the culture-dependent method never detected this microorganism; this was attributed to the growth of interfering flora on the BCYE agar plates used for analysis [
24]. Similar issues were observed in the monitoring of three active sludge plants in Germany receiving domestic and/or industrial effluents, due to the growth of contaminating flora on the GVPC agar plates [
25]. Similar results were also observed in other studies [
8,
26].
Considering the mean concentration obtained using the molecular method,
Legionella spp. ranged from 5.02 log G.U./L to 7.25 log G.U./L. The highest value was found in an influent sample (B), while the lowest value was detected in an effluent sample intended for reuse (F) (
Figure 2).
The levels of
Legionella spp. recorded at the influent of the wastewater treatment plant (A) were similar to those observed in another study that detected maximum concentrations of 7.2 log G.U./L at the influent of an Italian wastewater treatment plant [
8]. Conversely, lower contamination levels were found at the influent of three German WWTPs where a maximum concentration of 3.5 log G.U./L was observed [
25].
The
Legionella spp. levels monitored in the effluent samples from the WWTP were similar to those reported by other authors that monitored two different WWTPs in Italy [
8,
17]. Higher concentrations of
Legionella spp. were observed in the effluents of two Norwegian treatment plants (6.9 and 9.1 log G.U./L) [
19] and in the effluents of a WWTP in France (range 6–7.5 log UG/L) [
26]. On the contrary, lower values were recorded in the effluents of the treatment plants analysed in another study [
27], with a maximum concentration of 3.4 log. A possible explanation for these differences could be related to the type of treatment applied: the wastewater treatment plant analysed in the aforementioned study [
27] performs a more efficient treatment compared to those in the other studies. In particular, disinfection with performic acid enhances the effect of UV treatment, resulting in a greater reduction in
Legionella concentrations.
In our study, the influent samples (A) and those collected during the early stages of wastewater treatment (B and C) showed statistically significant higher levels of Legionella spp. contamination compared to the effluent samples (D, E and F) (p < 0.05). The data indicated a significant reduction in contamination starting from the effluent samples after secondary sedimentation, whereas no significant reduction was observed during the early stages of the treatment process or between the effluent samples (E—effluent vs. F—reuse effluent). This trend highlights that secondary treatments are necessary to reduce Legionella spp. contamination, while tertiary treatments, although required for achieving high-quality effluent, do not appear to significantly reduce the contamination levels.
Furthermore, the results showed that none of the intermediate stages of treatment seemed to lead to a significant increase in
Legionella spp. concentration. This finding contrasts with some studies in the literature, although knowledge on the progression of
Legionella contamination levels in relation to the treatment process stages is still limited. Indeed, in the study conducted by Caicedo et al., a significant increase in the presence of
Legionella was observed at the end of the secondary treatments [
25]. Similarly, the Kulkarni group observed an increase in
Legionella during the intermediate stages of the treatment process, with concentrations higher than those in the influent samples [
15]. An increase in
Legionella concentrations after primary and secondary treatments was also highlighted in a further study [
28]. The differences observed in
Legionella proliferation in the intermediate stages of the treatment process at the WWTP investigated in this study, compared to those reported in other works, could be partly related to the physicochemical parameters characterising the process and effluent (e.g., temperature < 25 °C; low levels of COD, 16–329 mg/L), as well as the type of treatment used (sand filtration + hydrogen peroxide + UV treatment vs. chlorination) or the microbiological characteristics of the effluent. In fact, some studies have highlighted that temperatures between 30 and 40 °C, high COD concentrations, and significant contamination by protozoa and amoebae in bacterial biofilms may create ideal conditions for
Legionella proliferation [
16,
25,
29].
Regarding
L. pneumophila, the contamination level was rather low in all stages of the treatment plant (
Figure 2). In fact, the positive samples showed concentrations often below the method quantification limit. These results align with those observed in other studies. Brissaud et al. detected
L. pneumophila values below the quantification limit in most of the samples analysed [
30]. Similar results were obtained by Bonetta et al., who observed values below the quantification limit in all samples that tested positive for the pathogen (17 out of 21 influent samples and 4 out of 21 effluent samples) [
8]. In the study by Lund et al., analysing effluents from Norwegian treatment plants, the concentrations of
L. pneumophila were above the quantification limit but still relatively low, ranging from 2.4 to 5.3 log G.U./L [
26].
Regarding the contamination levels observed at different treatment stages, statistical analysis showed a significant decrease in contamination only for the effluent samples treated for reuse (F), while no significant differences in contamination were observed between the other stages of the treatment plant. The positives found in the various samples were also sporadic, especially at certain points, making it difficult to trace a clear trend during the treatment process, in contrast to Legionella spp., which was present in all the analysed samples.
These results are consistent with those observed in other studies. The Kulkarni group observed a significant reduction in the concentration of
L. pneumophila only in the effluent samples from the treatment plant [
15]. Similarly, in the study conducted by Caicedo et al., a significant reduction in
L. pneumophila concentration was observed in the final stages of the treatment process, although the values were still higher than those recorded in this study [
25].
Moreover, considering
Legionella contamination across the different sampling sessions, no statistically significant variation among the monthly samples was observed using either method (CFU/L or G.U./L). This result is consistent with findings reported by other authors [
26].
4. Conclusions
One of the possible solutions to water shortage induced by climate change could be the reuse of treated wastewater, a technique that presents numerous advantages, both economic and environmental. However, there are also various critical issues to consider, particularly from a public health perspective. In addition to the chemical issues, it is undeniable that potential risks also include those of microbial origin. Indeed, if treated effluents are reused for agricultural purposes, it is necessary to consider that the irrigation method used (e.g., spray irrigation) could promote the dispersion of aerosols containing pathogens such as Legionella. The results obtained in this study showed that none of the stages of the investigated wastewater treatment plant seem to promote the proliferation of Legionella. However, considering that the chemical–physical parameters characterising the process and the effluent (pH, temperature), as well as the microbiological characteristics of the effluent, may favour Legionella growth in certain treatment stages, there is a need to further investigate how Legionella spp. proliferate and persist in wastewater treatment plants, with the aim of developing effective preventive and control strategies. Although no increase in Legionella concentrations was observed across the treatment stages in our data, future studies should include a parallel assessment of protozoan and amoebae populations, especially in biological treatment units, to better elucidate potential microbial interactions that may affect pathogen dynamics.
Regarding the possible reuse of treated wastewater, the effluent samples, analysed using the cultural method as required by current regulations, were found to be suitable for irrigation uses, including spray irrigation. However, the presence of high concentrations of Legionella spp. DNA detected by qPCR in the same samples raises the need for further investigation. Moreover, since current regulations require the assessment of Legionella presence solely through the cultural method, it will also be necessary to further explore the methodological issues related to Legionella spp. detection to avoid underestimating health risks to the population due to the presence of this pathogen in refined water sent for reuse.
While quantitative PCR (qPCR) offers high sensitivity for the detection of Legionella DNA, it may also overestimate actual health risks by detecting non-viable cells. To address this limitation and enhance the accuracy of risk assessment, future strategies might incorporate viability-based PCR techniques to differentiate between live and dead cells or integrate molecular findings within quantitative microbial risk assessment (QMRA) frameworks. Furthermore, updating regulatory approaches to consider multiple lines of evidence—encompassing culture-based methods, molecular analyses, and environmental data—could strengthen the reliability of water reuse safety evaluations. In this context, investing such complementary methodologies would help to close the existing methodological gap and support more evidence-based public health decision making. Based on the findings of this study, further investigation into the occurrence and distribution of Legionella in other wastewater treatment plants is warranted, as part of a preventive approach to better characterise potential public health risks.