Process Limit of Detection for Salmonella Typhi, Vibrio cholerae, Rotavirus, and SARS-CoV-2 in Surface Water and Wastewater
Abstract
1. Background
2. Materials and Methods
2.1. Source of Wastewater and Surface Water Samples
2.2. S. Typhi, V. cholerae, Rotavirus, and SARS-CoV-2 Sources
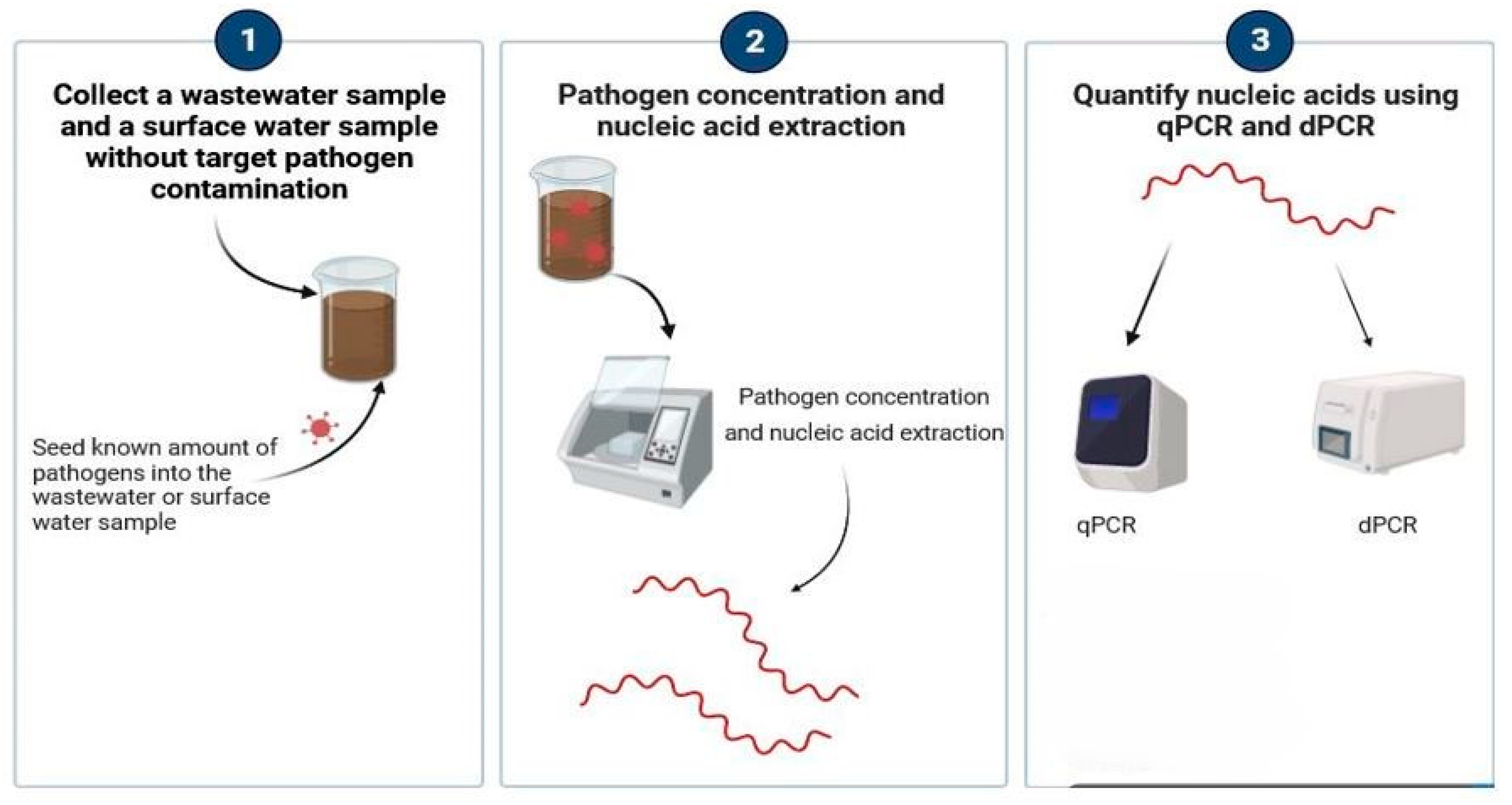
2.3. PLOD Seeding Experimental Workflow
2.4. Automated KingFisher™ Apex Workflow
2.5. Quantitative Real-Time PCR Method
2.6. RT-dPCR and dPCR Method
2.7. Recovery Efficiency
2.8. Data Analyses
3. Results
3.1. S. Typhi Process Limit of Recovery Efficiencies in Surface Water and Wastewater
| Genome Copies/ 10 mL Surface Water | No. of Positive /No. of Experiments | qPCR Ct Mean (SD †) | No. of Positive /No. of Experiments | dPCR Mean Partitions (SD) | Mean Recovery Rate (%) (SD) |
|---|---|---|---|---|---|
| 5.6 × 106 | 3/3 | 27.3 (2.0) | 3/3 | 911.3 (753.8) | 4.3 (3.5) |
| 4.6 × 105 | 3/3 | 29.6 (2.6) | 3/3 | 215.2 (155.6) | 10.4 (7.5) |
| 3.6 × 104 | 3/3 | 33.7 (3.2) | 3/3 | 15.5 (17.0) | 7.4 (8.1) |
| 2.6 × 103 | 3/3 | 37.3 (1.2) | 3/3 | 1.3 (1.6) | 6.4 (7.8) |
| 2.6 × 102 | 0/3 | - | 1/3 | - | - |
| Genome copies /10 mL wastewater | No. of positive /No. of experiments | qPCR Ct mean (SD) | No. of positive /No. of experiments | dPCR mean partition (SD) | Mean recovery rate (%) (SD) |
| 5.6 × 106 | 3/3 | 25.3 (1.0) | 3/3 | 7492.4 (3579.2) | 36.8 (14.6) |
| 4.6 × 105 | 3/3 | 29.2 (1.9) | 3/3 | 628.3 (344.4) | 26.5 (12.8) |
| 3.6 × 104 | 3/3 | 32.2 (1.5) | 3/3 | 87.3 (76.6) | 40.1 (28.7) |
| 2.6 × 103 | 3/3 | 35.9 (1.3) | 3/3 | 6.5 (7.2) | 29.8 (29.6) |
| 2.6 × 102 | 2/3 | 38.7 | 2/3 | - | - |
3.2. V. cholerae Process Limit of Detection and Recovery Efficiencies in Surface Water and Wastewater
| Genome Copies/ 10 mL Surface Water | No. of Positive /No. of Experiments | qPCR Ct Mean (SD †) | No. of Positive /No. of Experiments | dPCR Mean Partitions (SD) | Mean Recovery Rate (%) (SD) |
|---|---|---|---|---|---|
| 5.8 × 106 | 3/3 | 26.9 (1.4) | 3/3 | 1590.0 (1779.8) | 5.9 (6.6) |
| 4.8 × 105 | 3/3 | 29.0 (0.4) | 3/3 | 247.7 (98.5) | 9.3 (3.9) |
| 3.8 × 104 | 3/3 | 33.1 (0.2) | 3/3 | 15.8 (6.7) | 6.0 (2.6) |
| 2.8 × 103 | 3/3 | 36.5 (1.2) | 3/3 | 1.5 (1.0) | 5.8 (4.1) |
| 190 | 1/3 | - | 1/3 | - | - |
| Genome copies /10 mL wastewater | No. of positive /No. of experiments | qPCR Ct mean (SD) | No. of positive /No. of experiments | dPCR mean partition (SD) | Mean recovery rate (%) |
| 5.8 × 106 | 3/3 | 25.0 (1.8) | 3/3 | 4485.8 (2997.0) | 50.6 (34.0) |
| 4.8 × 105 | 3/3 | 27.5 (0.9) | 3/3 | 519.2 (275.7) | 58.8 (31.6) |
| 3.8 × 104 | 3/3 | 31.4 (0.7) | 3/3 | 37.2 (17.5) | 42.0 (19.2) |
| 2.8 × 103 | 3/3 | 34.4 (1.1) | 3/3 | 5.0 (2.5) | 48.0 (19.3) |
| 190 | 2/3 | 36.9 (0.5) | 1/3 | - | - |
3.3. Rotavirus Process Limit of Detection and Recovery Efficiencies in Surface Water and Wastewater
| Genome Copies/ 10 mL Surface Water | No. of Positive /No. of Experiments | qPCR Ct Mean (SD †) | No. of Positive /No. of Experiments | dPCR Mean Partitions (SD) | Mean Recovery Rate (%) (SD) |
|---|---|---|---|---|---|
| 5.1 × 106 | 3/3 | 26.8 (0.6) | 3/3 | 2977.8 (621.5) | 2.9 (0.6) |
| 4.1 × 105 | 3/3 | 30.5 (0.9) | 3/3 | 247.0 (31.7) | 3.5 (0.3) |
| 3.1 × 104 | 3/3 | 34.9 (1.5) | 3/3 | 8.2 (9.1) | 0.8 (0.9) |
| 2.1 × 103 | 3/3 | 38.1 (2.0) | 3/3 | 2.1 (2.6) | 2.1 (2.6) |
| 110 | 0/3 | - | 0/3 | - | - |
| Genome copies /10 mL wastewater | No. of positive /No. of experiments | qPCR Ct mean (SD) | No. of positive /No. of experiments | dPCR mean partition (SD) | Mean recovery rate (%) |
| 5.1 × 106 | 3/3 | 27.5 (1.1) | 3/3 | 1977.2 (680.0) | 1.9 (0.7) |
| 4.1 × 105 | 3/3 | 30.7 (0.8) | 3/3 | 171.8 (37.9) | 1.7 (0.4) |
| 3.1 × 104 | 3/3 | 33.7 (0.6) | 3/3 | 24.7 (10.3) | 2.4 (1.0) |
| 2.1 × 103 | 3/3 | 40.4 (0.9) | 3/3 | 1.5 (1.0) | 1.5 (1.0) |
| 110 | 2/3 | 36.9 (0.5) | 1/3 | - | - |
3.4. SARS-CoV-2 Process Limit Detection and Recovery Efficiencies in Surface Water and Wastewater
| Genome Copies/ 10 mL Surface Water | No. of Positive /No. of Experiments | qPCR Ct Mean (SD †) | No. of Positive /No. of Experiments | dPCR Mean Partitions (SD) | Mean Recovery Rate (%) (SD) |
|---|---|---|---|---|---|
| 4.9 × 106 | 3/3 | 28.3 (0.1) | 3/3 | 263.8 (57.4) | 2.3 (0.5) |
| 3.9 × 105 | 3/3 | 33.0 (0.4) | 3/3 | 16.7 (8.4) | 1.5 (0.7) |
| 2.9 × 104 | 3/3 | 36.2 (0.8) | 3/3 | 2.5 (1.9) | 2.2 (1.7) |
| 1.9 × 103 | 2/3 | 39.5 (0.1) | 2/3 | - | - |
| 60 | 0/3 | - | 1/3 | - | - |
| Genome copies /10 mL wastewater | No. of positive /No. of experiments | qPCR Ct mean (SD) | No. of positive /No. of experiments | dPCR mean partition (SD) | Mean recovery rate (%) |
| 4.9 × 106 | 3/3 | 29.4 (0.7) | 3/3 | 1046.5 (201.6) | 1.9 (0.7) |
| 3.9 × 105 | 3/3 | 32.5 (0.8) | 3/3 | 110.8 (32.3) | 1.7 (0.4) |
| 2.9 × 104 | 3/3 | 36.4 (0.8) | 3/3 | 9.5 (3.1) | 2.4 (1.0) |
| 1.9 × 103 | 0/3 | 40.5 (1.2) | 3/3 | 0.5 (0.6) | 1.5 (1.0) |
| 60 | 0/3 | - | 0/3 | - | - |
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Amin, N.; Liu, P.; Foster, T.; Rahman, M.; Miah, M.R.; Ahmed, G.B.; Kabir, M.; Raj, S.; Moe, C.L.; Willetts, J. Pathogen flows from on-site sanitation systems in low-income urban neighborhoods, Dhaka: A quantitative environmental assessment. Int. J. Hyg. Environ. Health 2020, 230, 113619. [Google Scholar] [CrossRef]
- Barrett, T.J.; Blake, P.A.; Morris, G.K.; Puhr, N.D.; Bradford, H.B.; Wells, J.G. Use of Moore swabs for isolating Vibrio cholerae from sewage. J. Clin. Microbiol. 1980, 11, 385–388. [Google Scholar] [CrossRef]
- Tao, Z.; Wang, H.; Xu, A.; Zhang, Y.; Song, L.; Zhu, S.; Li, Y.; Yan, D.; Liu, G.; Yoshida, H.; et al. Isolation of a recombinant type 3/type 2 poliovirus with a chimeric capsid VP1 from sewage in Shandong, China. Virus Res. 2010, 150, 56–60. [Google Scholar] [CrossRef]
- Kazama, S.; Masago, Y.; Tohma, K.; Souma, N.; Imagawa, T.; Suzuki, A.; Liu, X.; Saito, M.; Oshitani, H.; Omura, T. Temporal dynamics of norovirus determined through monitoring of municipal wastewater by pyrosequencing and virological surveillance of gastroenteritis cases. Water Res. 2016, 92, 244–253. [Google Scholar] [CrossRef]
- Giron-Guzman, I.; Cuevas-Ferrando, E.; Barranquero, R.; Diaz-Reolid, A.; Puchades-Colera, P.; Falco, I.; Perez-Cataluna, A.; Sanchez, G. Urban wastewater-based epidemiology for multi-viral pathogen surveillance in the Valencian region, Spain. Water Res. 2024, 255, 121463. [Google Scholar] [CrossRef]
- Jahan, F.; Nasim, M.I.; Wang, Y.; Kamrul Bashar, S.M.; Hasan, R.; Suchana, A.J.; Amin, N.; Haque, R.; Hares, M.A.; Saha, A.; et al. Integrating wastewater surveillance and meteorological data to monitor seasonal variability of enteric and respiratory pathogens for infectious disease control in Dhaka city. Int. J. Hyg. Environ. Health 2025, 267, 114591. [Google Scholar] [CrossRef]
- Mueller, J.E.; Bessaud, M.; Huang, Q.S.; Martinez, L.C.; Barril, P.A.; Morel, V.; Balanant, J.; Bocacao, J.; Hewitt, J.; Gessner, B.D.; et al. Environmental poliovirus surveillance during oral poliovirus vaccine and inactivated poliovirus vaccine use in Cordoba Province, Argentina. Appl. Environ. Microbiol. 2009, 75, 1395–1401. [Google Scholar] [CrossRef]
- Zoni, R.; Mezzetta, S.; Affanni, P.; Colucci, M.E.; Fiore, S.; Fontana, S.; Bracchi, M.; Capobianco, E.; Veronesi, L. Poliovirus and non-polio-enterovirus environmental surveillance in Parma within the “Global Polio Eradication Program” (GPEI). Acta Biomed. Atenei Parm. 2019, 90, 95–97. [Google Scholar]
- Lodder, W.J.; Buisman, A.M.; Rutjes, S.A.; Heijne, J.C.; Teunis, P.F.; de Roda Husman, A.M. Feasibility of quantitative environmental surveillance in poliovirus eradication strategies. Appl. Environ. Microbiol. 2012, 78, 3800–3805. [Google Scholar] [CrossRef]
- de Oliveira Pereira, J.S.; da Silva, L.R.; de Meireles Nunes, A.; de Souza Oliveira, S.; da Costa, E.V.; da Silva, E.E. Environmental Surveillance of Polioviruses in Rio de Janeiro, Brazil, in Support to the Activities of Global Polio Eradication Initiative. Food Environ. Virol. 2016, 8, 27–33. [Google Scholar] [CrossRef]
- Ozawa, H.; Yoshida, H.; Usuku, S. Environmental Surveillance Can Dynamically Track Ecological Changes in Enteroviruses. Appl. Environ. Microbiol. 2019, 85, e01604-19. [Google Scholar] [CrossRef]
- Liu, P.; Guo, L.; Cavallo, M.; Cantrell, C.; Hilton, S.P.; Nguyen, A.; Long, A.; Dunbar, J.; Barbero, R.; Barclay, R.; et al. Comparison of Nanotrap((R)) Microbiome A Particles, membrane filtration, and skim milk workflows for SARS-CoV-2 concentration in wastewater. Front. Microbiol. 2023, 14, 1215311. [Google Scholar] [CrossRef]
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J.; et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020, 728, 138764. [Google Scholar] [CrossRef]
- Liu, P.; Ibaraki, M.; Kapoor, R.; Amin, N.; Das, A.; Miah, R.; Mukhopadhyay, A.K.; Rahman, M.; Dutta, S.; Moe, C.L. Development of Moore Swab and Ultrafiltration Concentration and Detection Methods for Salmonella Typhi and Salmonella Paratyphi A in Wastewater and Application in Kolkata, India and Dhaka, Bangladesh. Front. Microbiol. 2021, 12, 684094. [Google Scholar] [CrossRef]
- Philo, S.E.; Ong, A.Q.W.; Keim, E.K.; Swanstrom, R.; Kossik, A.L.; Zhou, N.A.; Beck, N.K.; Meschke, J.S. Development and Validation of the Skimmed Milk Pellet Extraction Protocol for SARS-CoV-2 Wastewater Surveillance. Food Environ. Virol. 2022, 14, 355–363. [Google Scholar] [CrossRef]
- Wurtzer, S.; Marechal, V.; Mouchel, J.M.; Maday, Y.; Teyssou, R.; Richard, E.; Almayrac, J.L.; Moulin, L. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. Eurosurveillance 2020, 25, 2000776. [Google Scholar] [CrossRef]
- Fores, E.; Bofill-Mas, S.; Itarte, M.; Martinez-Puchol, S.; Hundesa, A.; Calvo, M.; Borrego, C.M.; Corominas, L.L.; Girones, R.; Rusinol, M. Evaluation of two rapid ultrafiltration-based methods for SARS-CoV-2 concentration from wastewater. Sci. Total Environ. 2021, 768, 144786. [Google Scholar] [CrossRef]
- Resch, T.K.; Wang, Y.; Moon, S.; Jiang, B. Serial Passaging of the Human Rotavirus CDC-9 Strain in Cell Culture Leads to Attenuation: Characterization from In Vitro and In Vivo Studies. J. Virol. 2020, 94, 10-1128. [Google Scholar] [CrossRef]
- Jothikumar, N.; Kang, G.; Hill, V.R. Broadly reactive TaqMan assay for real-time RT-PCR detection of rotavirus in clinical and environmental samples. J. Virol. Methods 2009, 155, 126–131. [Google Scholar] [CrossRef]
- Huang, J.; Zhu, Y.; Wen, H.; Zhang, J.; Huang, S.; Niu, J.; Li, Q. Quadruplex real-time PCR assay for detection and identification of Vibrio cholerae O1 and O139 strains and determination of their toxigenic potential. Appl. Environ. Microbiol. 2009, 75, 6981–6985. [Google Scholar] [CrossRef]
- Karkey, A.; Jombart, T.; Walker, A.W.; Thompson, C.N.; Torres, A.; Dongol, S.; Tran Vu Thieu, N.; Pham Thanh, D.; Tran Thi Ngoc, D.; Voong Vinh, P.; et al. The Ecological Dynamics of Fecal Contamination and Salmonella Typhi and Salmonella Paratyphi A in Municipal Kathmandu Drinking Water. PLoS Negl. Trop. Dis. 2016, 10, e0004346. [Google Scholar] [CrossRef]
- Igloi, Z.; Leven, M.; Abdel-Karem Abou-Nouar, Z.; Weller, B.; Matheeussen, V.; Coppens, J.; Koopmans, M.; Molenkamp, R. Comparison of commercial realtime reverse transcription PCR assays for the detection of SARS-CoV-2. J. Clin. Virol. 2020, 129, 104510. [Google Scholar] [CrossRef]
- Corman, V.M.; Drosten, C. Authors’ response: SARS-CoV-2 detection by real-time RT-PCR. Eurosurveillance 2020, 25, 2001035. [Google Scholar] [CrossRef]
- Kudo, E.; Israelow, B.; Vogels, C.B.F.; Lu, P.; Wyllie, A.L.; Tokuyama, M.; Venkataraman, A.; Brackney, D.E.; Ott, I.M.; Petrone, M.E.; et al. Detection of SARS-CoV-2 RNA by multiplex RT-qPCR. PLoS Biol. 2020, 18, e3000867. [Google Scholar] [CrossRef]
- Hayes, E.K.; Gouthro, M.T.; LeBlanc, J.J.; Gagnon, G.A. Simultaneous detection of SARS-CoV-2, influenza A, respiratory syncytial virus, and measles in wastewater by multiplex RT-qPCR. Sci. Total Environ. 2023, 889, 164261. [Google Scholar] [CrossRef]
- Rashid, R.B.; Ferdous, J.; Tulsiani, S.; Jensen, P.K.M.; Begum, A. Development and Validation of a Novel Real-time Assay for the Detection and Quantification of Vibrio cholerae. Front. Public Health 2017, 5, 109. [Google Scholar] [CrossRef]
- Ahmed, W.; Bivins, A.; Metcalfe, S.; Smith, W.J.M.; Verbyla, M.E.; Symonds, E.M.; Simpson, S.L. Evaluation of process limit of detection and quantification variation of SARS-CoV-2 RT-qPCR and RT-dPCR assays for wastewater surveillance. Water Res. 2022, 213, 118132. [Google Scholar] [CrossRef]
- Ahuja, S.; Tallur, S.; Kondabagil, K. Simultaneous microbial capture and nucleic acid extraction from wastewater with minimal pre-processing and high recovery efficiency. Sci. Total Environ. 2024, 918, 170347. [Google Scholar] [CrossRef]
- Ahmed, W.; Smith, W.J.M.; Sirikanchana, K.; Kitajima, M.; Bivins, A.; Simpson, S.L. Influence of membrane pore-size on the recovery of endogenous viruses from wastewater using an adsorption-extraction method. J. Virol. Methods 2023, 317, 114732. [Google Scholar] [CrossRef]
- Vadde, K.K.; Al-Duroobi, H.; Phan, D.C.; Jafarzadeh, A.; Moghadam, S.V.; Matta, A.; Kapoor, V. Assessment of Concentration, Recovery, and Normalization of SARS-CoV-2 RNA from Two Wastewater Treatment Plants in Texas and Correlation with COVID-19 Cases in the Community. Acs Es&T Water 2022, 2, 2060–2069. [Google Scholar]
- Zheng, X.; Deng, Y.; Xu, X.; Li, S.; Zhang, Y.; Ding, J.; On, H.Y.; Lai, J.C.C.; In Yau, C.; Chin, A.W.H.; et al. Comparison of virus concentration methods and RNA extraction methods for SARS-CoV-2 wastewater surveillance. Sci. Total Environ. 2022, 824, 153687. [Google Scholar] [CrossRef]
- Betancourt, W.Q.; Schmitz, B.W.; Innes, G.K.; Prasek, S.M.; Pogreba Brown, K.M.; Stark, E.R.; Foster, A.R.; Sprissler, R.S.; Harris, D.T.; Sherchan, S.P.; et al. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021, 779, 146408. [Google Scholar] [CrossRef]
- Cao, Y.; Raith, M.R.; Griffith, J.F. Droplet digital PCR for simultaneous quantification of general and human-associated fecal indicators for water quality assessment. Water Res. 2015, 70, 337–349. [Google Scholar] [CrossRef]
- Falzone, L.; Musso, N.; Gattuso, G.; Bongiorno, D.; Palermo, C.I.; Scalia, G.; Libra, M.; Stefani, S. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int. J. Mol. Med. 2020, 46, 957–964. [Google Scholar] [CrossRef]
- Liu, C.; Shi, Q.; Peng, M.; Lu, R.; Li, H.; Cai, Y.; Chen, J.; Xu, J.; Shen, B. Evaluation of droplet digital PCR for quantification of SARS-CoV-2 Virus in discharged COVID-19 patients. Aging 2020, 12, 20997–21003. [Google Scholar] [CrossRef]
- Tedim, A.P.; Almansa, R.; Dominguez-Gil, M.; Gonzalez-Rivera, M.; Micheloud, D.; Ryan, P.; Mendez, R.; Blanca-Lopez, N.; Perez-Garcia, F.; Bustamante, E.; et al. Comparison of real-time and droplet digital PCR to detect and quantify SARS-CoV-2 RNA in plasma. Eur. J. Clin. Investig. 2021, 51, e13501. [Google Scholar] [CrossRef]
- Liu, X.; Feng, J.; Zhang, Q.; Guo, D.; Zhang, L.; Suo, T.; Hu, W.; Guo, M.; Wang, X.; Huang, Z.; et al. Analytical comparisons of SARS-CoV-2 detection by qRT-PCR and ddPCR with multiple primer/probe sets. Emerg. Microbes Infect. 2020, 9, 1175–1179. [Google Scholar] [CrossRef]
- Ahmed, W.; Bertsch, P.M.; Angel, N.; Bibby, K.; Bivins, A.; Dierens, L.; Edson, J.; Ehret, J.; Gyawali, P.; Hamilton, K.A.; et al. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: A surveillance tool for assessing the presence of COVID-19 infected travellers. J. Travel Med. 2020, 27, taaa116. [Google Scholar] [CrossRef]
- Ciesielski, M.; Blackwood, D.; Clerkin, T.; Gonzalez, R.; Thompson, H.; Larson, A.; Noble, R. Assessing sensitivity and reproducibility of RT-ddPCR and RT-qPCR for the quantification of SARS-CoV-2 in wastewater. J. Virol. Methods 2021, 297, 114230. [Google Scholar] [CrossRef]
- Sidstedt, M.; Jansson, L.; Nilsson, E.; Noppa, L.; Forsman, M.; Radstrom, P.; Hedman, J. Humic substances cause fluorescence inhibition in real-time polymerase chain reaction. Anal. Biochem. 2015, 487, 30–37. [Google Scholar] [CrossRef]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors—Occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef] [PubMed]
| Pathogen | Vendor | Classification | Source/Quantification Method |
|---|---|---|---|
| S. Typhi | ATCC (#19430) | Strain Ty2 | Culture/spectrophotometer |
| V. cholerae | ATCC (#39315) | EI Tor Inaba N16961 | Culture/spectrophotometer |
| Rotavirus | US CDC | Human strain Wa | Culture/immunospot assay |
| SARS-CoV-2 | ATCC (VR-1986HK) | 2019-nCoV/USA-WA1/2020 | Patient/dPCR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, P.; Sablon, O.; Nguyen, A.; Long, A.; Moe, C. Process Limit of Detection for Salmonella Typhi, Vibrio cholerae, Rotavirus, and SARS-CoV-2 in Surface Water and Wastewater. Water 2025, 17, 2077. https://doi.org/10.3390/w17142077
Liu P, Sablon O, Nguyen A, Long A, Moe C. Process Limit of Detection for Salmonella Typhi, Vibrio cholerae, Rotavirus, and SARS-CoV-2 in Surface Water and Wastewater. Water. 2025; 17(14):2077. https://doi.org/10.3390/w17142077
Chicago/Turabian StyleLiu, Pengbo, Orlando Sablon, Anh Nguyen, Audrey Long, and Christine Moe. 2025. "Process Limit of Detection for Salmonella Typhi, Vibrio cholerae, Rotavirus, and SARS-CoV-2 in Surface Water and Wastewater" Water 17, no. 14: 2077. https://doi.org/10.3390/w17142077
APA StyleLiu, P., Sablon, O., Nguyen, A., Long, A., & Moe, C. (2025). Process Limit of Detection for Salmonella Typhi, Vibrio cholerae, Rotavirus, and SARS-CoV-2 in Surface Water and Wastewater. Water, 17(14), 2077. https://doi.org/10.3390/w17142077






