Assessment of Antimicrobial Resistance Genes and Pathobiome Diversity in Domestic Wastewater of a Tropical Country
Abstract
1. Introduction
2. Materials and Methods
2.1. Wastewater Treatment Facilities
2.1.1. Sample Collection
2.1.2. Physicochemical Characterisation
2.2. Pretreatment of Wastewater Samples and DNA Extraction
2.3. Detection and Quantification of ARGs
2.4. Pathobiome Diversity Analyses
2.5. ARG and Plasmid Sequences Identification
2.6. Statistical Analysis
3. Results
3.1. Removal of Physicochemical and Microbiological Contaminants
3.2. Quantification of the Relative Abundance of ARGs in Wastewater and Sludge
3.3. Pathobiome Diversity Analyses of Sludge
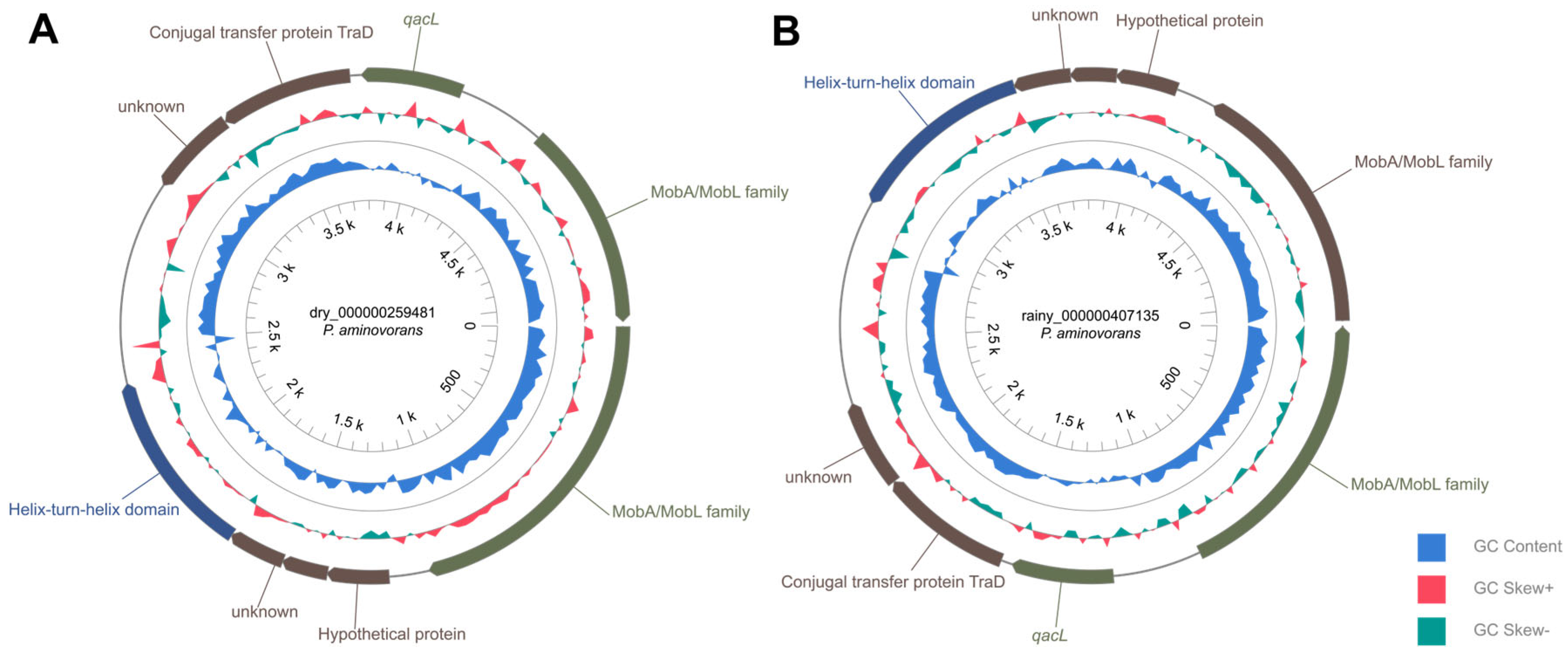
3.4. ARG and Plasmid Sequences Identification in Sludge Samples
4. Discussion
4.1. WWTP Performance
4.2. Antimicrobial Resistance Genes (ARGs) in Wastewater and Sludge Samples
4.3. Pathobiome Diversity Analysis in Sludge Samples
4.4. Predominant Chromosomal and Plasmid-Associated ARG Groups Found in Sludge
4.5. Plasmid-Associated Resistance Genes in Metagenomic Sludge Samples
4.6. Limitations of Our Study and Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| WWTPs | wastewater treatment plants |
| ARGs | antimicrobial resistance genes |
| QACs | quaternary ammonium compounds |
References
- Wu, N.-N.; Liu, S.; Xu, R.; Huang, Q.-Y.; Pan, Y.-F.; Li, H.-X.; Lin, L.; Hou, R.; Cheng, Y.-Y.; Xu, X.-R. New Insight into the Bioaccumulation and Trophic Transfer of Free and Conjugated Antibiotics in an Estuarine Food Web Based on Multimedia Fate and Model Simulation. J. Hazard. Mater. 2024, 465, 133088. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Collignon, P.; Athukorala, P.; Senanayake, S.; Khan, F. Antimicrobial Resistance: The Major Contribution of Poor Governance and Corruption to This Growing Problem. PLoS ONE 2015, 10, e0116746. [Google Scholar] [CrossRef]
- Barrantes Jiménez, K.; Chacón Jiménez, L.; Arias Andrés, M. The Impact of the Antibiotic Resistance on the Sustainable Development. Poblac. Salud Mesoam. 2022, 19, 305–329. [Google Scholar]
- Pärnänen, K.M.M.; Narciso-da-Rocha, C.; Kneis, D.; Berendonk, T.U.; Cacace, D.; Do, T.T.; Elpers, C.; Fatta-Kassinos, D.; Henriques, I.; Jaeger, T. Antibiotic Resistance in European Wastewater Treatment Plants Mirrors the Pattern of Clinical Antibiotic Resistance Prevalence. Sci. Adv. 2019, 5, eaau9124. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Guido, B.; Barrantes, K.; Rodríguez, C.; Rojas-Jimenez, K.; Arias-Andres, M. The Impact of Urban Pollution on Plasmid-Mediated Resistance Acquisition in Enterobacteria from a Tropical River. Antibiotics 2024, 13, 1089. [Google Scholar] [CrossRef]
- Brown, C.L.; Maile-Moskowitz, A.; Lopatkin, A.J.; Xia, K.; Logan, L.K.; Davis, B.C.; Zhang, L.; Vikesland, P.J.; Pruden, A. Selection and Horizontal Gene Transfer Underlie Microdiversity-Level Heterogeneity in Resistance Gene Fate during Wastewater Treatment. Nat. Commun. 2024, 15, 5412. [Google Scholar] [CrossRef] [PubMed]
- Macrì, M.; Bonetta, S.; Di Cesare, A.; Sabatino, R.; Corno, G.; Catozzo, M.; Pignata, C.; Mecarelli, E.; Medana, C.; Carraro, E. Antibiotic Resistance and Pathogen Spreading in a Wastewater Treatment Plant Designed for Wastewater Reuse. Environ. Pollut. 2024, 363, 125051. [Google Scholar] [CrossRef]
- Silvester, R.; Perry, W.B.; Webster, G.; Rushton, L.; Baldwin, A.; Pass, D.A.; Healey, N.; Craine, N.; Cross, G.; Kille, P. Metagenomics Unveils the Role of Hospitals and Wastewater Treatment Plants on the Environmental Burden of Antibiotic Resistance Genes and Opportunistic Pathogens. Sci. Total Environ. 2025, 961, 178403. [Google Scholar] [CrossRef]
- Naghavi, M.; Vollset, S.E.; Ikuta, K.S.; Swetschinski, L.R.; Gray, A.P.; Wool, E.E.; Aguilar, G.R.; Mestrovic, T.; Smith, G.; Han, C. Global Burden of Bacterial Antimicrobial Resistance 1990–2021: A Systematic Analysis with Forecasts to 2050. Lancet 2024, 404, 1199–1226. [Google Scholar] [CrossRef]
- Srathongneam, T.; Sresung, M.; Paisantham, P.; Ruksakul, P.; Singer, A.C.; Sukchawalit, R.; Satayavivad, J.; Mongkolsuk, S.; Sirikanchana, K. High Throughput QPCR Unveils Shared Antibiotic Resistance Genes in Tropical Wastewater and River Water. Sci. Total Environ. 2024, 908, 167867. [Google Scholar] [CrossRef]
- Leroy-Freitas, D.; Machado, E.C.; Torres-Franco, A.F.; Dias, M.F.; Leal, C.D.; Araújo, J.C. Exploring the Microbiome, Antibiotic Resistance Genes, Mobile Genetic Element, and Potential Resistant Pathogens in Municipal Wastewater Treatment Plants in Brazil. Sci. Total Environ. 2022, 842, 156773. [Google Scholar] [CrossRef] [PubMed]
- Devarajan, N.; Köhler, T.; Sivalingam, P.; van Delden, C.; Mulaji, C.K.; Mpiana, P.T.; Ibelings, B.W.; Poté, J. Antibiotic Resistant Pseudomonas spp. in the Aquatic Environment: A Prevalence Study under Tropical and Temperate Climate Conditions. Water Res. 2017, 115, 256–265. [Google Scholar] [CrossRef] [PubMed]
- American Public Health Association. Standard Methods for the Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 1926; Volume 6. [Google Scholar]
- Li, X.; Luo, X.; Mai, B.; Liu, J.; Chen, L.; Lin, S. Occurrence of Quaternary Ammonium Compounds (QACs) and Their Application as a Tracer for Sewage Derived Pollution in Urban Estuarine Sediments. Environ. Pollut. 2014, 185, 127–133. [Google Scholar] [CrossRef]
- Hsu, S.-C.; Chiu, T.-H.; Pang, J.-C.; Hsuan-Yuan, C.-H.; Chang, G.-N.; Tsen, H.-Y. Characterisation of Antimicrobial Resistance Patterns and Class 1 Integrons among Escherichia Coli and Salmonella Enterica Serovar Choleraesuis Strains Isolated from Humans and Swine in Taiwan. Int. J. Antimicrob. Agents 2006, 27, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk v2: Memory Friendly Classification with the Genome Taxonomy Database. Bioinformatics 2022, 38, 5315–5316. [Google Scholar] [CrossRef]
- Lu, J.; Breitwieser, F.P.; Thielen, P.; Salzberg, S.L. Bracken: Estimating Species Abundance in Metagenomics Data. PeerJ. Comput. Sci. 2017, 3, e104. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Mikheenko, A.; Saveliev, V.; Gurevich, A. MetaQUAST: Evaluation of Metagenome Assemblies. Bioinformatics 2016, 32, 1088–1090. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.K.; Fogarty, E.C.; Eren, A.M. Diverse Plasmid Systems and Their Ecology across Human Gut Metagenomes Revealed by PlasX and MobMess. Nat. Microbiol. 2024, 9, 830–847. [Google Scholar] [CrossRef] [PubMed]
- Eren, A.M.; Kiefl, E.; Shaiber, A.; Veseli, I.; Miller, S.E.; Schechter, M.S.; Fink, I.; Pan, J.N.; Yousef, M.; Fogarty, E.C. Community-Led, Integrated, Reproducible Multi-Omics with Anvi’o. Nat. Microbiol. 2021, 6, 3–6. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N. The COG Database: An Updated Version Includes Eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Seemann, T. Mass Screening of Contigs for Antimicrobial and Virulence Genes. 2019. Available online: https://github.com/tseemann/abricate (accessed on 21 January 2025).
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N. CARD 2017: Expansion and Model-Centric Curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Marshall, J. Pysam. Available online: https://github.com/pysam-developers/pysam (accessed on 5 January 2025).
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M. Twelve Years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Menzel, P.; Ng, K.L.; Krogh, A. Fast and Sensitive Taxonomic Classification for Metagenomics with Kaiju. Nat. Commun. 2016, 7, 11257. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Sherrill-Mix, S. Package Taxonomizr. Available online: https://github.com/sherrillmix/taxonomizr/ (accessed on 28 February 2025).
- Bartlett, A.; Padfield, D.; Lear, L.; Bendall, R.; Vos, M. A Comprehensive List of Bacterial Pathogens Infecting Humans. Microbiology 2022, 168, 001269. [Google Scholar] [CrossRef] [PubMed]
- Barnett, D.J.M.; Arts, I.C.W.; Penders, J. MicroViz: An R Package for Microbiome Data Visualization and Statistics. J. Open Source Softw. 2021, 6, 3201. [Google Scholar] [CrossRef]
- Li, Y.; Feng, X.; Chen, X.; Yang, S.; Zhao, Z.; Chen, Y.; Li, S.C. PlasmidScope: A Comprehensive Plasmid Database with Rich Annotations and Online Analytical Tools. Nucleic Acids Res. 2025, 53, D179–D188. [Google Scholar] [CrossRef]
- Decreto N° 33601-S; Reglamento de Vertido y Reuso de Aguas Residuales. MINAE: San José, Costa Rica, 2007. Available online: https://pgrweb.go.cr/scij/Busqueda/Normativa/Normas/nrm_articulo.aspx?param1=NRA&nValor1=1&nValor2=59524&nValor3=126254&nValor4=-1&nValor5=2&nValor6=09/08/2006&strTipM=FA (accessed on 25 December 2024).
- Askari, S.S.; Giri, B.S.; Basheer, F.; Izhar, T.; Ahmad, S.A.; Mumtaz, N. Enhancing Sequencing Batch Reactors for Efficient Wastewater Treatment across Diverse Applications: A Comprehensive Review. Environ. Res. 2024, 260, 119656. [Google Scholar] [CrossRef]
- Colomer Llinàs, J.; Wong, A.; Coma Bech, M.; Puig Broch, S.; Colprim Galceran, J. Qualitative Estimation of SBR Biological Nutrient Removal Performance for Wastewater Treatment. J. Chem. Technol. Biotechnol. 2013, 88, 1305–1313. [Google Scholar] [CrossRef]
- Hardy, K.; Buckley, S.; Collins, M.J.; Estalrrich, A.; Brothwell, D.; Copeland, L.; García-Tabernero, A.; García-Vargas, S.; De La Rasilla, M.; Lalueza-Fox, C. Neanderthal Medics? Evidence for Food, Cooking, and Medicinal Plants Entrapped in Dental Calculus. Naturwissenschaften 2012, 99, 617–626. [Google Scholar] [CrossRef]
- Cuetero-Martínez, Y.; Villamizar-Ojeda, K.N.; Hernández-Santiago, M.J.; De los Cobos-Vasconcelos, D.; Aguirre-Garrido, J.F.; López-Vidal, Y.; Noyola, A. Removal of IntI1, ARGs, and SARS-CoV-2 and Changes in Bacterial Communities in Four Sewage Treatment Facilities. Sci. Total Environ. 2023, 903, 165984. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Yu, S.; Rysz, M.; Luo, Y.; Yang, F.; Li, F.; Hou, J.; Mu, Q.; Alvarez, P.J.J. Prevalence and Proliferation of Antibiotic Resistance Genes in Two Municipal Wastewater Treatment Plants. Water Res. 2015, 85, 458–466. [Google Scholar] [CrossRef]
- Liu, B.; Xu, Y.; Liu, F.; Li, B.; Li, X.; Zha, R.; Wang, S.; Qiu, Y. Occurrence and Removal Prediction of Pharmaceuticals Positively Correlated with Antibiotic Resistance Genes in Wastewater Treatment Processes. Environ. Technol. Innov. 2023, 32, 103425. [Google Scholar] [CrossRef]
- Zhang, L.; Adyari, B.; Hou, L.; Yang, X.; Gad, M.; Wang, Y.; Ma, C.; Sun, Q.; Tang, Q.; Zhang, Y. Mass-Immigration Shapes the Antibiotic Resistome of Wastewater Treatment Plants. Sci. Total Environ. 2024, 908, 168193. [Google Scholar] [CrossRef]
- Petrovich, M.L.; Rosenthal, A.F.; Griffin, J.S.; Wells, G.F. Spatially Resolved Abundances of Antibiotic Resistance Genes and IntI1 in Wastewater Treatment Biofilms. Biotechnol. Bioeng. 2019, 116, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Niestępski, S.; Harnisz, M.; Ciesielski, S.; Korzeniewska, E.; Osińska, A. Environmental Fate of Bacteroidetes, with Particular Emphasis on Bacteroides Fragilis Group Bacteria and Their Specific Antibiotic Resistance Genes, in Activated Sludge Wastewater Treatment Plants. J. Hazard. Mater. 2020, 394, 122544. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Fu, J.; Zhao, K.; Yang, S.; Li, C.; Penttinen, P.; Ao, X.; Liu, A.; Hu, K.; Li, J. Class 1 Integron Carrying QacEΔ1 Gene Confers Resistance to Disinfectant and Antibiotics in Salmonella. Int. J. Food Microbiol. 2023, 404, 110319. [Google Scholar] [CrossRef]
- Lu, Z.; Mahony, A.K.; Arnold, W.A.; Marshall, C.W.; McNamara, P.J. Quaternary Ammonia Compounds in Disinfectant Products: Evaluating the Potential for Promoting Antibiotic Resistance and Disrupting Wastewater Treatment Plant Performance. Environ. Sci. Adv. 2024, 3, 208–226. [Google Scholar] [CrossRef]
- Guo, J.; Li, J.; Chen, H.; Bond, P.L.; Yuan, Z. Metagenomic Analysis Reveals Wastewater Treatment Plants as Hotspots of Antibiotic Resistance Genes and Mobile Genetic Elements. Water Res. 2017, 123, 468–478. [Google Scholar] [CrossRef]
- Murray, L.M.; Hayes, A.; Snape, J.; Kasprzyk-Hordern, B.; Gaze, W.H.; Murray, A.K. Co-Selection for Antibiotic Resistance by Environmental Contaminants. npj Antimicrob. Res. 2024, 2, 9. [Google Scholar] [CrossRef]
- Lu, S.-Y.; Zhang, Y.-L.; Geng, S.-N.; Li, T.-Y.; Ye, Z.M.; Zhang, D.S.; Zou, F.; Zhou, H.W. High Diversity of Extended-Spectrum Beta-Lactamase-Producing Bacteria in an Urban River Sediment Habitat. Appl. Environ. Microbiol. 2010, 76, 5972–5976. [Google Scholar] [CrossRef]
- Arias-Andres, M.; Klümper, U.; Rojas-Jimenez, K.; Grossart, H.-P. Microplastic Pollution Increases Gene Exchange in Aquatic Ecosystems. Environ. Pollut. 2018, 237, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Malham, S.K.; Rajko-Nenow, P.; Howlett, E.; Tuson, K.E.; Perkins, T.L.; Pallett, D.W.; Wang, H.; Jago, C.F.; Jones, D.L.; McDonald, J.E. The Interaction of Human Microbial Pathogens, Particulate Material and Nutrients in Estuarine Environments and Their Impacts on Recreational and Shellfish Waters. Environ. Sci. Process. Impacts 2014, 16, 2145–2155. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Environmental Factors Influencing the Development and Spread of Antibiotic Resistance. FEMS Microbiol. Rev. 2018, 42, fux053. [Google Scholar] [CrossRef]
- Avery, L.M.; Williams, A.P.; Killham, K.; Jones, D.L. Survival of Escherichia Coli O157:H7 in Waters from Lakes, Rivers, Puddles and Animal-Drinking Troughs. Sci. Total Environ. 2008, 389, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Sowani, H.; Kulkarni, M.; Zinjarde, S. An Insight into the Ecology, Diversity and Adaptations of Gordonia Species. Crit. Rev. Microbiol. 2018, 44, 393–413. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kang, S.-J.; Kim, W.; Yoon, J.-H. Gordonia Hankookensis sp. Nov., Isolated from Soil. Int. J. Syst. Evol. Microbiol. 2009, 59, 3172–3175. [Google Scholar] [CrossRef]
- Srinivasan, S.; Park, G.; Yang, H.; Hwang, S.; Bae, Y.; Jung, Y.-A.; Kim, M.K.; Lee, M. Gordonia Caeni sp. nov., Isolated from Sludge of a Sewage Disposal Plant. Int. J. Syst. Evol. Microbiol. 2012, 62, 2703–2709. [Google Scholar] [CrossRef] [PubMed]
- Kusada, H.; Tamaki, H.; Kamagata, Y.; Hanada, S.; Kimura, N. A Novel Quorum-Quenching N-Acylhomoserine Lactone Acylase from Acidovorax sp. Strain MR-S7 Mediates Antibiotic Resistance. Appl. Environ. Microbiol. 2017, 83, e00080-17. [Google Scholar] [CrossRef]
- Snaidr, J.; Amann, R.; Huber, I.; Ludwig, W.; Schleifer, K.-H. Phylogenetic Analysis and in Situ Identification of Bacteria in Activated Sludge. Appl. Environ. Microbiol. 1997, 63, 2884–2896. [Google Scholar] [CrossRef]
- Huggins, T.M.; Haeger, A.; Biffinger, J.C.; Ren, Z.J. Granular Biochar Compared with Activated Carbon for Wastewater Treatment and Resource Recovery. Water Res. 2016, 94, 225–232. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhou, J.; Zhao, S.; Qu, J.; Liu, S.; Zhai, M.; Chen, T.; Zhao, L.; Huang, F. Synergistic Degradation Efficiency of Nitrogen, Phosphorus and Microbial Dynamic Succession Driven by a Novel Strain Gordonia sp. D4 during Synthetic Wastewater Treatment. J. Environ. Chem. Eng. 2024, 12, 112837. [Google Scholar] [CrossRef]
- Chen, J.; Zhan, P.; Koopman, B.; Fang, G.; Shi, Y. Bioaugmentation with Gordonia Strain JW8 in Treatment of Pulp and Paper Wastewater. Clean Technol. Environ. Policy 2012, 14, 899–904. [Google Scholar] [CrossRef]
- Horcajada, J.P.; Milagro, M.; Antonio, O.; Luisa, S.; Sònia, L.; Silvia, G.-Z.; Natividad, B.; Santiago, G. Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas Aeruginosa Infections. Clin. Microbiol. Rev. 2019, 32, 4. [Google Scholar] [CrossRef]
- Cai, T.-M.; Guan, L.-B.; Chen, L.-W.; Cai, S.; Li, X.-D.; Cui, Z.-L.; Li, S.-P. Enhanced Biological Phosphorus Removal with Pseudomonas Putida GM6 from Activated Sludge. Pedosphere 2007, 17, 624–629. [Google Scholar] [CrossRef]
- Lin, B.; Lyu, J.; Lyu, X.; Yu, H.; Hu, Z.; Lam, J.C.W.; Lam, P.K.S. Characterization of Cefalexin Degradation Capabilities of Two Pseudomonas Strains Isolated from Activated Sludge. J. Hazard. Mater. 2015, 282, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Esteban, J.; García-Coca, M. Mycobacterium Biofilms. Front. Microbiol. 2018, 8, 2651. [Google Scholar] [CrossRef]
- Marutescu, L.G.; Popa, M.; Gheorghe-Barbu, I.; Barbu, I.C.; Rodríguez-Molina, D.; Berglund, F.; Blaak, H.; Flach, C.-F.; Kemper, M.A.; Spießberger, B.; et al. Wastewater Treatment Plants, an “Escape Gate” for ESCAPE Pathogens. Front. Microbiol. 2023, 14, 1193907. [Google Scholar] [CrossRef] [PubMed]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. In Virulence Mechanisms of Bacterial Pathogens; ASM Press: Washington, DC, USA, 2016; pp. 481–511. [Google Scholar]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal Gene Transfer: Building the Web of Life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef]
- Daw Elbait, G.; Daou, M.; Abuoudah, M.; Elmekawy, A.; Hasan, S.W.; Everett, D.B.; Alsafar, H.; Henschel, A.; Yousef, A.F. Comparison of QPCR and Metagenomic Sequencing Methods for Quantifying Antibiotic Resistance Genes in Wastewater. PLoS ONE 2024, 19, e0298325. [Google Scholar] [CrossRef]
- Courvalin, P.; Ounissi, H.; Arthur, M. Multiplicity of Macrolide–Lincosamide–Streptogramin Antibiotic Resistance Determinants. J. Antimicrob. Chemother. 1985, 16, 91–100. [Google Scholar] [CrossRef]
- Jangra, V.; Sharma, N.; Chhillar, A.K. Therapeutic Approaches for Combating Pseudomonas Aeruginosa Infections. Microbes Infect. 2022, 24, 104950. [Google Scholar] [CrossRef]
- Garneau-Tsodikova, S.; Labby, K.J. Mechanisms of Resistance to Aminoglycoside Antibiotics: Overview and Perspectives. MedChemComm 2016, 7, 11–27. [Google Scholar] [CrossRef]
- Chen, W.-Y.; Lee, C.-P.; Pavlović, J.; Pangallo, D.; Wu, J.-H. Characterization of Microbiome, Resistome, Mobilome, and Virulome in Anoxic and Oxic Wastewater Treatment Processes in Slovakia and Taiwan. Heliyon 2024, 10, e38723. [Google Scholar] [CrossRef]
- Kelly, S.A.; O’Connell, N.H.; Thompson, T.P.; Dillon, L.; Wu, J.; Creevey, C.; Kiely, P.; Slevin, B.; Powell, J.; Gilmore, B.F.; et al. Large-Scale Characterization of Hospital Wastewater System Microbiomes and Clinical Isolates from Infected Patients: Profiling of Multi-Drug-Resistant Microbial Species. J. Hosp. Infect. 2023, 141, 152–166. [Google Scholar] [CrossRef] [PubMed]
- Hurst-Hess, K.R.; Saxena, A.; Rudra, P.; Yang, Y.; Ghosh, P. Mycobacterium Abscessus HelR Interacts with RNA Polymerase to Confer Intrinsic Rifamycin Resistanceresistance. Mol. Cell 2022, 82, 3166–3177.e5. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.; Bollmann, A.; Seitz, W.; Schwartz, T. Microbiological Characterization of Aquatic Microbiomes Targeting Taxonomical Marker Genes and Antibiotic Resistance Genes of Opportunistic Bacteria. Sci. Total Environ. 2015, 512–513, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Sczesny, S.; Nau, H.; Hamscher, G. Residue Analysis of Tetracyclines and Their Metabolites in Eggs and in the Environment by HPLC Coupled with a Microbiological Assay and Tandem Mass Spectrometry. J. Agric. Food Chem. 2003, 51, 697–703. [Google Scholar] [CrossRef]
- Grossman, T.H. Tetracycline Antibiotics and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025387. [Google Scholar] [CrossRef]
- Göbel, A.; Thomsen, A.; McArdell, C.S.; Joss, A.; Giger, W. Occurrence and Sorption Behavior of Sulfonamides, Macrolides, and Trimethoprim in Activated Sludge Treatment. Environ. Sci. Technol. 2005, 39, 3981–3989. [Google Scholar] [CrossRef]
- Spongberg, A.L.; Witter, J.D.; Acuña, J.; Vargas, J.; Murillo, M.; Umaña, G.; Gómez, E.; Perez, G. Reconnaissance of Selected PPCP Compounds in Costa Rican Surface Waters. Water Res. 2011, 45, 6709–6717. [Google Scholar] [CrossRef]
- Sköld, O. Sulfonamide Resistance: Mechanisms and Trends. Drug Resist. Updates 2000, 3, 155–160. [Google Scholar] [CrossRef]
- Mulder, I.; Siemens, J.; Sentek, V.; Amelung, W.; Smalla, K.; Jechalke, S. Quaternary Ammonium Compounds in Soil: Implications for Antibiotic Resistance Development. Rev. Environ. Sci. Biotechnol. 2018, 17, 159–185. [Google Scholar] [CrossRef]
- Yadav, B.; Tyagi, R.D. 11—Development of Molecular Methods to Detect and Control Emerging Drug-Resistance Pathogens. In Current Developments in Biotechnology and Bioengineering; Tyagi, R.D., Sellamuthu, B., Tiwari, B., Yan, S., Drogui, P., Zhang, X., Pandey, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 377–419. [Google Scholar]
- Chacón, L.; Arias-Andres, M.; Mena, F.; Rivera, L.; Hernández, L.; Achi, R.; Garcia, F.; Rojas-Jimenez, K. Short-Term Exposure to Benzalkonium Chloride in Bacteria from Activated Sludge Alters the Community Diversity and the Antibiotic Resistance Profile. J. Water Health 2021, 19, 895–906. [Google Scholar] [CrossRef]
- Chacon, L.; Rojas-Jimenez, K.; Arias-Andres, M. Bacterial Communities in Residential Wastewater Treatment Plants are Physiologically Adapted to High Concentrations of Quaternary Ammonium Compounds. Clean 2023, 51, 2300056. [Google Scholar] [CrossRef]
- Arnold, W.A.; Blum, A.; Branyan, J.; Bruton, T.A.; Carignan, C.C.; Cortopassi, G.; Datta, S.; DeWitt, J.; Doherty, A.-C.; Halden, R.U.; et al. Quaternary Ammonium Compounds: A Chemical Class of Emerging Concern. Environ. Sci. Technol. 2023, 57, 7645–7665. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Tian, Z.; Luan, X.; Zhang, H.; Zhang, Y.; Yang, M. Distribution of Antibiotic Resistance Genes on Chromosomes, Plasmids and Phages in Aerobic Biofilm Microbiota Under Antibiotic Pressure. J. Environ. Sci. 2025, 156, 647–659. [Google Scholar] [CrossRef]
- Monzingo, A.F.; Ozburn, A.; Xia, S.; Meyer, R.J.; Robertus, J.D. The Structure of the Minimal Relaxase Domain of MobA at 2.1 A Resolution. J. Mol. Biol. 2007, 366, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and Barriers to, Horizontal Gene Transfer between Bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef]
- Huang, H.; Feng, G.; Wang, M.; Liu, C.; Wu, Y.; Dong, L.; Feng, L.; Zheng, X.; Chen, Y. Nitric Oxide: A Neglected Driver for the Conjugative Transfer of Antibiotic Resistance Genes among Wastewater Microbiota. Environ. Sci. Technol. 2022, 56, 6466–6478. [Google Scholar] [CrossRef]
- Liu, C.; Goh, S.G.; You, L.; Yuan, Q.; Mohapatra, S.; Gin, K.Y.-H.; Chen, B. Low Concentration Quaternary Ammonium Compounds Promoted Antibiotic Resistance Gene Transfer via Plasmid Conjugation. Sci. Total Environ. 2023, 887, 163781. [Google Scholar] [CrossRef]



| Dry Season | Rainy Season | ||||||
|---|---|---|---|---|---|---|---|
| Parameters | Influent | Effluent | Efficiency (%) | Influent | Effluent | Efficiency (%) | Limits in National Regulation [38] |
| Flow rate(m3/d) | (415.59 ± 28.65) | N/A | (449.90 ± 30.77) | N/A | |||
| Settleable solids (mL/L) | (0.30 ± 0.20) | (0.07 ± 0.06) | 77.8 | (0.20 ± 0.06) | (0.07 ± 0.06) | 60.0 | 5.00 |
| Total suspended solids (mg/L) | (82.43 ± 23.31) | (44.60 ± 1.98) | 45.7 | (57.00 ± 3.47) | (39.00 ± 0.99) | 31.7 | 100.00 |
| pH | (6.70 ± 0.58) | (6.70 ± 0.58) | N/A | (7 ± 0) | (7 ± 0) | N/A | 6.00–9.00 |
| Chemical oxygen demand (mg/L) | (133.60 ± 48.99) | (7.00 ± 9.64) | 94.7 | (180.00 ± 25.87) | (22.67 ± 21.78) | 87.4 | 150.00 |
| NO3-N (mg/L) | (0.63 ± 0.71) | (5.63 ± 9.24) | N/A | (1.10 ± 1.91) | (8.17 ± 7.41) | N/A | N/A |
| NH4-N (mg/L) | (31.36 ± 18.19) | (2.01 ± 2.18) | 93.6 | (26.90 ± 8.36) | (3.87 ± 3.40) | 85.6 | N/A |
| PO4 -P(mg/L) | (20.37 ± 12.62) | (1.49 ± 1.88) | 92.7 | (25.50 ± 4.02) | (16.93 ± 12.77) | 33.7 | N/A |
| Faecal coliforms (log10NMP/100 mL) | (5.40 ± 0.59) | (2.24 ± 1.28) | 58.8 | (6.60 ± 0.14) | (3.2 ± 0) | 51.3 | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molina-Ospina, F.; Mendoza-Guido, B.; Quesada-Gonzalez, A.; Chacon, L.; Barrios-Hernandez, M.L. Assessment of Antimicrobial Resistance Genes and Pathobiome Diversity in Domestic Wastewater of a Tropical Country. Water 2025, 17, 1574. https://doi.org/10.3390/w17111574
Molina-Ospina F, Mendoza-Guido B, Quesada-Gonzalez A, Chacon L, Barrios-Hernandez ML. Assessment of Antimicrobial Resistance Genes and Pathobiome Diversity in Domestic Wastewater of a Tropical Country. Water. 2025; 17(11):1574. https://doi.org/10.3390/w17111574
Chicago/Turabian StyleMolina-Ospina, Fernando, Bradd Mendoza-Guido, Andrea Quesada-Gonzalez, Luz Chacon, and Mary Luz Barrios-Hernandez. 2025. "Assessment of Antimicrobial Resistance Genes and Pathobiome Diversity in Domestic Wastewater of a Tropical Country" Water 17, no. 11: 1574. https://doi.org/10.3390/w17111574
APA StyleMolina-Ospina, F., Mendoza-Guido, B., Quesada-Gonzalez, A., Chacon, L., & Barrios-Hernandez, M. L. (2025). Assessment of Antimicrobial Resistance Genes and Pathobiome Diversity in Domestic Wastewater of a Tropical Country. Water, 17(11), 1574. https://doi.org/10.3390/w17111574








