1. Introduction
Water contamination caused by lipid residues constitutes a significant environmental concern, particularly in urban and industrial areas where wastewater contains high concentrations of oils and fats. These compounds, mainly derived from the food industry and domestic activities, form oily films on the water surface, hindering oxygen transfer and sunlight penetration, which severely disrupts aquatic ecosystems [
1]. Moreover, the accumulation of these residues generates persistent emulsions and greasy sediments that clog sewage systems and pollute natural water bodies [
2].
Various studies have determined that a single liter of lipid waste can contaminate up to one million liters of potable water, generating severe ecological impacts [
3]. This issue worsens when oils and fats are exposed to high temperatures, as even those considered healthy can form toxic compounds when heated [
2,
4,
5]. Conventional technologies for wastewater treatment, such as flotation, grease traps, and the use of solvents, present limitations due to their high costs, low efficiency at elevated lipid concentrations, and associated environmental impact [
6,
7]. In this context, bioremediation using lipolytic enzymes has emerged as a sustainable, efficient, and environmentally friendly alternative [
5].
Lipases (EC 3.1.1.3) are enzymes that catalyze the hydrolysis of triglycerides in the presence of water, releasing glycerol and free fatty acids [
1]. This mechanism facilitates the degradation of complex lipid compounds, promoting their removal in contaminated aquatic systems. Indeed, several studies have demonstrated that lipases, particularly those of fungal origin, are highly effective in degrading greasy residues in wastewater [
1,
2].
Among microbial sources of lipases, fungi stand out for their robustness in adverse environmental conditions and their ability to secrete extracellular enzymes on a large scale. Specifically, species of the genus
Penicillium have proven to be particularly effective in producing lipases with high biotechnological potential [
2,
4,
7,
8,
9]. Fungal lipases have been successfully employed in various industrial applications, such as biodiesel production through the transesterification of vegetable oils [
10,
11], as well as in the food industry to improve cheese maturation and enhance the flavor of various products [
12,
13,
14]. Additionally, these enzymes are incorporated into detergent formulations to improve grease stain removal and are used in the treatment of oily industrial waste.
Notably, the lipase-mediated transformation of used cooking oil appears as a promising method for the cost-effective production of free fatty acids, providing an efficient alternative to conventional physical and chemical treatments [
8,
15]. Approximately 1.6 × 10
6 tons of free fatty acids are estimated to be produced annually from various plant and animal sources through lipase-catalyzed processes [
16,
17].
Penicillium strains isolated from the Paranaense jungle, including
Penicillium rubens LBM 081, have been reported to exhibit remarkable lipolytic activity, attributed to the adaptation of these microorganisms to organic matter-rich environmental conditions [
9,
18]. This remarkable lipolytic activity suggests that these species could hold great potential for the bioremediation of lipid-rich wastewater.
Considering this context, the present study aimed to evaluate the biotechnological potential of the lipase produced by P. rubens LBM 081 for the hydrolysis of lipid-rich wastewater, emphasizing its application as an effective tool for the sustainable treatment of contaminated effluents.
2. Materials and Methods
2.1. Fungal Material and Maintenance
The Penicillium sp. LBM 081 strain was isolated from decomposing fruits collected in the Paranaense tropical rainforest of Misiones, Argentina. Initial identification was performed based on morphological and microscopic characteristics. This strain was deposited in the collection of the Laboratorio de Biotecnología Molecular, part of the Instituto de Biotecnología Misiones “Dra. María Ebe Reca” (InBioMis) at the Universidad Nacional de Misiones.
The selection of
Penicillium sp. LBM 081 was based on a previous screening study that evaluated the lipolytic capacity of various
Penicillium strains. In that study, this strain demonstrated remarkable lipase production [
9]. The strain was maintained through monthly subcultures in Petri dishes containing malt extract (12.7 g L
−1) and agar (17 g L
−1) (MEA, Biokar
® Diagnostics, Allonne, France) at 4 °C to ensure long-term viability and genetic stability.
2.2. Molecular Identification
The molecular identification of the
Penicillium sp. LBM 081 strain was performed following the protocol described by Fonseca et al. (2018) [
19]. To verify the quality of the isolated genomic DNA, its integrity was assessed by 1% agarose gel electrophoresis and quantified by spectrophotometry at 260 nm (Shimadzu Corporation, Kyoto, Japan).
Partial sequences of three genetic regions commonly used for the molecular identification of
Penicillium species were amplified: the ITS1-5.8S-ITS2 region using the ITS1 and ITS4 primers [
20]; the β-tubulin (Bt) gene using the Bt2a and Bt2b primers [
21]; and the rpb2 gene using the brpb2-5F and brpb2-7.1R primers [
22].
PCR reactions were performed in a final volume of 20 µL containing PCR buffer (5 mM KCl and 10 mM Tris-HCl, pH 8.4), 200 µM of dNTPs, 2.5 mM MgCl2, 10 pmol of each primer, 0.5 U of Taq DNA polymerase (Invitrogen™, Thermo Fisher Scientific, Waltham, MA, USA), and 50 ng of genomic DNA. The amplification program included an initial denaturation at 94 °C for 4 min, followed by 30 cycles of 94 °C for 40 s, 52 °C for ITS primers or 55 °C for Bt and rpb2 primers for 40 s, and 72 °C for 40 s, with a final extension at 72 °C for 10 min.
The amplified products were sequenced using the automated sequencing service provided by MACROGEN (Korea). Bioinformatic analysis was conducted using the BLASTn tool available on the NCBI platform (
https://www.ncbi.nlm.nih.gov/) (accessed on 12 November 2024) and the Fungal Barcoding Database. To confirm the identity of the LBM 081 strain, the obtained sequences were compared to those corresponding to the
Penicillium genus stored in the GenBank database (NCBI) (
Table S1).
The sequences obtained were aligned using the ClustalW tool in MEGA software (version 7.0, Center for Evolutionary Medicine and Informatics, Tempe, AZ, USA), and assembled into a concatenated alignment. Subsequently, a phylogenetic tree was constructed using the Neighbor-Joining (NJ) method with the Kimura two-parameter substitution model. The reliability of the tree was assessed using the bootstrap test with 1000 replicates in MEGA 7.0.
2.3. Evaluation of the Effect of Different Oils on Lipase Activity
The influence of different oils on lipase production by P. rubens LBM 081 was evaluated to identify the most suitable substrate for inducing enzymatic activity.
A liquid culture medium composed of 0.5% meat peptone (Britania®, Buenos Aires, Argentina), 0.3% yeast extract (Britania®, Buenos Aires, Argentina), and 0.3% yeast extract, and 0.3% meat extract (Britania®, Buenos Aires, Argentina), adjusted to pH 7, was used as the basal medium. This medium was individually supplemented with different oils at a concentration of 2%, including corn oil (Mazola®, Aceitera General Deheza, Córdoba, Argentina), peanut oil (Maní King®, General Cabrera, Córdoba, Argentina), sunflower oil and cod liver oil (Parafarm®, Buenos Aires, Argentina), chia oil (Tecnobotánica®, Córdoba, Argentina), canola oil (Krol®, Buenos Aires, Argentina), and olive oil (Cocinero Extra Virgen®, Molinos Río de la Plata, Buenos Aires, Argentina). Control cultures without oil supplementation were included for each evaluated condition.
Each culture was inoculated with a spore suspension (1 × 106 spores mL−1) in the presence of 0.1% (v v−1) Tween® 80 (Merck KGaA, Darmstadt, Germany) as a dispersing agent to promote homogeneous distribution of spores in the medium. Cultures were incubated at 30 °C with constant agitation at 140 rpm for 8 days, following conditions previously reported as optimal for lipase production in Penicillium species.
Aliquots were taken from each culture on days 4, 6, and 8 of incubation. The samples were subsequently centrifuged at 8000× g for 15 min to separate the mycelium from the supernatant. The resulting supernatant was used in lipase activity assays.
2.4. Lipase Activity Determination
Lipase activity was determined following the protocol described by Ortellado et al. (2021) [
9], using 4-nitrophenyl palmitate (4-NPP; Sigma-Aldrich, St. Louis, MO, USA) as a model substrate. This method was chosen for its precision and sensitivity in detecting lipase activity in fungal samples. 4-NPP is a chromogenic substrate widely used in enzymatic assays since its hydrolysis releases 4-nitrophenol (4-NP), a product that can be easily quantified by spectrophotometry.
The reaction mixture (1000 µL) was prepared by combining 100 µL of solution A (0.0015 g of 4-NPP dissolved in 4 mL of isopropanol; Biopack, Buenos Aires, Argentina), 800 µL of solution B (0.1 g of gum arabic, Biopack, Buenos Aires, Argentina), 0.4 mL of Triton® X-100 (Merck KGaA, Darmstadt, Germany), and 100 mL of 0.1 M phosphate buffer, pH 7, prepared with KH2PO4 and Na2HPO4 (Biopack, Buenos Aires, Argentina) and 100 µL of the enzymatic supernatant. The resulting mixture was incubated at 37 °C for 10 min. The amount of 4-NP released was subsequently measured at 405 nm using a spectrophotometer (Model UV-3600, Shimadzu, Kyoto, Japan).
Enzyme activity was expressed in international units (IU), where 1 unit was defined as the amount of enzyme that releases 1 µmol of 4-NP per minute under the assay conditions.
2.5. Effect of Different Olive Oil Concentrations on Lipase Activity
The effect of various olive oil concentrations as an inducer of lipase production was evaluated, given its high triglyceride content, which acts as an ideal substrate to promote lipolytic activity in filamentous fungi, including P. rubens.
The tested concentrations were 1%, 1.5%, 3%, 3.5%, 4%, and 4.5%. Cultures were incubated for 8 days at 30 °C with constant agitation at 140 rpm, conditions previously selected for their effectiveness in inducing lipase activity in Penicillium species.
On days 4 and 6, aliquots were taken from each culture and centrifuged at 8000× g for 15 min to remove residual mycelium. At the end of the assay (day 8), the mycelium was separated from the supernatant by filtration using a Büchner funnel equipped with glass fiber filters (pore size 1.2 µm; Macherey-Nagel, Düren, Germany). The resulting supernatant was used to determine lipase activity.
2.6. Effect of Different Nitrogen Sources on Lipase Activity
The effect of five nitrogen sources, selected for their potential to induce the production of hydrolytic enzymes—a key factor in lipase synthesis by filamentous fungi—was evaluated. The nitrogen sources assessed were meat extract (Britania®, Buenos Aires, Argentina), ammonium sulfate (Biopack®, Buenos Aires, Argentina), meat peptone (Britania®, Buenos Aires, Argentina), soybean peptone (Sigma-Aldrich®, St. Louis, MO, USA), and yeast extract (Britania®, Buenos Aires, Argentina).
Each nitrogen source was tested at three concentrations (0.3%, 1%, and 2%) and supplemented with the olive oil concentration that showed the highest lipase-inducing capacity in the previous assays (Cocinero Extra Virgen®, Molinos Río de la Plata, Buenos Aires, Argentina).
The incubation conditions, aliquot collection, centrifugation, and filtration procedures were identical to those described in the previous section. The resulting supernatant was used to determine lipase activity.
2.7. Effect of Different Inoculum Concentrations and Agitation on Lipase Activity
To optimize the growth conditions and lipase production, three inoculum concentrations were evaluated: 105, 106, and 107 spores mL−1. These concentrations were selected based on previously reported ranges for the production of hydrolytic enzymes in filamentous fungi.
Additionally, the effect of agitation at three different speeds—100, 140, and 160 rpm—was assessed to determine the optimal oxygenation conditions for lipase production. Agitation was carried out in an orbital shaker (model ES-20/60, Biosan, Riga, Latvia).
The cultures were conducted in sterilized Erlenmeyer flasks (Duran®, Mainz, Germany) (121 °C for 20 min) containing the optimized culture medium described previously, which included the nitrogen source and olive oil concentration that exhibited the highest lipase-inducing capacity.
Samples were incubated at 30 ± 1 °C for 8 days. Aliquots were taken on days 4, 6, and 8 under aseptic conditions for enzyme activity assessment.
All experiments were performed in triplicate to ensure the reproducibility of the results.
2.8. Effect of pH and Temperature on Lipase Activity
The effect of pH on lipase activity was evaluated by measuring enzymatic activity at a constant temperature of 30 °C, using sodium phosphate buffer solutions adjusted to pH values of 7, 7.5, 8, and 8.5. This range was selected as it falls within typical values reported in industrial and domestic effluents, where the enzyme is expected to maintain its activity to be effective in bioremediation processes.
The effect of temperature was evaluated by incubating the reaction mixture (prepared according to the lipase activity protocol) at the optimal pH previously determined in this experiment. The evaluated temperatures were 4, 25, 30, 40, and 55 °C, selected to cover a broad range of environmental and industrial process conditions. All other experimental conditions remained constant.
Enzymatic activity was measured at 405 nm, recording the release of 4-nitrophenol (4-NP). Relative activity was expressed as a percentage, considering 100% as the maximum value obtained under optimal conditions.
2.9. Thermostability and pH Stability of Lipase Activity Produced in the Optimized Medium
The stability of lipase activity under variable conditions characteristic of industrial and domestic effluents was determined.
Thermal stability was evaluated by preincubating the enzymatic extract obtained under optimal lipase production conditions at temperatures of 4, 25, 30, 40, and 50 °C.
Meanwhile, pH stability was determined by preincubating the enzymatic extract with sodium phosphate buffer solutions at pH values of 7, 7.5, 8, and 8.5, at a constant temperature of 30 °C.
In both assays, aliquots were withdrawn at 1, 6, 12, 48, and 72 h intervals and residual lipase activity was measured according to the previously described protocol.
Residual activity was expressed as a percentage, considering the initial enzymatic activity value as 100%.
2.10. Effluent Treatment
The effluent treatment was conducted to evaluate the capacity of the optimized enzymatic extract to reduce the pollutant load in lipid-rich wastewater. Two representative types of effluents were prepared: 1. Synthetic Olive Oil Effluent: This effluent was obtained by mixing olive oil and non-potable water at a 2:1 ratio, simulating a high-lipid-load effluent typical of the food industry. 2. Domestic Effluent: This effluent was prepared by combining non-potable water, sunflower oil, and olive oil in a 2:1:1 ratio, representing typical household waste. This effluent was obtained after two frying cycles at 185 °C for 12 min, followed by filtration through a Büchner funnel to remove solid particles.
Assays were prepared in 250 mL Erlenmeyer flasks with optimized supernatant in different proportions: 26% (26% optimized supernatant and 74% domestic effluent) and 44% (44% optimized supernatant and 56% domestic effluent). Each assay was incubated for 6, 24, and 48 h at 30 °C and 140 rpm to ensure proper dispersion of the supernatant in the medium [
23].
The pH and temperature parameters were measured at the beginning and end of each assay. For the determination of total oils and fats, the Soxhlet extraction method was employed using petroleum ether (Biopack
®, Buenos Aires, Argentina) and a Soxhlet apparatus (VELP Scientifica
®, Usmate Velate, Italy). The chemical oxygen demand (COD) was determined using the open reflux method as specified in the Standard Methods for the Examination of Water and Wastewater (APHA, 2017) [
24].
The percentage of removal was calculated using the following equation:
where
2.11. Gas Chromatography–Mass Spectrometry (GC/MS)
The fatty acid profile present in the samples was analyzed using gas chromatography coupled to mass spectrometry (GC/MS) with a Shimadzu GC/MS QP2010 system (Shimadzu Corporation, Kyoto, Japan). The analysis was performed following the protocols established in ISO 12966-2:2017 and ISO 12966-4:2015 standards, ensuring the standardization and reproducibility of the procedure [
25,
26].
Prior to analysis, the samples underwent a methylation process to obtain fatty acid methyl esters (FAMEs) as specified in the mentioned ISO standards using analytical-grade reagents (methanol, sulfuric acid, and hexane; Biopack®, Buenos Aires, Argentina). Results were expressed in grams of fatty acids per 100 g of methyl esters.
This methodology enabled the accurate identification and quantification of the lipid composition of the samples, highlighting the predominant fatty acids and their possible relationship with the hydrolysis processes catalyzed by the lipase.
The analyses were conducted by the Technical Directorate of Analytical Services and the Food Operations Sub-management at INTI (Instituto Nacional de Tecnología Industrial, San Martín, Buenos Aires, Argentina), ensuring compliance with analytical quality standards.
2.12. Toxicity Study
To evaluate the toxicity of the treated effluents, a batch of chemically untreated
Lactuca sativa seeds (La Semillería
®, Buenos Aires, Argentina) with an initial germination rate of 97% was used. The seeds were stored in a dry environment at 4 °C until use. Two parameters were assessed: the relative growth index (RGI) and the germination index (GI). As quality controls, germination rates above 90% and a coefficient of variation below 30% in the control group were established [
27,
28,
29].
The assays were conducted in 90 mm diameter Petri dishes (Britania
®, Buenos Aires, Argentina) lined with filter paper (Cytiva, Marlborough, MA, USA), with 10 seeds per plate. Each plate received 4 mL of treated effluent or distilled water in the case of the control. Each assay was performed in triplicate, and measurements were recorded after 5 days of incubation in darkness at 22 °C, following standardized protocols [
27,
28,
29,
30].
The RGI was calculated using the following equation [
29]:
where
The RGI values were categorized according to the observed toxic effects:
The germination index (GI) was calculated using the following equation:
where
2.13. Statistical Analysis
The data obtained in this study were statistically analyzed using Statgraphics Centurion XVI.I (Statgraphics Technologies, The Plains, VA, USA) and GraphPad Prism version 7.0 for Windows (GraphPad Software, San Diego, CA, USA). The Anderson–Darling test was used to assess data normality, while Bartlett’s test was applied to verify the homogeneity of variances. Data that met the assumptions of normality and homogeneity were analyzed using one-way ANOVA, followed by Tukey’s test for multiple comparisons between groups. In cases where normal distribution was not achieved, non-parametric tests such as the Kruskal–Wallis test or the Mann–Whitney U test were applied as appropriate.
All assays were performed in triplicate, and results were expressed as mean ± standard deviation (SD). Differences were considered statistically significant at p < 0.05. Statistical tests were applied to the variables analyzed in this study, including lipase activity, contaminant removal, and toxicity parameters.
3. Results and Discussion
3.1. Molecular Identification
In a previous study, the production of lipases was evaluated in twenty strains of the
Penicillium genus with the aim of identifying those with potential for treating used cooking oil residues. In that analysis, the isolate
P. rubens LBM 081 stood out as the best lipase producer, displaying superior enzymatic activity compared to the other analyzed strains [
9]. This finding prompted further investigation into its biotechnological potential, considering that different isolates of the same species commonly exhibit significant variations in enzyme production. This approach is crucial for identifying strains with novel properties in terms of enzyme efficiency and yield [
31,
32,
33]. Such strategies are widely employed in the enzyme industry to meet the increasing demand for more efficient biocatalysts, an area that has not been extensively explored.
The PCR products obtained from the ITS1-5.8S-ITS2 region, the β-tubulin (Bt) gene, and the rpb2 gene region generated fragments of approximately 477, 397, and 453 bp, respectively. These regions were selected for their well-established reliability in the molecular identification of fungal species due to their high conservation and specificity within the
Penicillium genus [
31,
33].
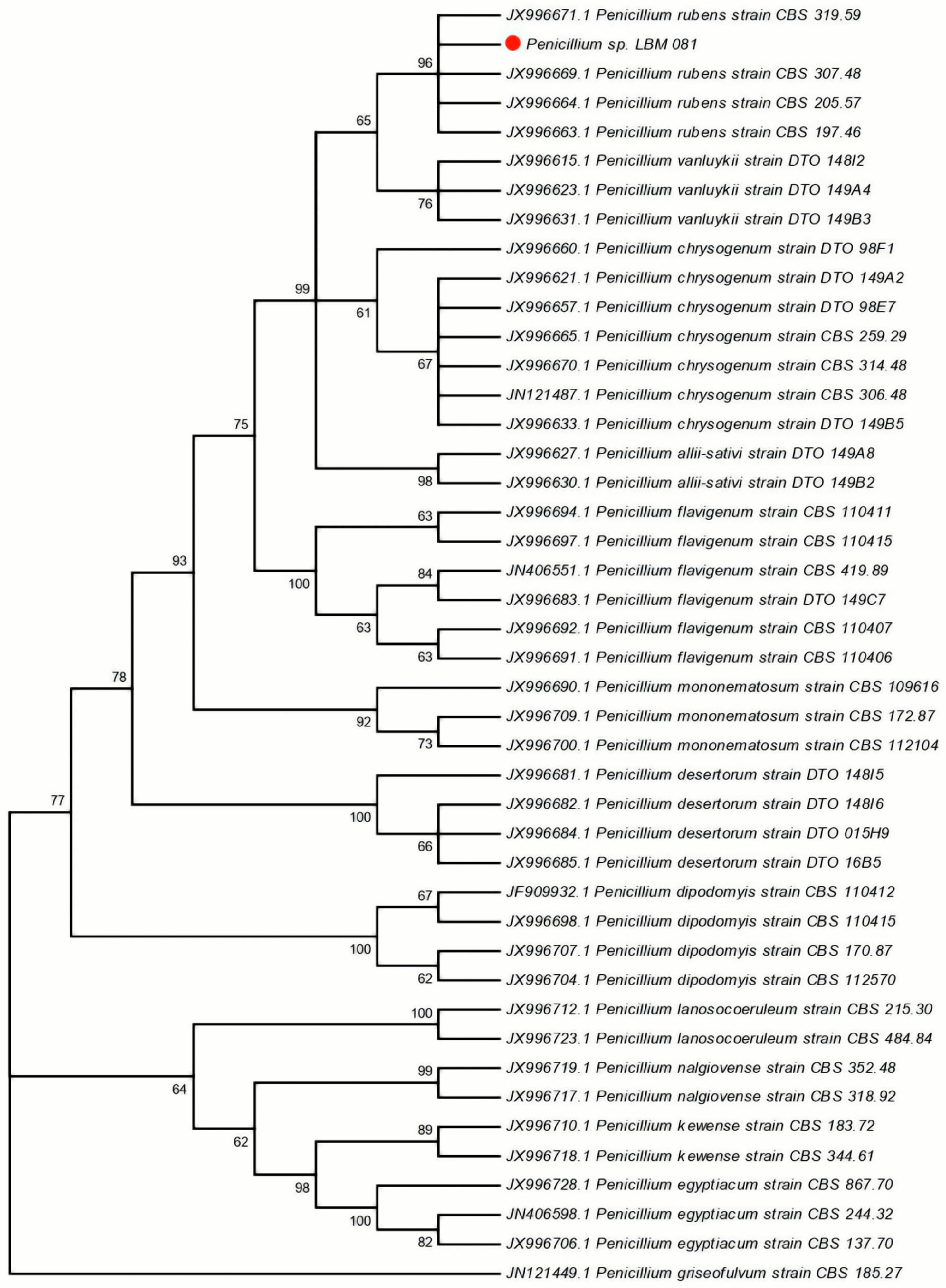
The sequences obtained were deposited in the NCBI database (
GenBank accession numbers: MT952959, MT955993, MT955992). Phylogenetic analysis using the Neighbor-Joining (NJ) method and Kimura’s two-parameter substitution model positioned the LBM 081 isolate within the clade corresponding to
P. rubens, with a bootstrap value of 96% (
Figure 1).
These results confirmed the molecular identity of the isolate and established its phylogenetic relationship with other species in the
Penicillium genus. This precise identification is crucial for understanding the biotechnological potential of the isolate, as
Penicillium species are well known for their ability to produce a wide variety of industrial enzymes, including lipases, which are recognized for their stability under extreme pH and temperature conditions [
34]. Specifically, species such as
Penicillium simplissicimum, Penicillium ochrochloron, and
Penicillium digitatum have demonstrated significant potential in industrial processes [
35,
36,
37]. The high reliability of the phylogenetic analysis, supported by the bootstrap value of 96%, ensures that the properties attributed to
P. rubens LBM 081 correspond specifically to this species, minimizing the risk of contamination or misidentification of other organisms present in the sample.
3.2. Evaluation of the Effect of Different Oils on Lipase Activity
The analysis of the effect of different oils on lipase production by P. rubens LBM 081 revealed a marked influence of the lipid source on enzymatic activity. Seven different oils—sunflower oil, cod liver oil, peanut oil, canola oil, corn oil, chia oil, and olive oil—were evaluated at a concentration of 2%.
The results showed that olive oil promoted the highest lipase activity, reaching 1148 U mL
−1 on day 4 of cultivation (
p < 0.05) and 1283 U mL
−1 on day 6 (
p < 0.05), maintaining 1230 U mL
−1 afterward (
Figure 2). Similarly, chia oil showed a comparable trend, albeit with lower values, reaching 808 U mL
−1 on day 4, 891 U mL
−1 on day 6, and 871 U mL
−1 at the end of the cultivation period (
p < 0.05).
Conversely, corn oil, cod liver oil, and canola oil resulted in considerably lower increases in lipase activity, with values of 252, 223, and 138 U mL
−1, respectively, which were significantly lower than those obtained with olive and chia oils (
p < 0.05) (
Figure 2).
The primary factor influencing the expression of lipase activity is the presence of carbon sources in the form of oils, such as olive oil, corn oil, or sunflower oil, among others [
1]. In this regard, olive oil is a rich source of long-chain triglycerides, particularly oleic acid (C18:1), a potent inducer of gene expression associated with lipase production in fungi [
38,
39]. Additionally, oleic acid has been reported to stabilize the structural conformation of lipases, enhancing both their secretion and prolonged enzymatic activity in culture media [
40].
The stimulation observed with olive oil aligns with previous studies highlighting its effectiveness as a lipase inducer in filamentous fungi. Lima et al. (2003) [
38] reported this same behavior in
Penicillium aurantiogriseum, where olive oil promoted greater lipase production compared to sunflower, soybean, and corn oils.
On the other hand, the behavior observed with chia oil, although inferior to that of olive oil, is also noteworthy. This oil is rich in polyunsaturated fatty acids such as α-linolenic acid (C18:3), which can also induce fungal lipase activity [
41]. However, its lower relative efficiency could be attributed to the greater susceptibility of α-linolenic acid to oxidation, which may reduce its stability under prolonged cultivation conditions.
In contrast, corn oil, cod liver oil, and canola oil, which are rich in saturated fatty acids or have a simpler structural composition, did not significantly promote lipase activity. This behavior is consistent with reports indicating that certain oils generate weaker stimulation of the enzymatic machinery, possibly due to low lipid bioavailability in the culture medium or reduced gene induction capacity [
39,
40].
Consequently, these results highlight that the selection of the type of oil is crucial in lipase production by P. rubens LBM 081, with olive oil proving to be the most effective inducer due to its favorable lipid profile and its ability to promote the enzyme’s structural stability during cultivation.
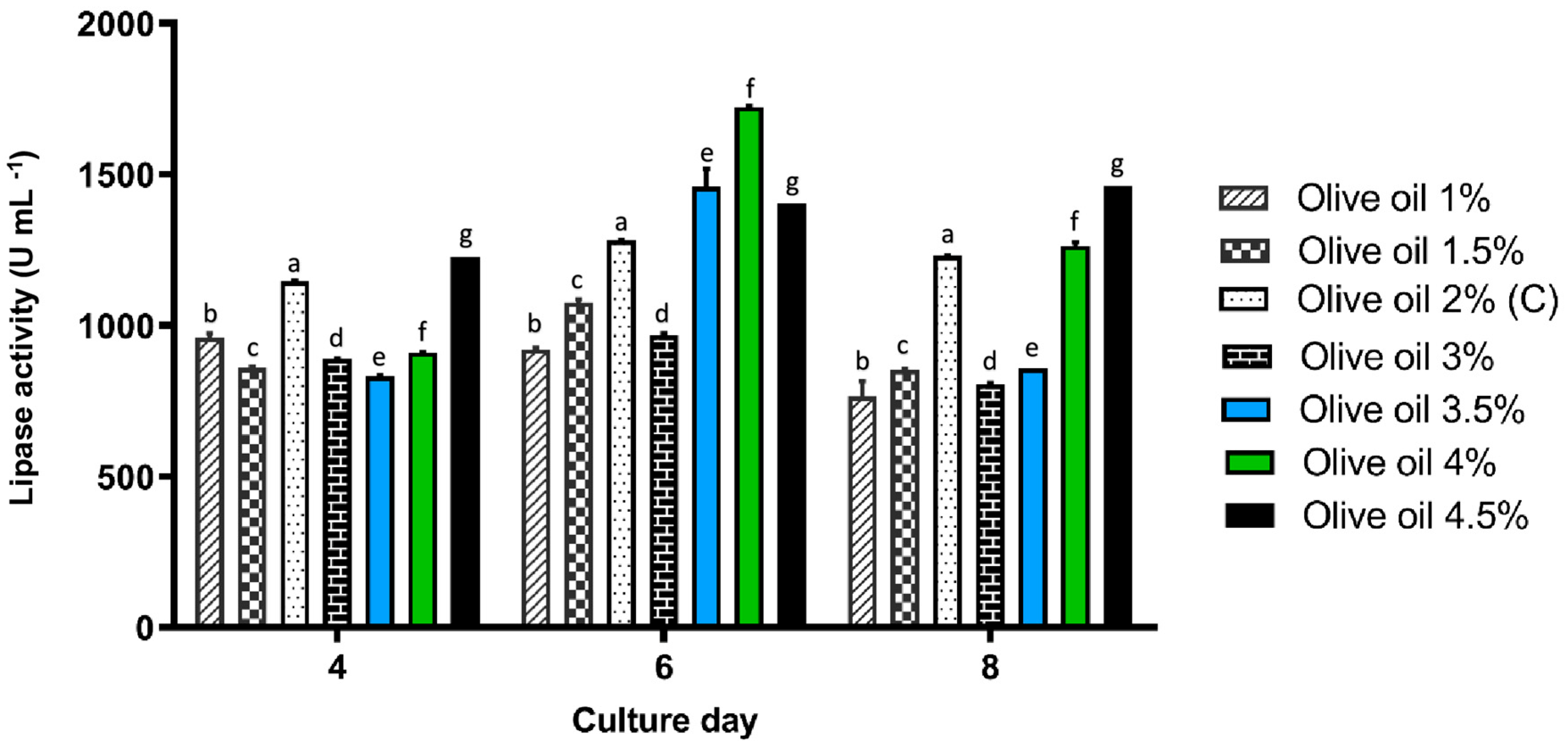
3.3. Effect of Different Olive Oil Concentrations on Lipase Activity
Since olive oil was identified as the most effective inducer of lipase activity in preliminary assays, different concentrations of this oil (1%, 1.5%, 3%, 3.5%, 4%, and 4.5%) were evaluated to determine the optimal concentration for maximizing enzyme production. The cultures were incubated for 8 days at 30 °C, with constant agitation at 140 rpm.
Lipase activity progressively increased with the rise in olive oil concentration, reaching its maximum on day 6 of cultivation with the addition of 4% olive oil (1722 U mL
−1,
p < 0.05). This concentration was significantly higher than the 3.5% treatment, which registered an activity of 1459 U mL
−1 (
Figure 3). On day 8, the highest activity was observed with 4.5% olive oil, reaching 1461 U mL
−1 (
p < 0.05), although this value did not significantly improve compared to the 4% concentration.
The progressive increase in lipase activity with increasing olive oil concentration can be explained by the inductive effect that this type of lipid exerts on the gene expression of fungal lipases. Previous studies have demonstrated that triglycerides rich in oleic acid (C18:1) promote the activation of genes that regulate lipase synthesis, thereby enhancing enzyme production [
42,
43]. This effect is attributed to oleic acid’s ability to act as an allosteric effector, activating transcription factors associated with lipase gene expression [
42].
This behavior is consistent with the findings of Das et al. (2016) [
44], who observed that a 2.5% concentration of coconut oil promoted the highest lipase activity in
Aspergillus tamarii JGIF06, while higher concentrations reduced activity due to oxygen diffusion interference and aeration limitations. This situation resembles what was observed in the present study, where the 4% olive oil concentration proved to be the most effective, while higher values did not significantly enhance enzymatic activity.
The decrease in enzymatic activity observed on day 8 for the 4% concentration suggests that excessive lipid accumulation in the medium may negatively affect enzyme stability or fungal viability. This phenomenon has been linked to the formation of oil layers on the culture surface, which interfere with oxygen transfer and reduce mycelial growth [
44]. Furthermore, elevated lipid concentrations may generate a competitive inhibitory effect, as unhydrolyzed triglycerides may occupy active enzyme sites, thereby decreasing catalytic efficiency [
45].
In this context, the 4% olive oil concentration proved to be the optimal condition for maximizing lipase production in P. rubens LBM 081. These results not only reinforce the importance of optimizing both the type and concentration of the oil used as an inducer but also provide valuable insights for designing biotechnological processes focused on large-scale lipase production.
3.4. Effect of Different Nitrogen Sources on Lipase Activity
Since the nitrogen source is a determining factor in fungal lipase production, the effect of five nitrogen sources on lipase activity in P. rubens LBM 081 was evaluated: meat extract, ammonium sulfate, meat peptone, soy peptone, and yeast extract.
The highest increase in lipase activity was achieved with the addition of 2% meat peptone, reaching 2881 U mL
−1 on day 6 of cultivation (
p < 0.05), followed by the treatment with 2% soy peptone, which reached 2393 U mL
−1 (
p < 0.05). On day 4 of cultivation, remarkable values were also observed with 0.3% meat peptone (2373 U mL
−1) and 2% soy peptone (2193 U mL
−1). However, on day 8, a significant decrease in lipase activity was recorded, reducing to 1813 U mL
−1 and 1653 U mL
−1 for the assays with 2% meat peptone and 2% soy peptone, respectively (
Figure 4).
The positive effect of peptone can be explained by its richness in short peptides, amino acids, and essential growth factors for fungal metabolism, which promote protein synthesis and the secretion of extracellular enzymes such as lipases [
10]. In particular, meat peptone has been recognized for facilitating fungal growth and stimulating the production of hydrolytic enzymes in filamentous fungi due to its balanced source of organic nitrogen, which is easily assimilated [
35].
The decrease in enzymatic activity observed on day 8 could be attributed to nutrient depletion in the culture medium, a characteristic phenomenon of the stationary phase, where the reduced rate of protein synthesis and the accumulation of secondary metabolites can negatively affect enzymatic activity [
45]. Additionally, the accumulation of inhibitory compounds derived from secondary metabolism in prolonged culture conditions has also been reported as a limiting factor in extracellular enzyme production.
On the other hand, inorganic sources such as ammonium sulfate did not significantly promote an increase in lipase activity. This behavior aligns with previous reports indicating that organic nitrogen sources are generally more effective for fungal lipase production since they provide a greater diversity of nitrogenous compounds that act as precursors in protein synthesis [
46]. Moreover, ammonium sulfate has been suggested to induce higher acidification of the culture medium, which could hinder mycelial growth and limit the production of extracellular enzymes.
Consequently, these results suggest that meat peptone, particularly at a concentration of 2%, was the most effective nitrogen source in this study for stimulating lipase production in P. rubens LBM 081. Nevertheless, this input may not be the only viable alternative, and it is therefore pertinent to explore other nitrogen sources that are more accessible and sustainable, and better suited for future large-scale applications.
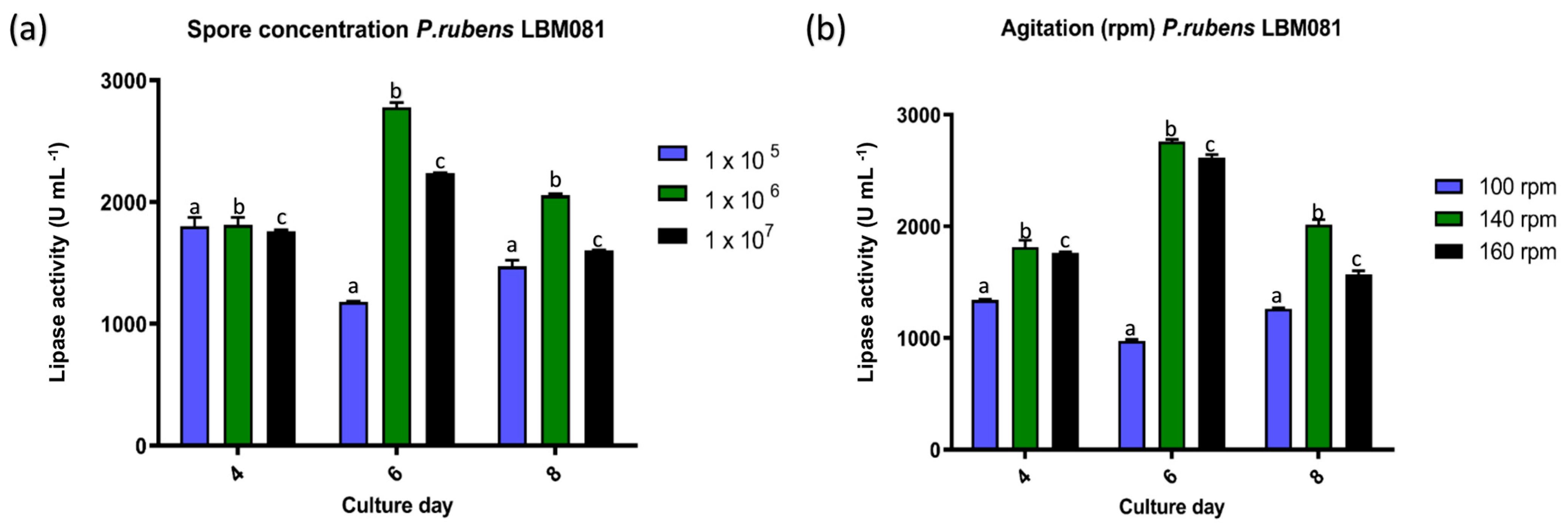
3.5. Effect of Different Inoculum Concentrations and Agitation on Lipase Activity
The combined effect of inoculum concentration and agitation conditions was crucial in the production of lipase by
P. rubens LBM 081. The highest enzymatic activity was achieved with a spore concentration of 1 × 10
6 spores mL
−1 and incubation at 140 rpm, reaching a maximum value of 2779 U mL
−1 on day 6 of cultivation (
p < 0.05) (
Figure 5).
The highest yield observed with a concentration of 1 × 10
6 spores mL
−1 aligns with studies indicating that a high inoculum density accelerates the logarithmic growth phase, optimizing fungal metabolism and promoting greater lipase secretion [
46]. This behavior is attributed to the fact that a higher initial cell density enables more efficient nutrient consumption during the early stages of cultivation. However, excessively high concentrations may induce an autoinhibition phenomenon due to cellular competition, which increases oxygen and nutrient demands, thus limiting growth and enzymatic production [
47]. Moreover, excessive concentrations can promote the accumulation of toxic metabolic by-products, which negatively affect enzymatic activity [
45].
Regarding agitation, a significant increase in lipase activity was observed when agitation rates of 140 rpm or 160 rpm were applied, compared to lower agitation at 100 rpm (
p > 0.05) (
Figure 5). This result relates to the positive effect of agitation on oxygen transfer and nutrient dispersion in submerged cultures, key factors for maximizing fungal enzyme production [
48].
In this study, agitation promoted the dispersion of oil micelles in the medium, increasing the contact surface between fungal cells and the lipid substrate, which enhanced lipase secretion [
48,
49]. Additionally, pellet formation was observed in cultures agitated at 140–160 rpm. This morphology may be advantageous since pellets increase the secretion rate of extracellular metabolites, including lipases, due to greater contact between the mycelium and the culture medium [
50,
51].
Pellet formation is considered a favorable morphology in submerged fermentation processes, as it promotes mycelial stability and improves nutrient diffusion. However, excessive agitation may generate shear forces that damage the mycelium structure, while insufficient agitation may induce the formation of cell aggregates that limit oxygen transfer and reduce extracellular enzyme production [
52,
53].
In this context, the combination of an inoculum concentration of 1 × 106 spores mL−1 and agitation at 140 rpm proved to be the optimal condition for maximizing lipase production in P. rubens LBM 081. This finding provides key information for the design of efficient submerged fermentation processes aimed at producing lipases for environmental and industrial applications.
3.6. Effect of Temperature and pH on Lipase Activity
The results revealed that the highest lipase activity was recorded in assays conducted at pH 7 and 7.5, and at a temperature of 30 °C (
p < 0.05), which corresponds to optimal conditions widely reported for fungal lipase production [
42].
Optimal behavior at neutral pH is characteristic of fungal lipases used in bioremediation since these enzymes exhibit greater stability and catalytic activity within pH ranges between 7 and 8.5 [
54]. This effect is attributed to the fact that, within this pH range, lipases maintain an adequate three-dimensional conformation, ensuring the integrity of the active site and enhancing substrate interaction [
55].
Regarding temperature, the optimal value recorded at 30 °C is consistent with reports indicating that most fungal lipases exhibit maximum catalytic performance within a range of 25–35 °C. This range is ideal for their application in biotechnological processes and effluent treatment under ambient conditions [
46] (
Figure 6).
3.7. Thermostability and pH Stability of Lipase Activity Produced in the Optimized Medium
Thermal stability and resistance to pH variations are fundamental properties for the industrial application of lipases, particularly in biotechnological processes such as bioremediation, detergent formulation, and the synthesis of pharmaceutical compounds [
56,
57]. In this study, the thermostability and pH stability of the lipase produced by
P. rubens LBM 081 were evaluated to characterize its potential for treating lipid-rich wastewater.
3.7.1. Thermal Stability
The results showed that the lipase from
P. rubens LBM 081 exhibited remarkable thermal stability under moderate conditions. At 30 °C, lipase activity was preserved at 94% after 6 h of incubation and 91% after 12 h. Although a progressive decrease was observed over time, residual activity remained at 61% and 46% after 48 and 72 h, respectively (
Figure 6).
At lower temperatures, such as 25 °C, a gradual loss of activity was observed, with a relative activity of approximately 87% after 6 h, which decreased to ~52% after 48 h and 41% after 72 h.
The high stability observed at 30 °C is particularly relevant for biotechnological applications since this temperature is common in environmental and bioremediation processes under real conditions [
42]. When compared to other fungal lipases, this favorable characteristic stands out: the lipase from
Aspergillus niger retained 85% of its activity after 6 h of incubation at 30 °C [
35], while the lipase from
Rhizopus oryzae retained only 60% of its activity after 72 h under similar conditions [
54,
58].
The progressive loss of activity over prolonged periods is an expected phenomenon in lipases and is commonly associated with thermal denaturation, active site modification, or proteolysis [
10]. In this context, the ability of the
P. rubens LBM 081 lipase to maintain 94% of its activity after 6 h at 30 °C represents a significant advantage for its implementation in prolonged environmental processes, where thermal stability is crucial to ensuring treatment efficiency.
3.7.2. pH Stability
The
P. rubens LBM 081 lipase also exhibited remarkable stability under slightly neutral and alkaline conditions. At pH 7, enzymatic activity was maintained at 95% after 6 h, gradually decreasing to 88% at 12 h and to 77% and 58% after 48 and 72 h, respectively. At pH 7.5, activity remained at 90% after 6 h and 84% after 12 h, while decreasing to 70% and 53% after 48 and 72 h, respectively (
Figure 6).
These results align with studies that emphasize the high stability of fungal lipases in neutral or slightly alkaline conditions, especially those applied in bioremediation and detergent processes [
59,
60]. This stability has been proposed to be linked to the conservation of salt bridges and optimal distribution of electrostatic charges in the active site, factors that maintain the enzyme’s structural conformation under alkaline conditions [
61].
The resistance of the
P. rubens LBM 081 lipase to these conditions is particularly advantageous for its application in the bioremediation of lipid-rich wastewater, as such effluents often exhibit a pH near neutrality or slightly alkaline due to the presence of detergents and other industrial compounds [
35]. Additionally, this property is highly favorable for the formulation of cleaning products and detergents, where alkaline conditions enhance the efficient removal of fats [
10].
3.8. Effluent Treatment Using the Supernatant of P. rubens LBM 081
The treatment of lipid-rich effluents represents a significant environmental challenge, as these residues generate a high organic load and hinder conventional wastewater treatment processes.
3.8.1. Reduction of Chemical Oxygen Demand (COD)
The results revealed that the highest COD removal efficiency was achieved with the application of 44% supernatant, reaching 50% efficiency after 24 h in the olive oil effluent. This positive effect persisted with a slight decrease, achieving 45% removal after 48 h.
Conversely, in the assays that utilized only 26% supernatant, COD removal was significantly lower, reaching 16% after 24 h and 23% after 48 h (
Figure 7a).
The improved performance observed with 44% supernatant is attributed to the greater availability of lipolytic enzymes, which enhanced catalytic efficiency in the hydrolysis of complex organic compounds. Previous studies have indicated that increasing enzyme concentration enhances reaction rates and the efficiency of the biodegradation process in wastewater treatment systems [
62,
63,
64].
Lipases are well known for efficiently breaking down triglycerides into organic acids and volatile fatty acids (VFAs), which are subsequently degraded into carbon dioxide and water via the β-oxidation pathway [
65]. In this context, the treatment with 44% supernatant promoted a higher availability of lipases to carry out this lipid degradation mechanism, thereby facilitating COD reduction.
In the case of the domestic effluent, COD removal was considerably lower than in the olive oil effluent. Treatment with 44% supernatant achieved 44% removal after 24 h, which decreased to 20% after 48 h. With 26% supernatant, only 4% removal was observed after 24 h and 2% after 48 h (
Figure 7a).
The lower efficiency in domestic effluent may be due to the presence of additional contaminants, such as detergents and diverse organic matter, which can hinder enzymatic activity [
66,
67,
68]. This behavior is consistent with previous reports indicating that lipases exhibit reduced efficiency in complex matrices with high surfactant content and other inhibitory substances [
69].
3.8.2. Removal of Total Oils and Greases
The results obtained for oil and grease removal followed a pattern similar to that observed for COD. In the olive oil effluent, treatment with 44% supernatant achieved 16% removal after 6 h of incubation, which significantly increased to 50% after 24 h and stabilized at 24% after 48 h.
On the other hand, the use of 26% supernatant showed lower efficacy, achieving only 4% removal in the first 6 h and 16% after 48 h of incubation (
Figure 7b).
The higher efficiency observed in the olive oil effluent is attributed to its high content of oleic acid and unsaturated triglycerides, which are more susceptible to enzymatic hydrolysis compared to saturated oils and more complex compounds found in domestic waste [
56].
In the case of the domestic effluent, fat removal was more limited. With 44% supernatant, a maximum of 29% removal was achieved after 24 h of incubation, while the 26% supernatant treatment achieved a maximum value of 23% after 48 h (
Figure 7b).
This behavior is associated with the presence of detergents and emulsifiers in the domestic effluent, which can form stable micelles that limit the enzyme’s access to lipid substrates [
35]. Additionally, the interaction between lipases and micelles has been reported to partially inhibit enzymatic activity, reducing the degradation rate [
69].
These results underscore the importance of using complementary parameters to evaluate the efficiency of enzymatic treatment. While the measurement of total oils and greases reflects the direct action of lipase on intact lipids, COD represents the overall reduction in the organic load, including intermediate compounds generated during hydrolysis. This combined approach provides a more comprehensive and accurate assessment of the effectiveness of the applied enzymatic treatment [
70,
71].
3.9. Free Fatty Acid Profile and Evaluation of Enzymatic Treatment via GC-MS
The chromatographic analysis of free fatty acids (FFAs) using GC-MS not only allowed the identification and quantification of the products generated after the enzymatic treatment of the effluents but also enabled the evaluation of the hydrolysis efficiency mediated by the
P. rubens LBM 081 supernatant. This technique is widely recognized for its sensitivity and precision in lipid compound characterization, facilitating the assessment of the degree of hydrolysis and the efficiency of the applied biotechnological treatment [
72].
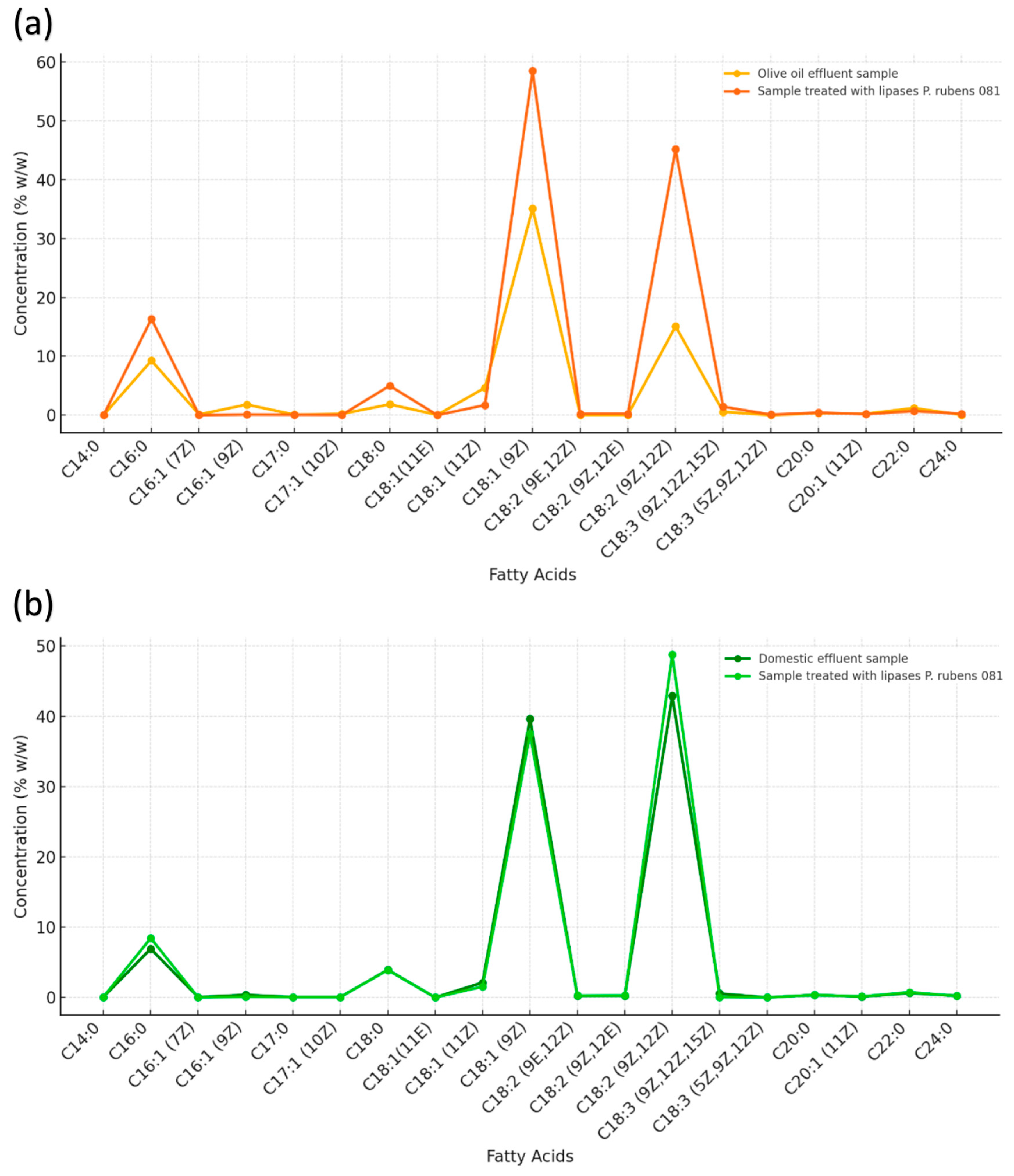
3.9.1. Olive Oil Effluent
In the olive oil-rich effluent, the predominant fatty acids were palmitic acid (C16:0), stearic acid (C18:0), oleic acid (C18:1(9Z)), linoleic acid (C18:2(9Z,12Z)), α-linolenic acid (C18:3(9Z,12Z,15Z)), arachidic acid (C20:0), and behenic acid (C22:0) (
Figure 8a,
Table S2).
The detection of these compounds in the initial effluent is consistent with the natural composition of olive oil, characterized by its high oleic acid content, which can account for up to 70% of the total fatty acids [
73].
After treatment with the P. rubens LBM 081 supernatant, a significant increase in the concentrations of these same fatty acids was observed. This increase indicates efficient hydrolysis, where the lipase catalyzed the release of free fatty acids from the triglycerides present in the effluent.
The rise in C16:0 and C18:0 levels is particularly relevant since these saturated fatty acids are more easily released during lipase-catalyzed hydrolysis due to their lower steric hindrance compared to long-chain unsaturated fatty acids [
74]. This behavior is characteristic of fungal lipases with high specificity for sn-1 and sn-3 bonds in triglycerides, a preference that facilitates the release of saturated fatty acids during the early stages of the hydrolytic process [
75].
3.9.2. Domestic Effluent
In the domestic effluent, six predominant fatty acids were identified: palmitic acid (C16:0), stearic acid (C18:0), vaccenic acid (C18:1(11Z)), oleic acid (C18:1(9Z)), linoleic acid (C18:2(9Z,12Z)), and behenic acid (C22:0) (
Figure 8b). This profile is characteristic of mixed lipid residues derived from vegetable oils combined with animal fats, which typically contain a higher proportion of saturated and trans fatty acids [
76].
Following treatment with the P. rubens LBM 081 supernatant, a significant increase in palmitic acid (C16:0) and linoleic acid (C18:2(9Z,12Z)) concentrations was observed.
The increase in linoleic acid is particularly noteworthy since this polyunsaturated fatty acid is highly susceptible to oxidation. This property facilitates its biodegradation during the final stages of effluent treatment, contributing to the reduction of persistent organic compounds that increase the system’s pollutant load [
77].
The ability of the
P. rubens supernatant to promote the release of free fatty acids, even in more complex matrices such as domestic effluent, demonstrates its effectiveness in degrading heterogeneous lipid residues. This activity is consistent with reports indicating that certain fungal lipases exhibit high specificity for triglycerides containing saturated and polyunsaturated fatty acids, favoring the hydrolysis of sn-1 and sn-3 bonds in complex lipid compounds [
75].
3.10. Toxicity Assessment in Lactuca sativa as a Biological Indicator
The toxicity assessment using Lactuca sativa bioassays was essential to determine the potential environmental impact of the P. rubens LBM 081 supernatant after its application in the treatment of lipid-rich effluents. This evaluation is crucial to ensure that this biotechnological alternative is safe and does not produce adverse phytotoxic effects on the environment.
The results showed that treatments with 44% supernatant, in both types of effluents (olive oil and domestic), exhibited germination indices above 50% after 24 h of treatment (
Figure 9,
Table S3). This value is considered acceptable according to international toxicity standards for germination bioassays [
78], suggesting that the supernatant does not induce severe phytotoxic effects during the early stages of plant development.
The fact that the germination index exceeded 50% indicates that, although some adverse effects were observed, they were mild and did not significantly compromise the viability of L. sativa seeds. This result is particularly encouraging since the lipolytic enzymes used in the treatment efficiently contribute to the degradation of complex organic compounds without causing significant environmental harm.
On the other hand, the slight decrease in the germination index observed in some treatments may be attributed to the presence of intermediate compounds formed during the enzymatic degradation of lipids, such as short-chain fatty acids. These compounds may present a certain degree of temporary toxicity until they are fully metabolized [
62,
79]. However, this effect has been reported to diminish with increased aeration time in the system or by optimizing the contact time between the enzymatic supernatant and the effluent [
80].
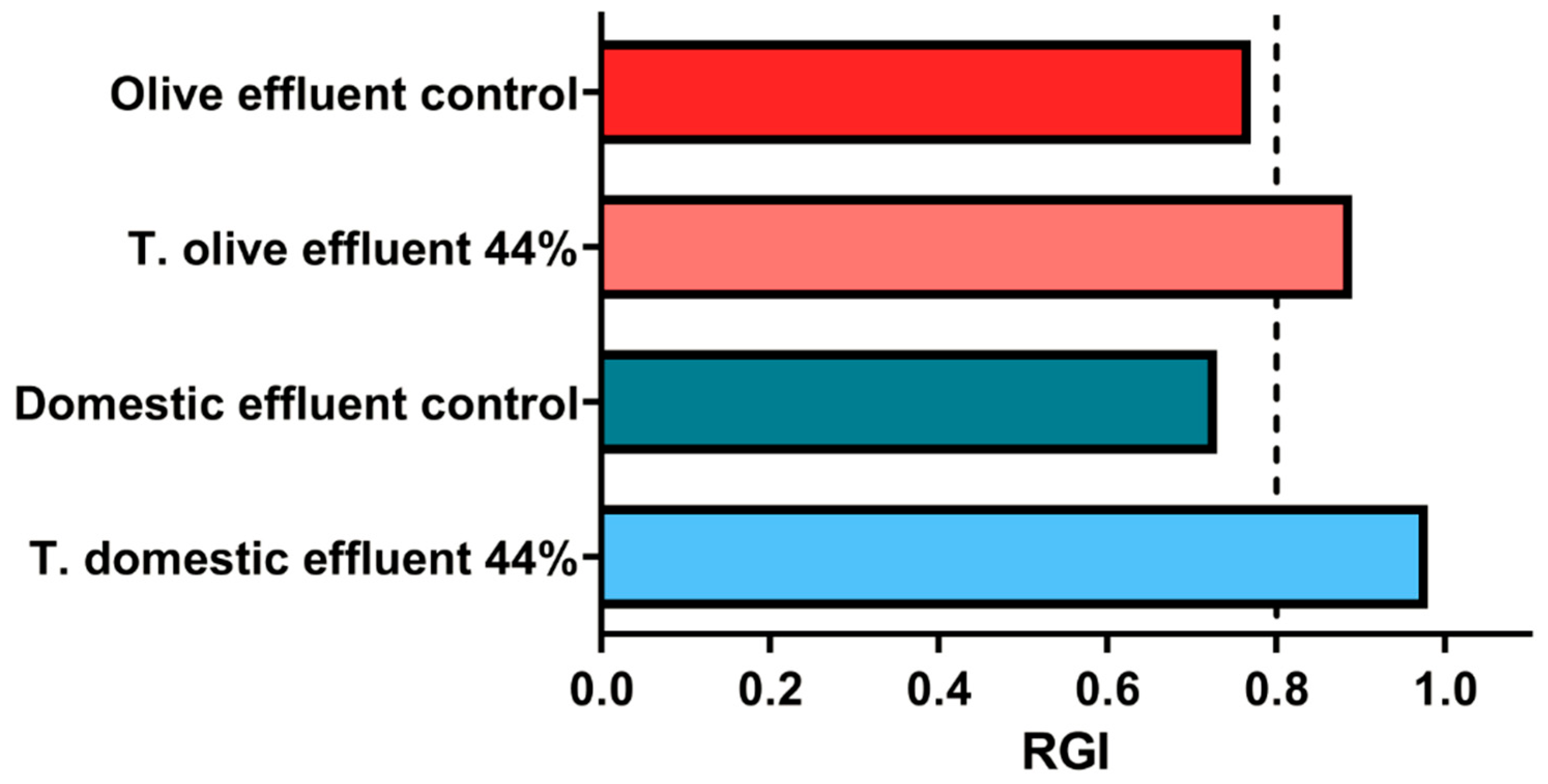
The values obtained from the calculation of the relative growth index (RGI) revealed that the controls showed indices below 0.8, indicating slight inhibition of root elongation in these groups (
Figure 9). This inhibition in the controls can be explained by the initial presence of lipid contaminants in the untreated effluents, which generate adverse conditions for root growth due to reduced oxygen availability and the accumulation of toxic compounds during the decomposition of organic matter [
81].
In contrast, in the treatments with 44% supernatant, RGI values above 0.8 were observed, indicating no significant adverse effects on root elongation (
Figure 9). This positive response is consistent with previous reports indicating that the application of lipolytic enzymes facilitates the degradation of toxic lipid compounds, thereby reducing the accumulation of intermediate products that interfere with plant development [
80].
These results demonstrate that the application of P. rubens LBM 081 supernatant in the treatment of lipid-rich effluents does not generate significant phytotoxic effects during the early stages of plant development. The combination of high contaminant removal efficiency and the absence of severe toxicity reinforces the potential of this biotechnological strategy as a safe alternative for the bioremediation of contaminated wastewater.
4. Future Perspectives
The results obtained in this study provide a solid foundation for the development of sustainable biotechnological strategies for the treatment of lipid-rich wastewater using microbial enzymes. In this context, addressing certain methodological and operational limitations is pertinent, as their analysis in future research will allow progress toward a more efficient implementation of the system at a real scale.
Firstly, it was demonstrated that the direct use of the culture medium supernatant preserved maximum enzymatic activity and confirmed the biocatalyst’s viability under simulated conditions. However, this methodological approach may be impractical for large-scale applications due to the high volume required and the potential additional load of COD, nitrogen, and phosphorus introduced into the system. For this reason, future efforts will focus on the development of solid or concentrated lipase formulations, as well as the design of continuous treatment systems that optimize both catalytic efficiency and the enzyme/effluent volume ratio. In this regard, the evaluation of solid enzymatic preparations—such as crude powdered lipase—also appears promising, as they may offer logistical and operational advantages in terms of storage, transport, and dosing, provided that sufficient catalytic activity is retained after the drying process. Future studies should explore these formulations with the aim of achieving an optimal balance between enzymatic stability, ease of application, and biodegradation efficiency when comparing the liquid and solid forms of crude lipase.
In this sense, evaluating enzyme immobilization technologies is also a priority, as these would allow the reuse of the biocatalyst and greater operational stability, thereby reducing process costs and improving technical feasibility. These strategies would help minimize the proportion observed in this study (44% supernatant to 56% effluent), facilitating broader-scale application.
An important aspect of this work is the recognition of olive oil and meat peptone as effective components in the culture medium, due to their high inductive capacity for lipase production. However, both inputs are high-value food products with growing demand in the global market, which hinders their sustainable implementation on a large scale. Consequently, future stages will prioritize the replacement of these components with fatty residues and nitrogen sources of agro-industrial origin or by-products not suitable for human consumption, thus promoting waste valorization within a circular economy framework.
Moreover, considering the moderate contaminant removal efficiency obtained (≤50%), the application of thermophilic microorganisms is proposed, as their enzymes exhibit higher catalytic activity at elevated temperatures. This approach could not only improve process performance but also allow operation under conditions in which the lower viscosity of lipid residues facilitates their degradation [
82]. In addition, it is recommended to explore the combination of the supernatant with complementary strategies, such as the addition of biodegradable surfactants to enhance fat emulsification or the implementation of bioaugmentation systems with synergistic microorganisms that boost enzymatic activity, promoting microbial synergies that optimize the overall treatment efficiency.
Finally, future research in this line should include kinetic modeling and optimization of the biodegradation system, as well as chronic toxicity assessments using bioindicator organisms, in order to evaluate the actual ecological impact of the proposed treatment. Taken together, these advances will help consolidate the use of fungal lipases as a versatile and sustainable tool for the treatment of high-lipid-load effluents.