Abstract
Numerous treatment modalities have been employed over the years to eradicate bacterial communities in industrial wastewater. Oxidizing agents and chemical additives, such as ozone, permanganate, glutaraldehyde, and chlorine, are effective in treating microbial contaminants that are typically found in domestic wastewater. However, the chemical complexity of water produced from fracking requires novel approaches, because the microbes have developed mechanisms to overcome typical disinfectants. In this work, we test the effectiveness of bacteriophages for the eradication of two model bacteria from produced water: Pseudomonas aeruginosa and Bacillus megaterium. These bacteria were grown in low salinity produced water and exposed to their corresponding phage. Overall, the total inactivation of the P. aeruginosa population was achieved, as well as the inactivation of B. megaterium. These promising results provide a potentially useful tool for bacterial elimination in overall PW treatment, at an industrial scale. Particularly, since phage treatment is a rapid and cost-effective alternative. Moreover, these results fall within the objectives proposed as part of the sustainable development goals adopted worldwide.
1. Introduction
The emergence of shale energy extraction has brought forth economic opportunities and environmental concerns, of which a majority pertain to the management of fresh water and wastewater resources. For instance, in 2017, the Permian Basin, the epicenter of shale energy extraction in the United States, reported an annual water demand of 1322.26 × 106 barrels (bbl) for production well stimulation, as well as an annual produced water volume of 1663.21 × 106 bbl []. Produced water (PW) is the major waste stream generated in oil and gas production. The composition of PW varies significantly according to the location of the well and the corresponding petroliferous strata of interest [,]. These waters are comprised of a wide array of major ions, including sodium, calcium, magnesium, barium, strontium, iron, chloride, and bromide [,,]. Additionally, naturally occurring radioactive material, including radium and uranium, has been detected at several locations [,]. Along with these minerals, volatile organic compounds (VOCs), including xylenes, benzene, and toluene [,,]; total petroleum hydrocarbons []; and other organic compounds contribute to the incredibly complex composition of PW.
These chemicals can provide an ideal environment for the growth and proliferation of different classes of bacteria, many of which can have deleterious effects on unconventional oil and gas development (UOG). These effects range from pipe corrosion and commodity souring to biofilm accumulation and a reduction in the formation’s porosity and permeability []. Microorganisms found in PW cover a wide spectrum, from aerobic to anaerobic bacteria. Examples of these are the sulfate-reducing bacteria Desulfomicrobium, acid-producing bacteria like Halanaerobium, iron-oxidizing bacteria such as Pseudomonas, halotolerant bacteria including Planococcus sp., and the alkali-tolerant bacteria Pannonibacter sp. [,,].
Several treatment modalities can be used in the sanitation of PW. These include the use of oxidizing agents, such as ozone, permanganate, and hydrogen peroxide []. Alternatively, glutaraldehyde, hypochlorite, peracetic acid, and other biocides are widely utilized in the field [,]. The efficacy of the treatment relies upon the permeability of the bacteria’s cell membrane. Santos et al. studied the response of bacteria to the toxic compounds characteristic of contaminated groundwater and PW and observed an increase in the saturated/unsaturated fatty acid ratio, which resulted in a decrease in membrane permeability []. This phenomenon is likely a survival mechanism that bacteria employ when subjected to a contaminated external environment; these processes render the use of traditional biocides less efficacious. These findings provide an impetus for the development of alternative methods of microbial eradication, such as the use of bacteriophages. The use of bacteriophages, or phages, provides a rapid, cost effective, and potentially renewable source of biocide that is devoid of any environmentally noxious chemicals. Phages are a form of virus that infect specific bacteria and convert the cell into a bacteriophage producing factory, before lysing the cell to release the bacteriophage progeny and killing the host in the process. The specificity of the phage attachment, often referred to as a ‘lock and key’ mechanism, primarily depends on the interaction of the phage with receptors found on the bacterial cell surface [,]. Examples of phage treatment implementation are clinical trials for bacterial infections [], use in food animal production [], and food safety [].
Historically, bacteriophages have demonstrated the ability to withstand atypical environments, such as low and elevated temperatures, pH, and salinity. For instance, the viability of phages isolated from hot springs in California (40–90 °C) was tested by Breitbart et al. The phages maintained 75% of their activity when incubated on ice and about 30% of their activity when boiled at 105 °C []. On the other hand, Jepson and March tested phage activity in the range of pH 2–14. Phage viability was only affected at the pH extremes of 11.8–14 and a pH ≤ 2 []. Moreover, Lu et al. found phage isolates in sauerkraut formation tanks (pH < 3.5) after 60 days []. Lastly, bacteriophages were recovered from soil samples in Oklahoma, where salt concentrations vary from 0.3–27% []. Taken as a whole, the specificity of phages, as well as their ability to survive in extreme conditions, support their use as an alternative biocide for PW sanitation.
Currently, methods ranging from physical methods, such as sedimentation and centrifugation; membrane filtration, including micro, nano, and ultrafiltration; to advanced oxidation processes, such as using hydrogen peroxide, are used to deplete microbial communities from PW []. Unfortunately, drawbacks including the size of the particles, operational costs, membrane fouling, byproducts formation, environmental persistence, and many others, are to be considered [,]. This study is the first attempt to evaluate the efficacy of phage treatment in controlling the proliferation of planktonic bacteria in PW, with a view to implementing this technology in above-ground storage tanks (ASTs). This study provides a novel and promising model of PW sanitation, which coupled with conventional polishing techniques, will provide high-quality water that meets the standards established for agriculture, domestic usage, and aquifer discharge.
Here, we present the use of bacteriophages to treat two prominent bacterial species found in produced water: Pseudomonas aeruginosa and Bacillus megaterium. The interaction of each individual bacteria with the corresponding phages were studied, as well as the treatment of B. megaterium with a combination of bacteriophages. These findings suggest that phage treatment can be utilized to simultaneously abate the presence of different bacterial communities in PW. Phage treatment of produced water provides a potentially more rapid, effective, and green alternative to current biocidal treatment modalities used in UOG development.
2. Materials and Methods
2.1. Produced Water Collection and Processing
The wastewater was collected at different steps in the treatment process at the Permian Basin (TX, USA), using sterile 500 mL capped bottles. The samples were filter sterilized through 0.2 µm Nalgene syringe filters (Thermo-Fisher Scientific, Rochester, NY, USA) and stored at 4 °C.
2.2. Model Strains and Culture Conditions
The Pseudomonas aeruginosa strain HER-1018 was obtained from ATCC (ATCC BAA-47). The Bacillus megaterium isolate was kindly provided by Mei Liu (Center for Phage Technology, College Station, TX, USA). Both hosts were cultured in tryptic soy (TS) broth (Corning, Christiansburg, VA, USA) for 7 ± 1 h, at 37 °C, in a corning LSE benchtop shaking incubator (Corning, Corning, NY, USA) and glycerol stocks (Sigma Aldrich, Saint Louis, MO, USA) were made, as required.
2.3. Bacteriophage Growth Conditions
The P. aeruginosa bacteriophage SN (NCBI accession no. NC_011756) was acquired from a research-based collaboration with Vadim Mesyanzhinov and the B. megaterium bacteriophages Slash (GenBank accession no. KF669661) and Palmer (GenBank accession no. KP411017) were donated by Mei Liu (Center for Phage Technology, College Station, TX, USA). P. aeruginosa cells were cultured aerobically with shaking (230 rpm) in tryptic soy broth at 37 °C. After 7 h (OD600 of 0.8 AU), 20 mL of the culture was infected with 500 µL of Pseudomonas bacteriophage SN (5.4 × 106 PFU/mL) and left to incubate for 8 h. The lysate was then centrifuged (Beckman coulter model Avanti j-E, Indianapolis, IN, USA) at 4 °C (10,000 rpm, 15 min) and filtrated through Nalgene syringe filters (Thermo-Fisher Scientific, Rochester, NY, USA) for sterilization. Titers for B. megaterium phages, Slash (3.4 × 104 PFU/mL) and Palmer (1.4 × 104 PFU/mL), were increased as described above. After filter sterilization, the filtrate was transferred to centrifugal filter units (10,000 MWCO; Millipore, Saint Louis, MO, USA) and centrifuged for 10 min at 6000 rpm. The remaining retentate was collected, mixed with equal parts of salt–magnesium (SM) buffer (NaCl 100 mM, MgSO4∙7H2O 8mM, Tris-HCl 50 mM, and gelatin 2% w/v) (all purchased from Sigma Aldrich, St. Louis, MO, USA), and stored at 4 °C.
2.4. Spot Assay and Bacteriophage Titer Determination
The initial screening was performed via spot tests []. The spot tests were performed on 1% tryptic soy agar (TSA) (Sigma Aldrich, MO, USA) plates, overlaid with 3 mL of soft nutrient agar (0.52% KCl, 3% TS broth, 0.6% agarose) (all purchased from Sigma Aldrich, St. Louis, MO, USA), inoculated with the host bacteria. Serial dilutions of phage stock were made, and 5 μL was put on six spots around the agar plate after the host-containing soft nutrient agar solidified.
Phage titer quantification was performed via plaque assays, using seven 1:10 serial dilutions of the phage in the SM buffer. For this, the host cells were cultured, as previously described, to an OD600 of 0.2 absorbance units (AU). CaCl2 (Sigma Aldrich, St. Louis, MO, USA) was added to the culture to a final concentration of 20 mM CaCl2. Then, 50 μL of the bacterial culture and 100 μL of the phage dilution were added into a conical tube containing 3 mL of nutrient agar (0.52% KCl, 3% TS broth, 0.6% agarose). The mixture was then poured onto 1% agarose TS plates (Sigma Aldrich, St. Louis, MO, USA), ensuring the surface was covered. The plates were then left to incubate overnight.
2.5. Strain Growth Curves in Produced Water
The growth curve of the P. aeruginosa and B. megaterium cells in produced water were studied by inoculating 20 mL of sterilized low-salinity PW with 500 microliters of P. aeruginosa and B. megaterium stocks, separately. Two hundred microliters of the infected water aliquot were transferred to 96-wells plate (Corning, Corning, NY, USA) and incubated aerobically by shaking (180 rpm) at 37 °C for 24 h. Absorbance measurements (OD600) were taken at 60 min intervals using a Varioskan Lux (Thermo-Fisher Scientific, Rochester, NY, USA). Three independent experiments were performed for each host cell.
2.6. Individual Phage Treatment Assays
The efficacy of the phage treatment was studied using both bacterial models and the corresponding phages. For P. aeruginosa, 500 μL of the host stock was mixed with 500 μL of P. aeruginosa phage SN, in 20 mL of the sterilized produced water sample. For each B. megaterium assay, 250 μL of the B. megaterium host was mixed with 250 μL of the slash phage, in 20 mL of the sterilized produced water sample. The same protocol was followed for the Palmer phages. For each assay, two controls were included, namely the positive bacterial control and the negative phage control, both in PW. All the assays were incubated aerobically via shaking (180 rpm) at 37 °C for 24 h and absorbance measurements were taken at 60 min intervals.
2.7. Phage Cocktail Assay Involving B. megaterium sp.
The efficacy of the phage cocktail was studied using the B. megaterium strain and the corresponding phages. Moreover, 500 μL of B. megaterium cells were mixed with 500 μL of the Slash phage and 500 μL of the Palmer phage, in 20 mL of the sterilized produced water sample. For each assay, two controls were included, namely the positive bacterial control and the negative phage control, both in PW. The mixtures were incubated aerobically by shaking (180 rpm) at 37 °C for 24 h and 100 μL aliquots were plated on 1% TSA at the following times: 0 h, 4 h, 12 h and 24 h.
2.8. Statistical Analysis
Statistical analysis was performed using Origin (Origin (pro), version 2022). The results obtained for the controls and assays were corrected with the blanks for each point of the replicates. Significance was assessed using a two-way two-sample t-test and non-parametric Wilcoxon test (p < 0.05). Standard deviation was calculated from the triplicates and plotted accordingly.
3. Results and Discussion
3.1. Selection of Model Organisms
A wide variety of bacteria can be found in produced water; however, for this initial study we pursued the use of P. aeruginosa due to its abundance in PW, as well as B. megaterium, both with negative impacts on the UOG []. For example, microbial-induced corrosion is caused by bacterial biofilms on metal surfaces. Previous studies have reported stainless steel corrosion induced by P. aeruginosa, an iron-oxidizing bacterium, which represents a serious problem for the longevity of pipelines and other metal-based infrastructure in the oil and gas industry []. On the other hand, B. megaterium has been demonstrated to be efficient in the decomposition of hydrocarbons from crude oil []. The experiments reproduced AST conditions, where the bacteria are in a planktonic state and phage treatment can be utilized. In ASTs, water goes in and out intermittently, lowering the chances of biofilm formation.
It is also worth mentioning that P. aeruginosa and B. megaterium were selected as model organisms for this study based on the commercial availability of their corresponding phage partners. The availability of phages currently limits their broad application for phage treatment of PW, particularly when more specialized extremophilic and halotolerant bacteria are found in PW.
In all proceeding experiments, bacteria models were cultured in sterilized low-salt PW (<1000 mg/L; pH = 8.98) from the Permian Basin, where healthy colonies were observed after overnight incubation. The phages were also viable after incubation in PW, demonstrating their higher resilience to adverse environmental conditions. Additionally, the multiplicity of infection (MOI), the ratio of phage particles to the number of host cells, is 10 unless specified.
3.2. Produced Water Processing and Growth Assays
The growth curve of the B. megaterium and P. aeruginosa cells in low-salinity sterilized PW revealed that healthy colonies can proliferate in these water samples during the tested period (48 h). An OD600 ranging from 0.01–0.35 AU was observed in the host cultures during the first 26 h (Figure 1). Afterwards, the OD600 slowly decreased until the 36th hour and more rapidly up to the 48th hour. Precipitation of cellular debris was observed with the decrease in OD600 suggesting cell death, probably due to insufficient nutrients in the water sample.
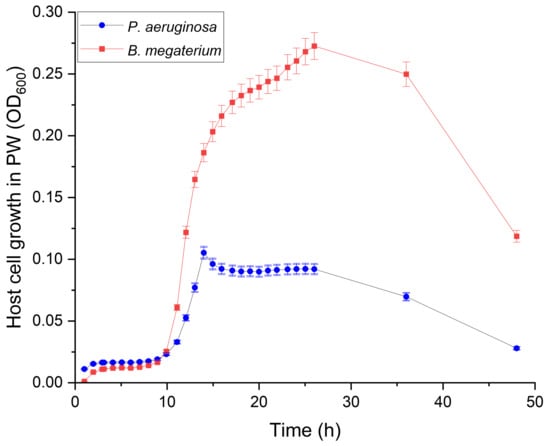
Figure 1.
The growth curves of P. aeruginosa HER-1018 strain and B. megaterium sp. cell growth in sterilized low-salinity produced water during 48 h of incubation at 37 °C. The error bars represent the standard deviations for the triplicates.
3.3. Phage Treatment Assays
3.3.1. Single Phage Treatment Involving P. aeruginosa
The spot test results provided an estimate of the dilution that should be employed in the phage titration. After titration, the phage titers were 1.2 × 109 PFU/mL for P. aeruginosa phage SN, Slash 1.6 × 109 PFU/mL for B. megaterium phage “Slash”, and 2.7 × 109 PFU/mL for B. megaterium phage “Palmer”. These phages were employed in the subsequent experiments.
The impact of the specific lytic phage SN on the growth of the P. aeruginosa strain HER-1018 in PW after 24 h of incubation at 37 °C is illustrated in Figure 2. Complete inhibition of P. aeruginosa growth is evident when the phage is co-incubated with the bacterium, contrasting with the normal growth observed in the absence of the phage. A noticeable sharp increase in P. aeruginosa colonies is observed in the control, indicating uninhibited bacterial growth. However, the addition of the phage treatment effectively hinders the growth of the host population, leading to the complete eradication of P. aeruginosa after 24 h of treatment. The Wilcoxon sign rank test confirmed the inhibitory effect of P. aeruginosa bacteriophages on the growth of the host cell. The median, post-phage treatment was significantly lower than the control (Z = −4.4447, p < 0.05) (see Table S1).
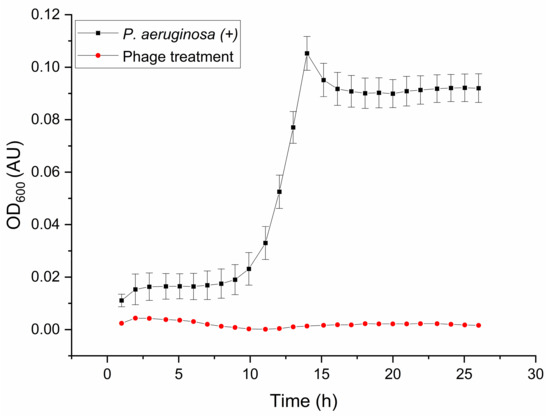
Figure 2.
Effect of P. aeruginosa phage SN treatment on the host cells grown in low-salinity PW, MOI = 10. A comparison between a normal growth curve of P. aeruginosa HER-1018 strain and a growth curve after inoculation with P. aeruginosa phage SN in sterilized low-salinity PW during 26 h of incubation at 37 °C. The error bars represent the standard deviations for the triplicates.
The interaction between P. aeruginosa and its phages in aquatic environments has been studied for decades. In 1991, KokJohn and Sayler concluded that phages play a significant role in the dynamic equilibrium of bacteria in natural aquatic ecosystems and could potentially be used to control the density and genetic diversity of the bacterial population []. Bacteriophages have been successful at reducing 50% of P. aeruginosa cells from anthracite and 99.9% from granular-activated charcoal clean wastewater filters, although bacteria removal efficiency decreased when biofilm was present in the filters []. Further studies have shown that the addition of disinfectants, such as chlorine, to the phage treatment is an effective method for the removal of pre-existing P. aeruginosa biofilms []. Recently, phages were used to treat P. aeruginosa biofilms, proving to be efficient at limiting the spread of biofilm and reducing planktonic bacteria by up to 90% of the original population []. Phage cocktails have been employed for P. aeruginosa biocontrol in water, demonstrating higher capabilities than single phage treatment at reducing the host population [] The results in this study certainly display that planktonic P. aeruginosa population growth in PW can be inactivated when co-incubated with the phage. Phage resistance was not observed within the duration of the experiments. Although bacteria may modify the receptor sites to overcome phage infection [], the composition of PW is a challenging environment for the host cells and may compromise the phage-resistance mutation efforts as the bacterium trades fitness over resistance acquisition [].
3.3.2. Single Phage Treatment Involving B. megaterium
The response of the B. megaterium host to the specific lytic phages, Slash and Palmer, in PW after 24 h of incubation at 37 °C is shown in Figure 3. The absorbance measurements of B. megaterium growth co-incubated with the Slash phage show inhibition of the host growth until the 8 h mark, when an increase in the OD600 values is observed. Interestingly, the absorbance quickly drops after 12 h of interaction with the phage, contrasting the increasing rate of the singly incubated host. A similar behavior is observed with B. megaterium when treated with the Palmer phage. These results indicate inactivation of the host cells. The statistical t-test for two independent samples corroborated that the Slash bacteriophage treatment (M = 0.2591, SD = 0.3914) does limit B. megaterium cell growth (M = 0.6238, SD = 0.6287) in PW, n = 19, t = 2.1471, p < 0.05 (see Table S2). Similarly, the Palmer phage treatment (M = 0.1541, SD = 0.2004) was also statistically significant in regard to the inactivation of B. megaterium cells (M = 0.6238, SD = 0.6287) in PW, n = 19, t = 3.1031, p > 0.05 (see Table S3).
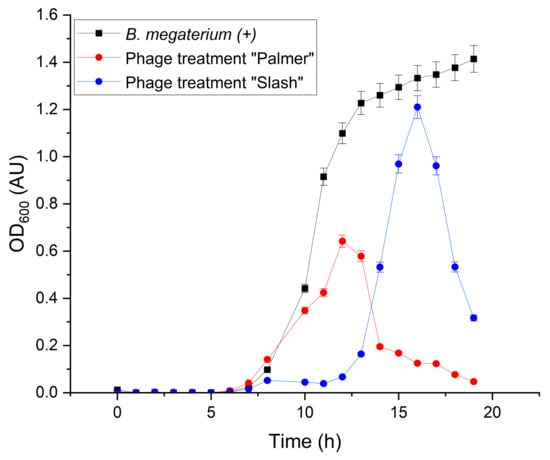
Figure 3.
Effect of the individual B. megaterium phage treatments (Slash and Palmer) on the host cell growth in sterilized low salinity PW, MOI = 10. A comparison between a normal growth curve of B. megaterium sp. and a growth curve after individual inoculation with the B. megaterium Slash and Palmer phages in low-salinity PW during 24 h of incubation at 37 °C. The error bars represent the standard deviations for the triplicates.
The previous results suggest that B. megaterium growth in PW could be controlled by the utilization of phage treatment. The delayed inactivation of the host observed in the Palmer treatment compared to the Slash treatment may be caused by the difference in phage classification. Palmer belongs to the Podoviridae family, whilst Slash is a Siphoviridae bacteriophage [,]. Podophages and Siphophages have distinct assembly pathways []. Additionally, phage infection relies on attachment to the host, an appropriate chemical, and a suitable environment, as well as complementary interactions with receptors on the host []. To our knowledge, there is not much available research involving B. megaterium bacteriophages for phage treatment in environmental applications.
3.3.3. Phage Cocktail Treatment Involving B. megaterium
To further demonstrate the efficacy of phages in eradicating bacteria from PW, the host’s ability to overcome phage infection was assessed by means of colony growth. The treatment of B. megaterium with a cocktail made of Palmer and Slash bacteriophages is illustrated in Figure 4. Host cell population growth is controlled by the phage cocktail, with maximum growth observed at 12 h, after which there is a significant decrease in the colonies. The inhibitory effect of the phage cocktail treatment (M = 0.97, SD = 1.27) on B. megaterium in PW (M = 4.18, SD = 2.25) was confirmed with a t-test for two independent samples, n = 4, t = 2.48, p < 0.05 (see Table S4).
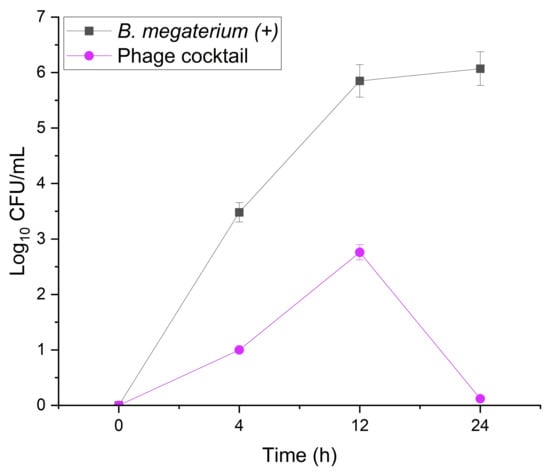
Figure 4.
Effect of the phage cocktail treatment (Slash and Palmer) on the host cell growth in sterilized low-salinity PW. Comparison between normal growth of B. megaterium sp. control against growth after inoculation with cocktail in low-salinity PW during 24 h of incubation at 37 °C.
Previous studies have provided evidence that phage treatment is a tool for limiting bacterial colonization []. Nale et al. have also concluded that the effectiveness of a particular phage relies on the sensitivity of the host. Sadeqi et al. achieved the reduction of eight harmful antibiotic resistant bacteria in hospital wastewater with a phage cocktail [], minimizing the risk of surface water cross-contamination. They stress the use of phage cocktails to prevent phage resistance. Phage treatment has been successful in reducing bacterial infection in Atlantic Salmon, without affecting the microbiota of the water []. These are important results as membrane bioreactors and other biological treatments are wastewater management techniques that are growing in popularity, particularly in regard to PW []. One of the main challenges of phage treatment in the unconventional oil and gas industry is the variable biogeochemical composition of the wastewater. An all-purpose phage cocktail is not up for discussion just yet, as more in-depth research is needed for industrial applications. Quality indicators, such as pH, temperature, biological oxygen demand, chemical oxygen demand, and microbial population, are to be considered in each phage treatment case []. Nevertheless, our results clearly demonstrate that P. aeruginosa is inactivated completely in PW. Additionally, our data shows the inhibition on B. megaterium growth in both single and cocktail phage applications. One of the contributions of the present work is that all the experiments were performed considering the intended industrial conditions of planktonic cell elimination from partially treated produced water. Further efforts will focus on the creation of a phage bank for the application in PW. Other outlooks in regard to the application of phage treatment in PW point toward genome engineering, as well as the combination of biocides with phages to maximize microbial reduction from wastewater [].
One of the transformations contemplated within the sustainable development goals is water accessibility, which includes water pollution and management. With an expected increase in water demand of 50% by the end of 2030 [], research like ours is necessary to continuously aid in the development of technologies and protocols that satisfy the global water demand in the next few decades. Treating different effluents for reuse in agriculture and by livestock is the most environmentally friendly approach to managing industrial waste. In matrices like produced water, containing a complex chemical and biological composition, every little step is significant. Particularly, when the estimated global volumes produced are 158,900 million barrels per day [].
4. Conclusions
As bacteria change their membrane composition to alter their permeability to the external environment, considerable challenges for traditional treatment modalities used in wastewater sanitation can be expected. This phenomenon effectively drives an ‘arms race’ between the bacteria and biocidal compounds, whereby more and more exogenous chemicals are required to eradicate the unwanted bacteria, which can have potentially deleterious effects on the peripheral ecosystem. Bacteriophage treatment offers a rapid and cost-effective alternative to this developed resistance, as phages operate with a high degree of specificity and efficacy. Additionally, phages have the robustness to withstand the PW environment, which can contain elevated levels of dissolved and suspended solids, a multitude of various metal ions, a myriad of hydrocarbons and chemical additives, and an extreme pH. However, the potential systemic application of phage treatment is plagued by the limited amount of commercially available specimens; thus, larger phage libraries are required to more comprehensively treat the breadth of microorganisms found in PW. Moreover, further characterization of bacteria/phage partners following the results presented here is required, as not all will behave as those examined in this study. The study of phage-resistance mechanisms demonstrated by differential bacterial species is a critical research topic, with potential widespread implications for various commercial and industrial applications.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/w16060797/s1, Table S1: Effect of P. aeruginosa phage SN on the P. aeruginosa strain HER-1018 population in low-salinity PW using Wilcoxon sign-rank test (n = 26); Table S2: Effect of B. megaterium phage Slash on the B. megaterium sp. population in low-salinity PW using a t-test for two independent samples (n = 19); Table S3: Effect of B. megaterium phage Palmer on the B. megaterium sp. population in low-salinity PW using a t-test for two independent samples (n = 19); Table S4: Effect of B. megaterium phage cocktail on the B. megaterium sp. population in low-salinity PW using a t-test for two independent samples (n = 4).
Author Contributions
Conceptualization, Z.L.H., R.A.B. and R.S.-R.; methodology, Z.L.H., R.A.B., R.S.-R. and K.A.S.; validation, R.S.-R., Z.L.H. and R.A.B.; formal analysis, R.S.-R., J.G. and V.R.; investigation, R.S.-R. and J.G.; resources, Z.L.H. and K.A.S.; data curation, R.S.-R. and J.G.; writing—original draft preparation, R.S.-R. and J.G.; writing—review and editing, Z.L.H., R.A.B. and K.A.S.; visualization, R.S.-R.; supervision, Z.L.H., R.A.B. and K.A.S.; project administration, Z.L.H. and K.A.S.; funding acquisition, Z.L.H. and K.A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Biota Solutions.
Data Availability Statement
Data are contained within the article and supplementary materials.
Acknowledgments
Support for this work was provided by Biota Solutions, Challenger Water Solutions, and Chevron Technical Ventures. We would also like to thank Aris Water for supplying the samples of treated produced water.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Scanlon, B.R.; Reedy, R.C.; Xu, P.; Engle, M.; Nicot, J.P.; Yoxtheimer, D.; Yang, Q.; Ikonnikova, S. Can We Beneficially Reuse Produced Water from Oil and Gas Extraction in the U.S.? Sci. Total Environ. 2020, 717, 137085. [Google Scholar] [CrossRef]
- Al-Ghouti, M.A.; Al-Kaabi, M.A.; Ashfaq, M.Y.; Da’na, D.A. Produced Water Characteristics, Treatment and Reuse: A Review. J. Water Process. Eng. 2019, 28, 222–239. [Google Scholar] [CrossRef]
- Igunnu, E.T.; Chen, G.Z. Produced Water Treatment Technologies. Int. J. Low-Carbon Technol. 2014, 9, 157–177. [Google Scholar] [CrossRef]
- Rodriguez, A.Z.; Wang, H.; Hu, L.; Zhang, Y.; Xu, P. Treatment of produced water in the permian basin for hydraulic fracturing: Comparison of different coagulation processes and innovative filter media. Water 2020, 12, 770. [Google Scholar] [CrossRef]
- Xiao, F. Characterization and Treatment of Bakken Oilfield Produced Water as a Potential Source of Value-Added Elements. Sci. Total Environ. 2021, 770, 145283. [Google Scholar] [CrossRef] [PubMed]
- Barbot, E.; Vidic, N.S.; Gregory, K.B.; Vidic, R.D. Spatial and Temporal Correlation of Water Quality Parameters of Produced Waters from Devonian-Age Shale Following Hydraulic Fracturing. Environ. Sci. Technol. 2015, 47, 41–59. [Google Scholar] [CrossRef]
- Lewandowski, C.M.; Co-investigator, N.; Lewandowski, C.M. Naturally Occurring Radioactive Materials (NORM) in Produced Water and Scale from Texas Oil, Gas, and Geothermal Wells: Geographic, Geologic, and Geochemical Controls. Vr. Lanscapes Texas 2015, 1, 1689–1699. [Google Scholar]
- McMahon, P.B.; Galloway, J.M.; Hunt, A.G.; Belitz, K.; Jurgens, B.C.; Johnson, T.D. Geochemistry and Age of Groundwater in the Williston Basin, USA: Assessing Potential Effects of Shale-Oil Production on Groundwater Quality. Appl. Geochem. 2021, 125, 104833. [Google Scholar] [CrossRef]
- Varona-Torres, E.; Carlton, D.D.; Hildenbrand, Z.L.; Schug, K.A. Matrix-Effect-Free Determination of BTEX in Variable Soil Compositions Using Room Temperature Ionic Liquid Co-Solvents in Static Headspace Gas Chromatography Mass Spectrometry. Anal Chim Acta 2018, 1021, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Hildenbrand, Z.L.; Santos, I.C.; Liden, T.; Carlton, D.D.; Varona-Torres, E.; Martin, M.S.; Reyes, M.L.; Mulla, S.R.; Schug, K.A. Characterizing Variable Biogeochemical Changes during the Treatment of Produced Oilfield Waste. Sci. Total Environ. 2018, 634, 1519–1529. [Google Scholar] [CrossRef] [PubMed]
- Abass, O.K.; Zhuo, M.; Zhang, K. Concomitant Degradation of Complex Organics and Metals Recovery from Fracking Wastewater: Roles of Nano Zerovalent Iron Initiated Oxidation and Adsorption. Chem. Eng. J. 2017, 328, 159–171. [Google Scholar] [CrossRef]
- De Oliveira, E.S.D.; Roseana, F.; Pereira, C.; Alice, M.; Lima, G.D.A. Study on Biofilm Forming Microorganisms Associated with the Biocorrosion of X80 Pipeline Steel in Produced Water from Oilfield. Mater. Res. 2021, 24. [Google Scholar] [CrossRef]
- Mohan, A.M.; Bibby, K.J.; Lipus, D.; Hammack, R.W.; Gregory, K.B. The Functional Potential of Microbial Communities in Hydraulic Fracturing Source Water and Produced Water from Natural Gas Extraction Characterized by Metagenomic Sequencing. PLoS ONE 2014, 9, e107682. [Google Scholar] [CrossRef] [PubMed]
- Kahrilas, G.A.; Blotevogel, J.; Stewart, P.S.; Borch, T. Biocides in Hydraulic Fracturing Fluids: A Critical Review of Their Usage, Mobility, Degradation, and Toxicity. Environ. Sci. Technol. 2015, 49, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Akyon, B.; Lipus, D.; Bibby, K. Glutaraldehyde Inhibits Biological Treatment of Organic Additives in Hydraulic Fracturing Produced Water. Sci. Total Environ. 2019, 666, 1161–1168. [Google Scholar] [CrossRef]
- Santos, I.C.; Chaumette, A.; Smuts, J.; Hildenbrand, Z.L.; Schug, K.A. Analysis of Bacteria Stress Responses to Contaminants Derived from Shale Energy Extraction. Environ. Sci. Process Impacts 2019, 21, 269–278. [Google Scholar] [CrossRef]
- Sinha, S.; Grewal, R.K.; Roy, S. Modeling Bacteria–Phage Interactions and Its Implications for Phage Therapy; Elsevier Inc.: Amsterdam, The Netherlands, 2018; Volume 103. [Google Scholar]
- Burrowes, B.H.; Abedon, S.T.; Burrowes, B.H.; McConville, M.L.; Harper, D.R. Bacteriophages: Biology, Technology; Springer: Berlin/Heidelberg, Germany, 2018; ISBN 9783319419855. [Google Scholar]
- Sahota, J.S.; Smith, C.M.; Radhakrishnan, P.; Winstanley, C.; Goderdzishvili, M.; Chanishvili, N.; Kadioglu, A.; O’Callaghan, C.; Clokie, M.R.J. Bacteriophage Delivery by Nebulization and Efficacy Against Phenotypically Diverse Pseudomonas Aeruginosa from Cystic Fibrosis Patients. J. Aerosol Med. Pulm. Drug Deliv. 2015, 28, 353–360. [Google Scholar] [CrossRef]
- Klopatek, S.; Callaway, T.R.; Wickersham, T.; Sheridan, T.G.; Nisbet, D.J. Bacteriophage Utilization in Animal Hygiene. In Bacteriophages; Springer: Cham, Switzerland, 2018; pp. 1–28. [Google Scholar] [CrossRef]
- Fieseler, L.; Loessner, M.J.; Hagens, S. Bacteriophages and Food Safety. In Protective Cultures, Antimicrobial Metabolites and Bacteriophages for Food and Beverage Biopreservation; Woodhead Publishing: Sawston, UK, 2011; pp. 161–178. [Google Scholar]
- Breitbart, M.; Wegley, L.; Leeds, S.; Schoenfeld, T.; Rohwer, F. Phage Community Dynamics in Hot Springs. Appl. Environ. Microbiol. 2004, 70, 1633–1640. [Google Scholar] [CrossRef]
- Jepson, C.D.; March, J.B. Bacteriophage Lambda Is a Highly Stable DNA Vaccine Delivery Vehicle. Vaccine 2004, 22, 2413–2419. [Google Scholar] [CrossRef]
- Lu, Z.; Breidt, F.; Plengvidhya, V.; Fleming, H.P. Bacteriophage Ecology in Commercial Sauerkraut Fermentations. Appl. Environ. Microbiol. 2003, 69, 3192–3202. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.; Caton, T.M.; Buchheim, J.A.; Buchheim, M.A.; Schneegurt, M.A.; Miller, R.V. DNA-Repair Potential of Halomonas spp. from the Salt Plains Microbial Observatory of Oklahoma. Microb. Ecol. 2004, 48, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Rosario, R.; Hildenbrand, Z.L. Produced Water Treatment and Valorization: A Techno-Economical Review. Energies 2022, 15, 4619. [Google Scholar] [CrossRef]
- Jiménez, S.; Micó, M.M.; Arnaldos, M.; Medina, F.; Contreras, S. State of the Art of Produced Water Treatment. Chemosphere 2018, 192, 186–208. [Google Scholar] [CrossRef] [PubMed]
- Chibani-Chennoufi, S.; Sidoti, J.; Bruttin, A.; Kutter, E.; Sarker, S.; Brüssow, H. In Vitro and in Vivo Bacteriolytic Activities of Escherichia Coli Phages: Implications for Phage Therapy. Antimicrob. Agents Chemother. 2004, 48, 2558–2569. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Yang, D.; Xu, D.; Gu, T. Anaerobic Corrosion of 304 Stainless Steel Caused by the Pseudomonas Aeruginosa Biofilm. Front. Microbiol. 2017, 8, 2335. [Google Scholar] [CrossRef] [PubMed]
- Alnuaimi, M.T.; Taher, T.A.; Aljanabi, Z.Z.; Adel, M.M. High-Resolution Gc/Ms Study of Biodegradation of Crude Oil by Bacillus Megaterium. Res. Crops 2020, 21, 650–657. [Google Scholar] [CrossRef]
- Kokjohn, T.A.; Sayler, G. Attachment and Replication of Pseudomonas Aeruginosa Bacteriophages under Conditions Simulating Aquatic Environments. Microbiology 1991, 137, 661–666. [Google Scholar] [CrossRef]
- Zhang, Y.; Hunt, H.K.; Hu, Z. Application of Bacteriophages to Selectively Remove Pseudomonas Aeruginosa in Water and Wastewater Filtrationsystems. Water Res. 2013, 47, 4507–4518. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Z. Combined Treatment of Pseudomonas Aeruginosa Biofilms with Bacteriophages and Chlorine. Biotechnol. Bioeng. 2013, 110, 286–295. [Google Scholar] [CrossRef]
- Magin, V.; Garrec, N.; Andrés, Y. Selection of Bacteriophages to Control in Vitro 24 h Old Biofilm of Pseudomonas Aeruginosa Isolated from Drinking and Thermal Water. Viruses 2019, 11, 749. [Google Scholar] [CrossRef]
- Kauppinen, A.; Siponen, S.; Pitkänen, T.; Holmfeldt, K.; Pursiainen, A.; Torvinen, E.; Miettinen, I.T. Phage Biocontrol of Pseudomonas Aeruginosa in Water. Viruses 2021, 13, 928. [Google Scholar] [CrossRef] [PubMed]
- Lenski, R.E.; Levin, B.R. Constraints on the coevolution of bacteria and virulent phage: A model, some experiments, and predictions for natural communities. Am. Nat. 1985, 125, 585–602. [Google Scholar] [CrossRef]
- Oechslin, F. Resistance Development to Bacteriophages Occurring during Bacteriophage Therapy. Viruses 2018, 10, 351. [Google Scholar] [CrossRef] [PubMed]
- DeCrescenzo, A.J.; Ritter, M.A.; Chamakura, K.R.; Kuty Everett, G.F. Complete Genome of Bacillus Megaterium Siphophage Slash. Genome Announc. 2013, 1, e00862-13. [Google Scholar] [CrossRef]
- Hargrove, E.C.; Lopez, M.S.; Hernandez, A.C.; Everett, G.F.K. Complete Genome Sequence of Bacillus Megaterium Podophage Palmer. Genome Announc. 2015, 3, e00358-15. [Google Scholar] [CrossRef]
- Huang, Y.; Sun, H.; Wei, S.; Cai, L.; Liu, L.; Jiang, Y.; Xin, J.; Chen, Z.; Que, Y.; Kong, Z.; et al. Structure and Proposed DNA Delivery Mechanism of a Marine Roseophage. Nat. Commun. 2023, 14, 3609. [Google Scholar] [CrossRef]
- Dennehy, J.J.; Abedon, S.T. Adsorption: Phage Acquisition of Bacteria. In Bacteriophages; Springer: Cham, Switzerland, 2021; pp. 93–117. [Google Scholar] [CrossRef]
- Nale, J.Y.; Spencer, J.; Hargreaves, K.R.; Trzepiński, P.; Douce, G.R.; Clokie, M.R.J. Bacteriophage Combinations Significantly Reduce Clostridium Difficile Growth in Vitro and Proliferation in Vivo. Antimicrob. Agents Chemother. 2016, 60, 968–981. [Google Scholar] [CrossRef] [PubMed]
- Sadeqi, S.; Shahraki, A.H.; Nikkhahi, F.; Javadi, A.; Mahmoud, S.; Marashi, A. Application of Bacteriophage Cocktails for Reducing the Bacterial Load of Nosocomial Pathogens in Hospital Wastewater. Iran. J. Microbiol. 2022, 14, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, A.W.; Gundersen, M.S.; Vo, T.P.; Almaas, E.; Vadstein, O.; Bakke, I. Phage Therapy Minimally Affects the Water Microbiota in an Atlantic Salmon (Salmo Salar) Rearing System While Still Preventing Infection. Sci. Rep. 2023, 13, 19145. [Google Scholar] [CrossRef]
- Abujayyab, M.A.; Hamouda, M.; Aly Hassan, A. Biological Treatment of Produced Water: A Comprehensive Review and Metadata Analysis. J. Pet. Sci. Eng. 2022, 209, 109914. [Google Scholar] [CrossRef]
- Beheshti Maal, K.; Delfan, A.S.; Salmanizadeh, S. Isolation and Identification of Two Novel Escherichia Coli Bacteriophages and Their Application in Wastewater Treatment and Coliform’s Phage Therapy. Jundishapur J. Microbiol. 2015, 8, e14945. [Google Scholar] [CrossRef]
- Mathieu, J.; Yu, P.; Zuo, P.; Da Silva, M.L.B.; Alvarez, P.J.J. Going Viral: Emerging Opportunities for Phage-Based Bacterial Control in Water Treatment and Reuse. Acc. Chem. Res. 2019, 52, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Sachs, J.D.; Schmidt-Traub, G.; Mazzucato, M.; Messner, D.; Nakicenovic, N.; Rockström, J. Six Transformations to Achieve the Sustainable Development Goals. Nat. Sustain. 2019, 2, 805–814. [Google Scholar] [CrossRef]
- Amakiri, K.T.; Ogolo, N.A.; Angelis-Dimakis, A.; Albert, O. Physicochemical Assessment and Treatment of Produced Water: A Case Study in Niger Delta Nigeria. Pet. Res. 2023, 8, 87–95. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).