Metagenomic Meta-Analysis of Antibiotic-Resistance Genes in Wastewater: A Perspective from the COVID-19 Pandemic
Abstract
:1. Introduction
2. Materials and Methods
2.1. Systematic Review and Metagenomic Data Retrieval
2.2. Metagenomic Sequences Pre-Processing
2.3. Identification and Abundance Estimation of ARGs
2.4. Statistical Analysis
3. Results
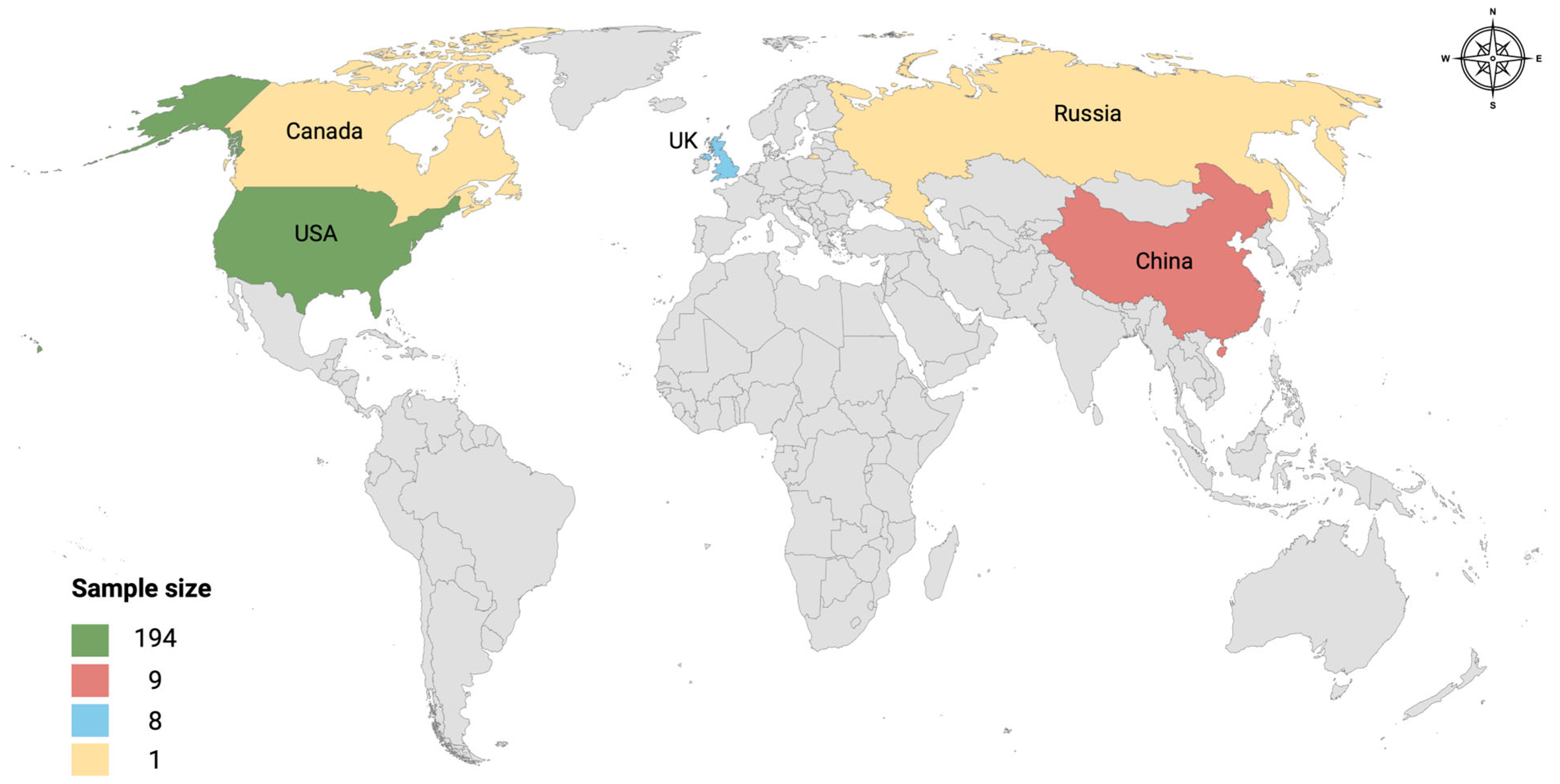
3.1. Retrieved Metagenomic Data
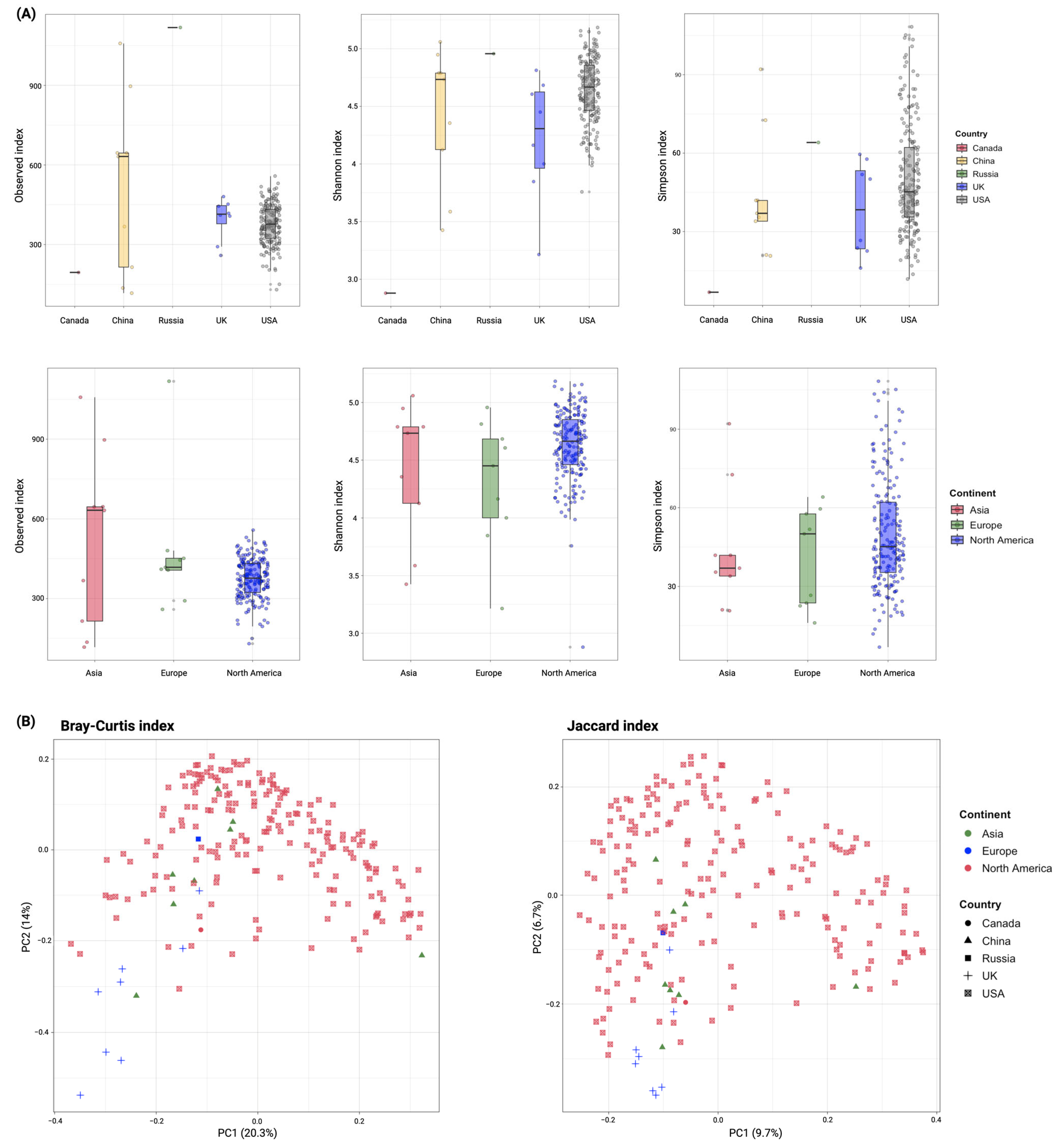
3.2. Diversity of ARGs in Global Wastewater
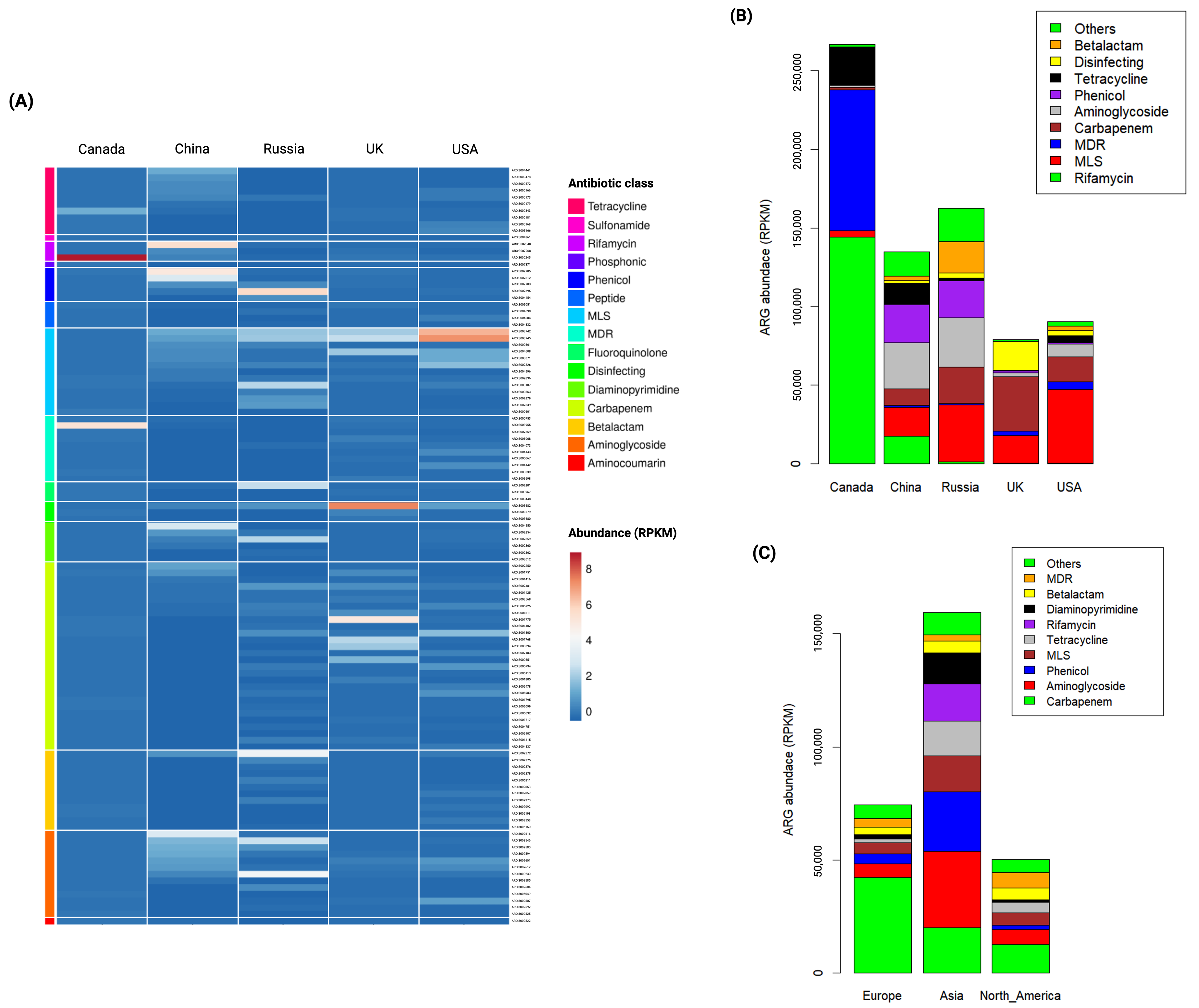
3.3. Abundance of ARGs in Global Wastewater
3.4. Country-Specific Indicator ARGs in Global Wastewater
3.5. Continent-Specific Indicator ARGs in Global Wastewater
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Russia | Canada | UK | China | ||||
|---|---|---|---|---|---|---|---|
| ARG | p-Value * | ARG | p-Value * | ARG | p-Value * | ARG | p-Value * |
| MCR-5.1 | 0.009 | CrcB | 0.011 | Rm3 | 0.006 | erm (46) | 0.039 |
| VEB-14 | 0.012 | OprA | 0.015 | THIN-B | 0.007 | tet (42) | 0.047 |
| MCR-5.2 | 0.013 | CMY-157 | 0.046 | ParS | 0.021 | ||
| Ccol_ACT_CHL | 0.014 | ||||||
| VEB-5 | 0.017 | ||||||
| aadA4 | 0.017 | ||||||
| cmlA5 | 0.018 | ||||||
| BES-1 | 0.019 | ||||||
| AAC6_IB_HZ | 0.02 | ||||||
| OXA-836 | 0.02 | ||||||
| dfrA13 | 0.027 | ||||||
| linG | 0.029 | ||||||
| VEB-1 | 0.03 | ||||||
| CMY-42 | 0.03 | ||||||
| lnuF | 0.031 | ||||||
| dfrA7 | 0.031 | ||||||
| OXA-928 | 0.037 | ||||||
| OXA-921 | 0.04 | ||||||
| PAC-1 | 0.043 | ||||||
| VEB-9 | 0.044 | ||||||
| ANT(2′′)-Ia | 0.045 | ||||||
| VEB-7 | 0.046 | ||||||
| China and Russia | Russia and Canada | Russia and USA | Russia and UK | ||||
|---|---|---|---|---|---|---|---|
| ARG | p-Value * | ARG | p-Value * | ARG | p-Value * | ARG | p-Value * |
| VEB-3 | 0.001 | AAC(2’)-Ic | 0.009 | OXA-504 | 0.002 | mexP | 0.003 |
| dfrA1 | 0.009 | mef(B) | 0.009 | OXA-780 | 0.007 | OXA-118 | 0.008 |
| aadA16 | 0.011 | Erm(38) | 0.01 | AxyX | 0.01 | ESP-1 | 0.012 |
| arr-3 | 0.015 | tap | 0.01 | MOX-2 | 0.024 | TriB | 0.018 |
| pp-flo | 0.015 | efpA | 0.01 | CMY-48 | 0.026 | OXA-198 | 0.033 |
| AAC(6′)-Ib9 | 0.017 | QnrVC4 | 0.014 | CepS | 0.03 | OXA-20 | 0.044 |
| dfrA27 | 0.019 | OXA-56 | 0.015 | MOX-13 | 0.033 | ||
| tet(59) | 0.02 | Rv2856 | 0.036 | OXA-724 | 0.035 | ||
| EreB | 0.024 | OXA-912 | 0.036 | ||||
| AAC(6′)-31 | 0.026 | OXA-7 | 0.04 | ||||
| dfrA17 | 0.026 | FosA8 | 0.043 | ||||
| CARB-12 | 0.029 | ||||||
| OXA-21 | 0.03 | ||||||
| AAC(6′)-Ib | 0.031 | ||||||
| EreA | 0.033 | ||||||
| tet(33) | 0.038 | ||||||
| sul4 | 0.038 | ||||||
| tet(L) | 0.039 | ||||||
| AAC(6′)-IIa | 0.044 | ||||||
| dfrA14 | 0.045 | ||||||
| Asia | Europe | North America | |||
|---|---|---|---|---|---|
| ARG | p-Value * | ARG | p-Value * | ARG | p-Value * |
| AAC(6′)-Ib’ | 0.001 | LAQ-1 | 0.001 | AAC(6′)-Ic | 0.019 |
| AAC(6′)-Ii | 0.001 | LRA-1 | 0.001 | CMY-79 | 0.001 |
| AAC(6′)-31 | 0.001 | FOX-9 | 0.003 | CepS | 0.001 |
| AAC(6′)-Ib8 | 0.001 | OXA-20 | 0.001 | CMY-48 | 0.001 |
| AAC6_30_AAC6_Ib | 0.001 | OXA-228 | 0.001 | SRT-2 | 0.033 |
| APH(2′′)-If | 0.001 | OXA-257 | 0.001 | CMY-105 | 0.039 |
| aphA15 | 0.001 | CPS-1 | 0.001 | CMY-41 | 0.042 |
| AAC_6_IB_Su | 0.001 | OXA-669 | 0.001 | OXA-724 | 0.001 |
| APH(3′)-IIa | 0.001 | OXA-274 | 0.001 | AMZ-1 | 0.017 |
| AAC(6′)-IIa | 0.001 | OXA-37 | 0.001 | cphA2 | 0.037 |
| AAC_3Ib_AAC_6Ib | 0.001 | Rm3 | 0.001 | QnrB47 | 0.005 |
| AAC(6′)-Ib9 | 0.001 | OXA-198 | 0.001 | AxyX | 0.001 |
| AAC6_Ie_APH2_Ia | 0.001 | THIN-B | 0.001 | OprZ | 0.001 |
| aadA16 | 0.001 | OXA-667 | 0.001 | golS | 0.033 |
| AAC(6′)-IIc | 0.002 | OXA-355 | 0.001 | MCR-3.4 | 0.034 |
| ANT(3′′)-IIa | 0.002 | OXA-229 | 0.001 | MCR-3.6 | 0.036 |
| aad(6) | 0.004 | SGM-6 | 0.001 | tet(X1) | 0.001 |
| ANT3II_ANT6II | 0.006 | OXA-668 | 0.001 | tet(41) | 0.014 |
| CrcB | 0.009 | ESP-1 | 0.001 | ||
| APH(6)-Ic | 0.011 | OXA-5 | 0.001 | ||
| SCO-1 | 0.001 | OXA-118 | 0.001 | ||
| TEM-72 | 0.001 | OXA-119 | 0.001 | ||
| TEM-116 | 0.001 | LEN-9 | 0.005 | ||
| CTX-M-101 | 0.002 | OXA-209 | 0.006 | ||
| CTX-M-42 | 0.002 | PDC-9 | 0.009 | ||
| IreK | 0.002 | PDC-133 | 0.012 | ||
| CTX-M-130 | 0.003 | OXA-513 | 0.026 | ||
| VEB-3 | 0.005 | PDC-62 | 0.026 | ||
| CTX-M-155 | 0.008 | TriB | 0.001 | ||
| EC-14 | 0.019 | TriA | 0.001 | ||
| TEM-183 | 0.033 | QnrB8 | 0.012 | ||
| EC-15 | 0.046 | cfrC | 0.001 | ||
| CARB-12 | 0.001 | ParS | 0.001 | ||
| GES-44 | 0.001 | mexP | 0.001 | ||
| OXA-45 | 0.001 | ParR | 0.001 | ||
| OXA-3 | 0.001 | opmE | 0.002 | ||
| OXA-21 | 0.001 | rosB | 0.009 | ||
| OXA-417 | 0.002 | tet(30) | 0.001 | ||
| OXA-1 | 0.002 | ||||
| GES-12 | 0.002 | ||||
| PER-4 | 0.003 | ||||
| OXA-320 | 0.003 | ||||
| OXA-496 | 0.005 | ||||
| OXA-96 | 0.005 | ||||
| RAD-1 | 0.006 | ||||
| OXA-926 | 0.006 | ||||
| ACT-34 | 0.007 | ||||
| OXA-282 | 0.014 | ||||
| GES-3 | 0.014 | ||||
| ACT-25 | 0.022 | ||||
| TEM-102 | 0.022 | ||||
| TEM-198 | 0.027 | ||||
| LAP-2 | 0.031 | ||||
| dfrB4 | 0.001 | ||||
| dfrA16 | 0.001 | ||||
| dfrA27 | 0.001 | ||||
| dfrA17 | 0.001 | ||||
| dfrA1 | 0.001 | ||||
| QnrD1 | 0.001 | ||||
| QnrS8 | 0.001 | ||||
| QnrS1 | 0.003 | ||||
| TLA-2 | 0.001 | ||||
| lsaA | 0.001 | ||||
| efrB | 0.001 | ||||
| efrA | 0.002 | ||||
| Abau_AmvA | 0.006 | ||||
| AAC(6’)-Ib-cr1 | 0.011 | ||||
| Erm(51) | 0.001 | ||||
| ErmC | 0.001 | ||||
| erm(46) | 0.001 | ||||
| lnuA | 0.001 | ||||
| Erm(47) | 0.001 | ||||
| ErmT | 0.001 | ||||
| msrC | 0.001 | ||||
| mef(F) | 0.001 | ||||
| msr(G) | 0.002 | ||||
| LnuP | 0.005 | ||||
| EreA | 0.008 | ||||
| ErmX | 0.009 | ||||
| EreB | 0.012 | ||||
| Erm(52) | 0.013 | ||||
| msrF | 0.018 | ||||
| ErmQ | 0.022 | ||||
| SAT-4 | 0.02 | ||||
| catB2 | 0.001 | ||||
| pp-flo | 0.001 | ||||
| cmlA4 | 0.005 | ||||
| catQ | 0.006 | ||||
| Abau_AbaF | 0.002 | ||||
| arr-3 | 0.001 | ||||
| sul3 | 0.001 | ||||
| sul4 | 0.001 | ||||
| tet(K) | 0.001 | ||||
| tet(59) | 0.001 | ||||
| tet(Z) | 0.001 | ||||
| tet(42) | 0.001 | ||||
| tet(33) | 0.001 | ||||
| tet(43) | 0.001 | ||||
| tet(L) | 0.001 | ||||
| tet(36) | 0.002 | ||||
| Asia and Europe | Europe and North America | Asia and North America | |||
|---|---|---|---|---|---|
| ARG | p-Value * | ARG | p-Value * | ARG | p-Value * |
| novA | 0.001 | aadA7 | 0.001 | APH(9)-Ic | 0.002 |
| aadA4 | 0.001 | OXA-780 | 0.001 | CMY-114 | 0.001 |
| AAC(3)-IIe | 0.001 | OXA-504 | 0.001 | CfxA3 | 0.03 |
| AAC(6’)-Ib | 0.001 | MOX-13 | 0.006 | CMY-116 | 0.031 |
| APH(3’)-Ib | 0.003 | OXA-726 | 0.01 | MIR-2 | 0.043 |
| AAC(3)-Ia | 0.004 | imiH | 0.03 | adeF | 0.039 |
| aadA27 | 0.004 | OXA-34 | 0.034 | mphF | 0.001 |
| aadA15 | 0.014 | OXA-681 | 0.04 | MCR-3.3 | 0.014 |
| ANT(9)-Ia | 0.016 | QnrB19 | 0.026 | Ecol_catII | 0.033 |
| RanA | 0.016 | MuxA | 0.001 | tet(D) | 0.001 |
| APH(3’)-VIa | 0.018 | MexV | 0.011 | tet(B) | 0.001 |
| aadA3 | 0.024 | MCR-9.1 | 0.001 | ||
| AAC(3)-IIb | 0.028 | MCR-3.17 | 0.041 | ||
| ANT(6)-Ib | 0.037 | ||||
| VEB-7 | 0.001 | ||||
| PJM-1 | 0.001 | ||||
| AIM-1 | 0.001 | ||||
| VEB-5 | 0.002 | ||||
| RAHN-1 | 0.011 | ||||
| CTX-M-88 | 0.013 | ||||
| VEB-9 | 0.017 | ||||
| BEL-1 | 0.017 | ||||
| FOX-3 | 0.019 | ||||
| VEB-1 | 0.024 | ||||
| VEB-14 | 0.025 | ||||
| LCR-1 | 0.031 | ||||
| SGM-1 | 0.001 | ||||
| OXA-296 | 0.001 | ||||
| JOHN-1 | 0.001 | ||||
| CARB-14 | 0.001 | ||||
| OXA-420 | 0.001 | ||||
| OXA-47 | 0.001 | ||||
| OXA-4 | 0.001 | ||||
| CARB-5 | 0.001 | ||||
| RCP-1 | 0.001 | ||||
| OXA-129 | 0.001 | ||||
| OXA-31 | 0.001 | ||||
| OXA-392 | 0.002 | ||||
| OXA-134 | 0.002 | ||||
| OXA-58 | 0.003 | ||||
| BKC-1 | 0.003 | ||||
| OXA-275 | 0.004 | ||||
| OXA-333 | 0.004 | ||||
| OXA-164 | 0.006 | ||||
| OXA-9 | 0.008 | ||||
| SHV-18 | 0.011 | ||||
| SHV-24 | 0.011 | ||||
| OXA-650 | 0.011 | ||||
| GES-14 | 0.012 | ||||
| blaF | 0.015 | ||||
| CGA-1 | 0.016 | ||||
| AER-1 | 0.02 | ||||
| GES-17 | 0.029 | ||||
| ORN-1 | 0.041 | ||||
| OXA-651 | 0.044 | ||||
| OXA-727 | 0.046 | ||||
| dfrB10 | 0.001 | ||||
| dfrA14 | 0.003 | ||||
| dfrA7 | 0.005 | ||||
| QepA4 | 0.001 | ||||
| QepA1 | 0.001 | ||||
| QepA2 | 0.002 | ||||
| Abau_AbaQ | 0.002 | ||||
| lfrA | 0.002 | ||||
| qnrE1 | 0.019 | ||||
| adeN | 0.001 | ||||
| aadT | 0.002 | ||||
| Rv2856 | 0.004 | ||||
| abeM | 0.004 | ||||
| EstT | 0.027 | ||||
| AAC(6’)-Ib-cr3 | 0.033 | ||||
| Erm(42) | 0.001 | ||||
| lnuF | 0.001 | ||||
| lnuG | 0.001 | ||||
| oleC | 0.001 | ||||
| linG | 0.001 | ||||
| msr(I) | 0.001 | ||||
| mef(J) | 0.001 | ||||
| lmrD | 0.001 | ||||
| lnuB | 0.004 | ||||
| lsaE | 0.008 | ||||
| vatB | 0.011 | ||||
| msrA | 0.027 | ||||
| Erm(38) | 0.03 | ||||
| lnuD | 0.039 | ||||
| vanS_in_vanO_cl | 0.001 | ||||
| vanR_in_vanO_cl | 0.001 | ||||
| LpsB | 0.001 | ||||
| ICR-Mo | 0.007 | ||||
| vanW_in_vanG_cl | 0.031 | ||||
| cmlA1 | 0.001 | ||||
| cmx | 0.001 | ||||
| floR | 0.001 | ||||
| cmlB1 | 0.004 | ||||
| Ccol_ACT_CHL | 0.008 | ||||
| catP | 0.028 | ||||
| catB11 | 0.047 | ||||
| cmlA5 | 0.048 | ||||
| FosXCC | 0.003 | ||||
| Nfar_rox | 0.001 | ||||
| Sven_rox | 0.001 | ||||
| rphA | 0.001 | ||||
| HelR | 0.001 | ||||
| rphB | 0.004 | ||||
| tet(Y) | 0.001 | ||||
| otr(A)S.rim | 0.001 | ||||
| tet(X6) | 0.001 | ||||
| tet(H) | 0.001 | ||||
| tet(S) | 0.001 | ||||
| tet(X5) | 0.001 | ||||
| tet(V) | 0.001 | ||||
| tap | 0.004 | ||||
| tetA(p) | 0.015 | ||||
References
- Sims, N.; Kannan, A.; Holton, E.; Jagadeesan, K.; Mageiros, L.; Standerwick, R.; Craft, T.; Barden, R.; Feil, E.J.; Kasprzyk-Hordern, B. Antimicrobials and antimicrobial resistance genes in a one-year city metabolism longitudinal study using wastewater-based epidemiology. Environ. Pollut. 2023, 333, 122020. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Kurittu, P.; Al-Mustapha, A.I.; Heljanko, V.; Johansson, V.; Thakali, O.; Mishra, S.K.; Lehto, K.-M.; Lipponen, A.; Oikarinen, S.; et al. Wastewater surveillance of antibiotic-resistant bacterial pathogens: A systematic review. Front. Microbiol. 2022, 13, 977106. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Antimicrobial Resistance Surveillance System Manual for Early Implementation; World Health Organization: Geneva, Switzerland, 2015; Available online: https://iris.who.int/bitstream/handle/10665/188783/9789241549400_eng.pdf (accessed on 2 September 2024).
- World Health Organization. Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation 2017–2018; World Health Organization: Geneva, Switzerland, 2018; Available online: https://iris.who.int/bitstream/handle/10665/279656/9789241515061-eng.pdf (accessed on 2 September 2024).
- Chau, K.K.; Goodall, T.; Bowes, M.; Easterbrook, K.; Brett, H.; Hughes, J.; Crook, D.W.; Read, D.S.; Walker, A.S.; Stoesser, N. High-resolution characterization of short-term temporal variability in the taxonomic and resistome composition of wastewater influent. Microb. Genom. 2023, 9, 000983. [Google Scholar] [CrossRef] [PubMed]
- Bijlsma, L.; Xu, L.; Gracia-Marín, E.; Pitarch, E.; Serrano, R.; Kasprzyk-Hordern, B. Understanding associations between antimicrobial agents usage and antimicrobial resistance genes prevalence at the community level using wastewater-based epidemiology: A Spanish pilot study. Sci. Total Environ. 2024, 926, 171996. [Google Scholar] [CrossRef] [PubMed]
- Clarke, L.M.; O’Brien, J.W.; Murray, A.K.; Gaze, W.H.; Thomas, K.V. A review of wastewater-based epidemiology for antimicrobial resistance surveillance. J. Environ. Expo. Assess. 2024, 3, 7. [Google Scholar] [CrossRef]
- Parkins, M.D.; Lee, B.E.; Acosta, N.; Bautista, M.; Hubert, C.R.J.; Hrudey, S.E.; Frankowski, K.; Pang, X.-L. Wastewater-based surveillance as a tool for public health action: SARS-CoV-2 and beyond. Clin. Microbiol. Rev. 2023, 37, e00103-22. [Google Scholar] [CrossRef]
- Gholipour, S.; Shamsizadeh, Z.; Halabowski, D.; Gwenzi, W.; Nikaeen, M. Combating antibiotic resistance using wastewater surveillance: Significance, applications, challenges, and future directions. Sci. Total Environ. 2023, 908, 168056. [Google Scholar] [CrossRef]
- Kang, Y.; Wang, J.; Li, Z. Meta-analysis addressing the characterization of antibiotic resistome in global hospital wastewater. J. Hazard. Mater. 2024, 466, 133577. [Google Scholar] [CrossRef]
- Liu, C.; Chen, J.; Shan, X.; Yang, Y.; Song, L.; Teng, Y.; Chen, H. Meta-analysis addressing the characterization and risk identification of antibiotics and antibiotic resistance genes in global groundwater. Sci. Total Environ. 2023, 860, 160513. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. COVID-19: U.S. Impact on Antimicrobial Resistance, Special Report 2022; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2022. Available online: https://www.cdc.gov/antimicrobial-resistance/media/pdfs/covid19-impact-report-508.pdf (accessed on 5 September 2024).
- Centers for Disease Control and Prevention. Antimicrobial Resistance Threats in the United States, 2021–2022; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2024. Available online: https://www.cdc.gov/antimicrobial-resistance/data-research/threats/update-2022.html (accessed on 5 September 2024).
- Leinonen, R.; Sugawara, H.; Shumway, M. The Sequence Read Archive. Nucleic Acids Res. 2010, 39, D19–D21. [Google Scholar] [CrossRef]
- Andrews, S. FastQC A Quality Control tool for High Throughput Sequence Data. Babraham Bioinformatics. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 23 September 2024).
- Lo, C.-C.; Chain, P.S.G. Rapid evaluation and quality control of next generation sequencing data with FaQCs. BMC Bioinform. 2014, 15, 366. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2022, 51, D690–D699. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner. In Proceedings of the 9th Annual Genomics of Energy & Environment Meeting, Walnut Creek, CA, USA, 20 March 2014; Available online: https://www.osti.gov/servlets/purl/1241166 (accessed on 23 September 2024).
- McCall, C.; Xagoraraki, I. Comparative study of sequence aligners for detecting antibiotic resistance in bacterial metagenomes. Lett. Appl. Microbiol. 2017, 66, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Lahti, L.; Shetty, S. Microbiome R Package. Available online: https://bioconductor.org/packages/release/bioc/html/microbiome.html (accessed on 24 October 2024).
- Oksanen, J.; Simpson, G.; Blanchet, F.; Kindt, R.; Legendre, P.; Minchin, P.; O’Hara, R.; Solymos, P.; Stevens, M.; Szoecs, E.; et al. vegan: Community Ecology Package [R package vegan version 2.6-6.1]. Comprehensive R Archive Network (CRAN). 2024. Available online: https://cran.r-project.org/package=vegan (accessed on 24 October 2024).
- De Cáceres, M.; Legendre, P. Associations between species and groups of sites: Indices and statistical inference. Ecology 2009, 90, 3566–3574. [Google Scholar] [CrossRef]
- R Core Team. R: The R Project for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2008; Available online: https://www.r-project.org/ (accessed on 24 October 2024).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis. Springer: New York, NY, USA, 2016. [Google Scholar] [CrossRef]
- Begmatov, S.; Beletsky, A.V.; Dorofeev, A.G.; Pimenov, N.V.; Mardanov, A.V.; Ravin, N.V. Metagenomic insights into the wastewater resistome before and after purification at large scale wastewater treatment plants in the Moscow city. Sci. Rep. 2024, 14, 6349. [Google Scholar] [CrossRef]
- Brumfield, K.D.; Leddy, M.; Usmani, M.; Cotruvo, J.A.; Tien, C.-T.; Dorsey, S.; Graubics, K.; Fanelli, B.; Zhou, I.; Registe, N.; et al. Microbiome Analysis for Wastewater Surveillance during COVID-19. mBio 2022, 13, e00591-22. [Google Scholar] [CrossRef]
- Fierer, N.; Holland-Moritz, H.; Alexiev, A.; Batther, H.; Dragone, N.B.; Friar, L.; Gebert, M.J.; Gering, S.; Henley, J.B.; Jech, S.; et al. A Metagenomic Investigation of Spatial and Temporal Changes in Sewage Microbiomes across a University Campus. mSystems 2022, 7, e00651-22. [Google Scholar] [CrossRef]
- Fu, S.; Yang, Q.; Sheng, Y.; Wang, Q.; Wu, J.; Qiu, Z.; Lan, R.; Wang, Y.; Liu, Y. Metagenomics combined with comprehensive validation as a public health risk assessment tool for urban and agricultural run-off. Water Res. 2021, 209, 117941. [Google Scholar] [CrossRef]
- Jankowski, P.; Gan, J.; Le, T.; McKennitt, M.; Garcia, A.; Yanaç, K.; Yuan, Q.; Uyaguari-Diaz, M. Metagenomic community composition and resistome analysis in a full-scale cold climate wastewater treatment plant. Environ. Microbiome 2022, 17, 3. [Google Scholar] [CrossRef]
- Lepper, H.C.; Perry, M.R.; Wee, B.A.; Wills, D.; Nielsen, H.; Otani, S.; Simon, M.; Aarestrup, F.M.; Woolhouse, M.E.J.; Van Bunnik, B.A.D. Distinctive hospital and community resistomes in Scottish urban wastewater: Metagenomics of a paired wastewater sampling design. Sci. Total Environ. 2023, 902, 165978. [Google Scholar] [CrossRef]
- Li, Z.; Yuan, D.; Kou, Y.; Li, X.; Du, C. Metagenome sequencing to unveil the occurrence and distribution of antibiotic resistome and in a wastewater treatment plant. Environ. Technol. 2023, 45, 1933–1942. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yao, J.; Ma, H.; Rukeya, A.; Liang, Z.; Du, W.; Chen, Y. Bacterial Hosts and Genetic Characteristics of Antibiotic Resistance Genes in Wastewater Treatment Plants of Xinjiang (China) Revealed by Metagenomics. Appl. Sci. 2022, 12, 3100. [Google Scholar] [CrossRef]
- Luo, L.; Yao, J.; Liu, W.; Yang, L.; Li, H.; Liang, M.; Ma, H.; Liu, Z.; Chen, Y. Comparison of bacterial communities and antibiotic resistance genes in oxidation ditches and membrane bioreactors. Sci. Rep. 2021, 11, 8955. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, B.; Zhao, J.; Tian, Y.; Qiu, Y. Occurrence and Distribution of Antibiotic Resistance Genes in Municipal Wastewater Treatment Plants with D-Type Filters. Water 2021, 13, 3398. [Google Scholar] [CrossRef]
- Barcellos, D.S.; Barquilha, C.E.R.; Oliveira, P.E.; Prokopiuk, M.; Etchepare, R.G. How has the COVID-19 pandemic impacted wastewater-based epidemiology? Sci. Total Environ. 2023, 892, 164561. [Google Scholar] [CrossRef]
- O’Keeffe, J. Wastewater-based epidemiology: Current uses and future opportunities as a public health surveillance tool. Environ. Health Rev. 2021, 64, 44–52. [Google Scholar] [CrossRef]
- Qiao, M.; Ying, G.-G.; Singer, A.C.; Zhu, Y.-G. Review of antibiotic resistance in China and its environment. Environ. Int. 2017, 110, 160–172. [Google Scholar] [CrossRef]
- Zhang, R.; Eggleston, K.; Rotimi, V.; Zeckhauser, R.J. Antibiotic resistance as a global threat: Evidence from China, Kuwait and the United States. Glob. Health 2006, 2, 6. [Google Scholar] [CrossRef]
- Zakharenkov, I.A.; Rachina, S.A.; Kozlov, R.S.; Belkova, Y.A. Consumption of systemic antibiotics in the Russian Federation in 2017–2021. Clin. Microbiol. Antimicrob. Chemother. 2022, 24, 220–225. [Google Scholar] [CrossRef]
- Buelow, E.; Rico, A.; Gaschet, M.; Lourenço, J.; Kennedy, S.P.; Wiest, L.; Ploy, M.-C.; Dagot, C. Hospital discharges in urban sanitation systems: Long-term monitoring of wastewater resistome and microbiota in relationship to their eco-exposome. Water Res. X 2020, 7, 100045. [Google Scholar] [CrossRef]
- Czatzkowska, M.; Wolak, I.; Harnisz, M.; Korzeniewska, E. Impact of Anthropogenic Activities on the Dissemination of ARGs in the Environment—A Review. Int. J. Environ. Res. Public Health 2022, 19, 12853. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Kaur, R.; Verma, S.; Singh, S. Antimicrobials and Antibiotic Resistance Genes in Water Bodies: Pollution, Risk, and Control. Front. Environ. Sci. 2022, 10, 830861. [Google Scholar] [CrossRef]
- Yin, Z.; Tan, J.; Huang, H.; Zhao, J.; Gong, X.; Li, J.; Chen, C.; Luo, F.; Huang, X.; Wang, H.; et al. Trends in the antimicrobial susceptibility among Chinese neonates from 2012 to 2021: A multicenter study. Antimicrob. Resist. Infect. Control. 2024, 13, 83. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Peng, Y.; Chan, C.-L.; On, H.; Wai, H.K.-F.; Shekhawat, S.S.; Gupta, A.B.; Varshney, A.K.; Chuanchuen, R.; Zhou, X.; et al. Metagenomic Survey Reveals More Diverse and Abundant Antibiotic Resistance Genes in Municipal Wastewater Than Hospital Wastewater. Front. Microbiol. 2021, 12, 712843. [Google Scholar] [CrossRef]
- Munk, P.; Brinch, C.; Møller, F.D.; Petersen, T.N.; Hendriksen, R.S.; Seyfarth, A.M.; Kjeldgaard, J.S.; Svendsen, C.A.; Van Bunnik, B.; Berglund, F.; et al. Genomic analysis of sewage from 101 countries reveals global landscape of antimicrobial resistance. Nat. Commun. 2022, 13, 7251. [Google Scholar] [CrossRef]
- Pazda, M.; Kumirska, J.; Stepnowski, P.; Mulkiewicz, E. Antibiotic resistance genes identified in wastewater treatment plant systems—A review. Sci. Total Environ. 2019, 697, 134023. [Google Scholar] [CrossRef]
- Karakonstantis, S.; Kritsotakis, E.I.; Gikas, A. Treatment options for K. pneumoniae, P. aeruginosa and A. baumannii co-resistant to carbapenems, aminoglycosides, polymyxins and tigecycline: An approach based on the mechanisms of resistance to carbapenems. Infection 2020, 48, 835–851. [Google Scholar] [CrossRef]
- Mutuku, C.; Gazdag, Z.; Melegh, S. Occurrence of antibiotics and bacterial resistance genes in wastewater: Resistance mechanisms and antimicrobial resistance control approaches. World J. Microbiol. Biotechnol. 2022, 38, 152. [Google Scholar] [CrossRef]
- Deshpande, A.S.; Ehasz, G.; Eramo, A.; Fahrenfeld, N. Changes in Prevalence but Not Hosts of Antibiotic Resistance Genes during the COVID-19 Pandemic versus Prepandemic in Wastewater Influent. ACS ES&T Water 2023, 3, 3626–3638. [Google Scholar] [CrossRef]
- Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2023, 12, 116. [Google Scholar] [CrossRef]
- Nandi, A.; Pecetta, S.; Bloom, D.E. Global antibiotic use during the COVID-19 pandemic: Analysis of pharmaceutical sales data from 71 countries, 2020–2022. EClinicalMedicine 2023, 57, 101848. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, N.; Bruyndonckx, R.; Versporten, A.; Hens, N.; Monnet, D.L.; Molenberghs, G.; Goossens, H.; Weist, K.; Coenen, S.; Strauss, R.; et al. Consumption of macrolides, lincosamides and streptogramins in the community, European Union/European Economic Area, 1997–2017. J. Antimicrob. Chemother. 2021, 76, ii30–ii36. [Google Scholar] [CrossRef] [PubMed]
- Sharma, E.; Chen, Y.; Kelso, C.; Sivakumar, M.; Jiang, G. Navigating the environmental impacts and analytical methods of last-resort antibiotics: Colistin and carbapenems. Soil Environ. Health 2024, 2, 100058. [Google Scholar] [CrossRef]
- Mancuso, G.; De Gaetano, S.; Midiri, A.; Zummo, S.; Biondo, C. The Challenge of Overcoming Antibiotic Resistance in Carbapenem-Resistant Gram-Negative Bacteria: “Attack on Titan”. Microorganisms 2023, 11, 1912. [Google Scholar] [CrossRef] [PubMed]
- Piotrowski, M.; Alekseeva, I.; Arnet, U.; Yücel, E. Insights into the Rising Threat of Carbapenem-Resistant Enterobacterales and Pseudomonas aeruginosa Epidemic Infections in Eastern Europe: A Systematic Literature Review. Antibiotics 2024, 13, 978. [Google Scholar] [CrossRef]
- Xu, W.; Zhang, G.; Li, X.; Zou, S.; Li, P.; Hu, Z.; Li, J. Occurrence and elimination of antibiotics at four sewage treatment plants in the Pearl River Delta (PRD), South China. Water Res. 2007, 41, 4526–4534. [Google Scholar] [CrossRef]
- Jiang, H.; Cheng, H.; Liang, Y.; Yu, S.; Yu, T.; Fang, J.; Zhu, C. Diverse Mobile Genetic Elements and Conjugal Transferability of Sulfonamide Resistance Genes (sul1, sul2, and sul3) in Escherichia coli Isolates from Penaeus vannamei and Pork from Large Markets in Zhejiang, China. Front. Microbiol. 2019, 10, 1787. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alhazmi, S.M.; BaniMustafa, A.; Alindonosi, A.R.; Almutairi, A.F. Metagenomic Meta-Analysis of Antibiotic-Resistance Genes in Wastewater: A Perspective from the COVID-19 Pandemic. Water 2024, 16, 3571. https://doi.org/10.3390/w16243571
Alhazmi SM, BaniMustafa A, Alindonosi AR, Almutairi AF. Metagenomic Meta-Analysis of Antibiotic-Resistance Genes in Wastewater: A Perspective from the COVID-19 Pandemic. Water. 2024; 16(24):3571. https://doi.org/10.3390/w16243571
Chicago/Turabian StyleAlhazmi, Shaima M., Ala’a BaniMustafa, Abrar R. Alindonosi, and Adel F. Almutairi. 2024. "Metagenomic Meta-Analysis of Antibiotic-Resistance Genes in Wastewater: A Perspective from the COVID-19 Pandemic" Water 16, no. 24: 3571. https://doi.org/10.3390/w16243571
APA StyleAlhazmi, S. M., BaniMustafa, A., Alindonosi, A. R., & Almutairi, A. F. (2024). Metagenomic Meta-Analysis of Antibiotic-Resistance Genes in Wastewater: A Perspective from the COVID-19 Pandemic. Water, 16(24), 3571. https://doi.org/10.3390/w16243571






