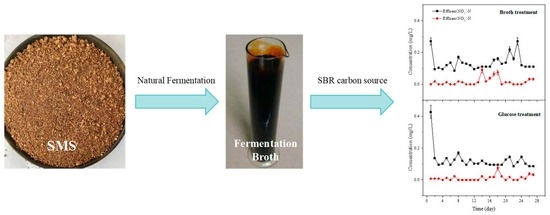
Microbiological Mechanisms for Nitrogen Removal Using Anaerobic Fermentation Liquid from Spent Mushroom Substrates as a Carbon Source
Abstract
:1. Introduction
2. Material and Methods
2.1. Preparation of SMS Fermentation Liquid
2.2. SBR Device and Operation
2.3. High through Sequencing
2.4. Functional Gene Quantification
2.5. Extracellular Polymeric Substance Extraction and Determination
2.6. Analytical Techniques and Statistical Analysis
3. Results and Discussion
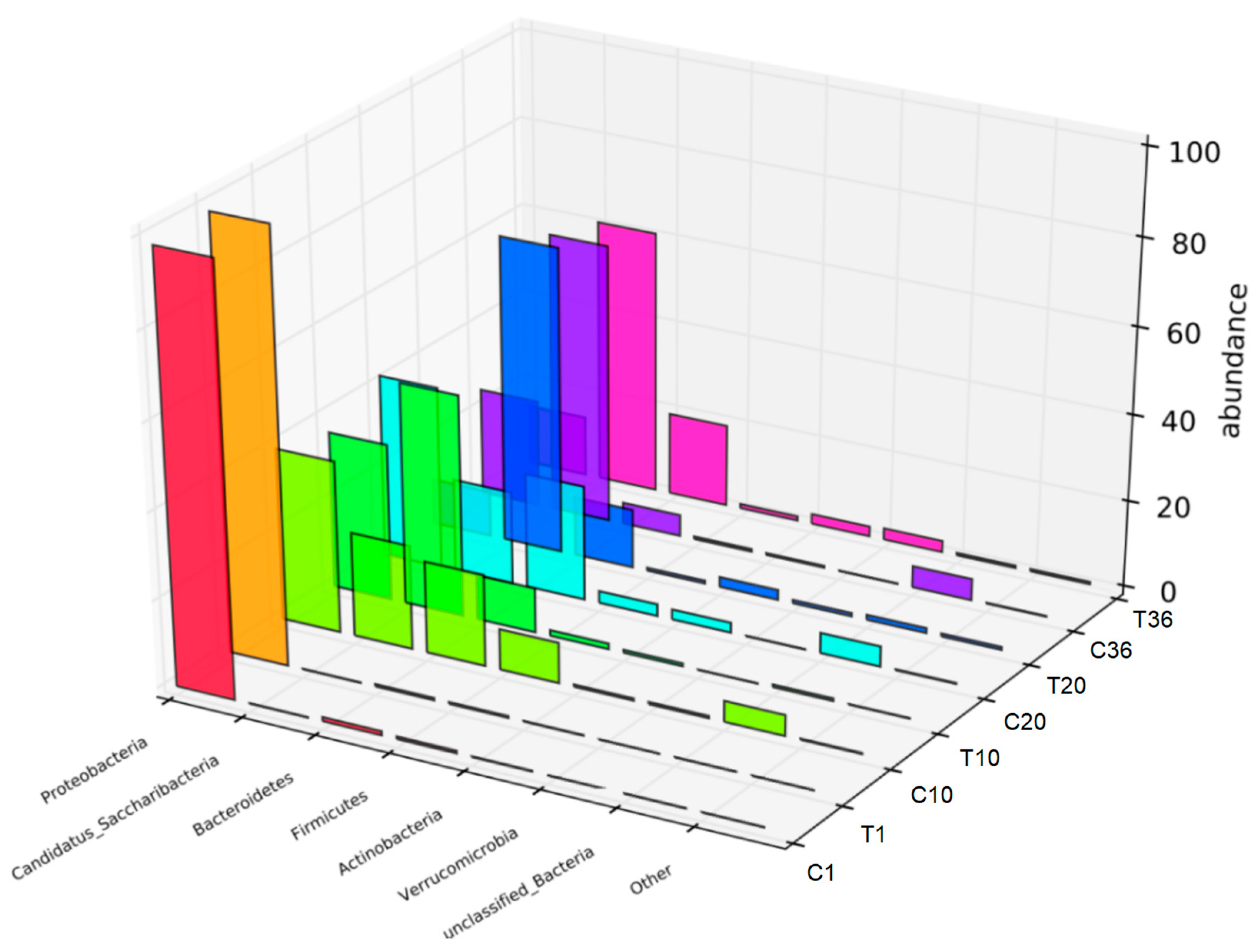
3.1. Composition and Microbial Community of Fermentation Broth
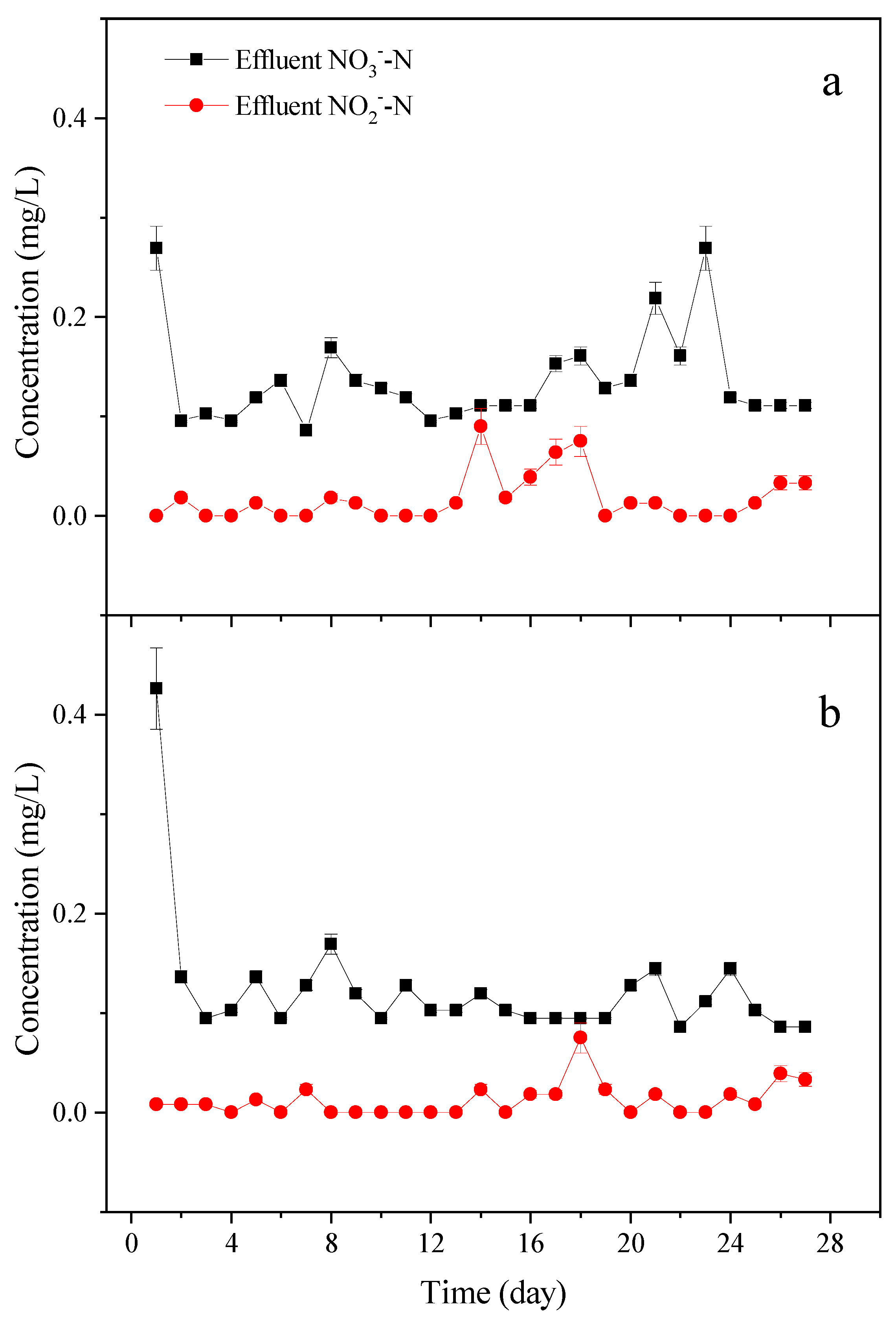
3.2. Denitrification Performance and COD Removal
3.3. Extracellular Polymeric Substance Composition
3.4. Absolute Abundance of Denitrification Functional Genes
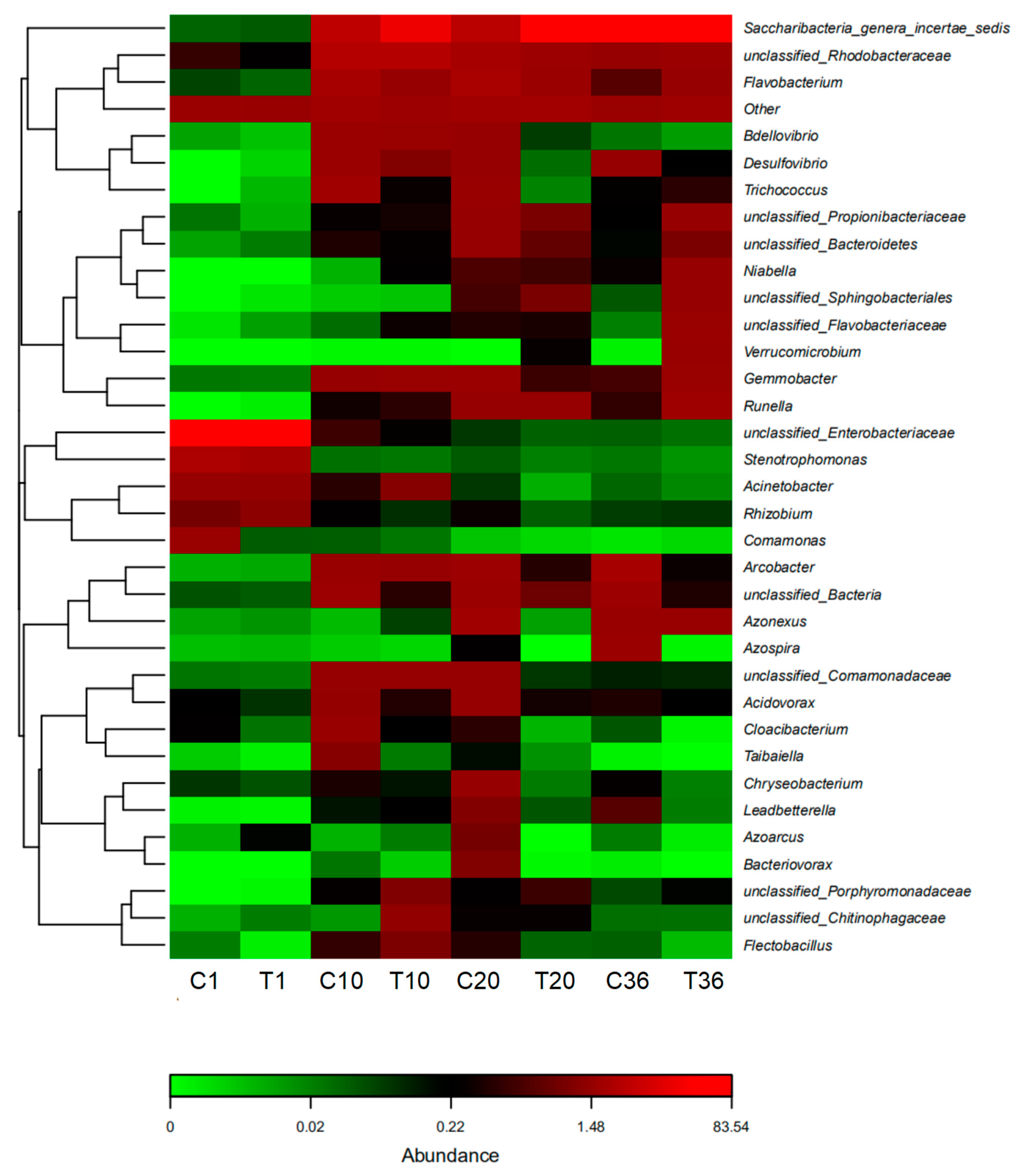
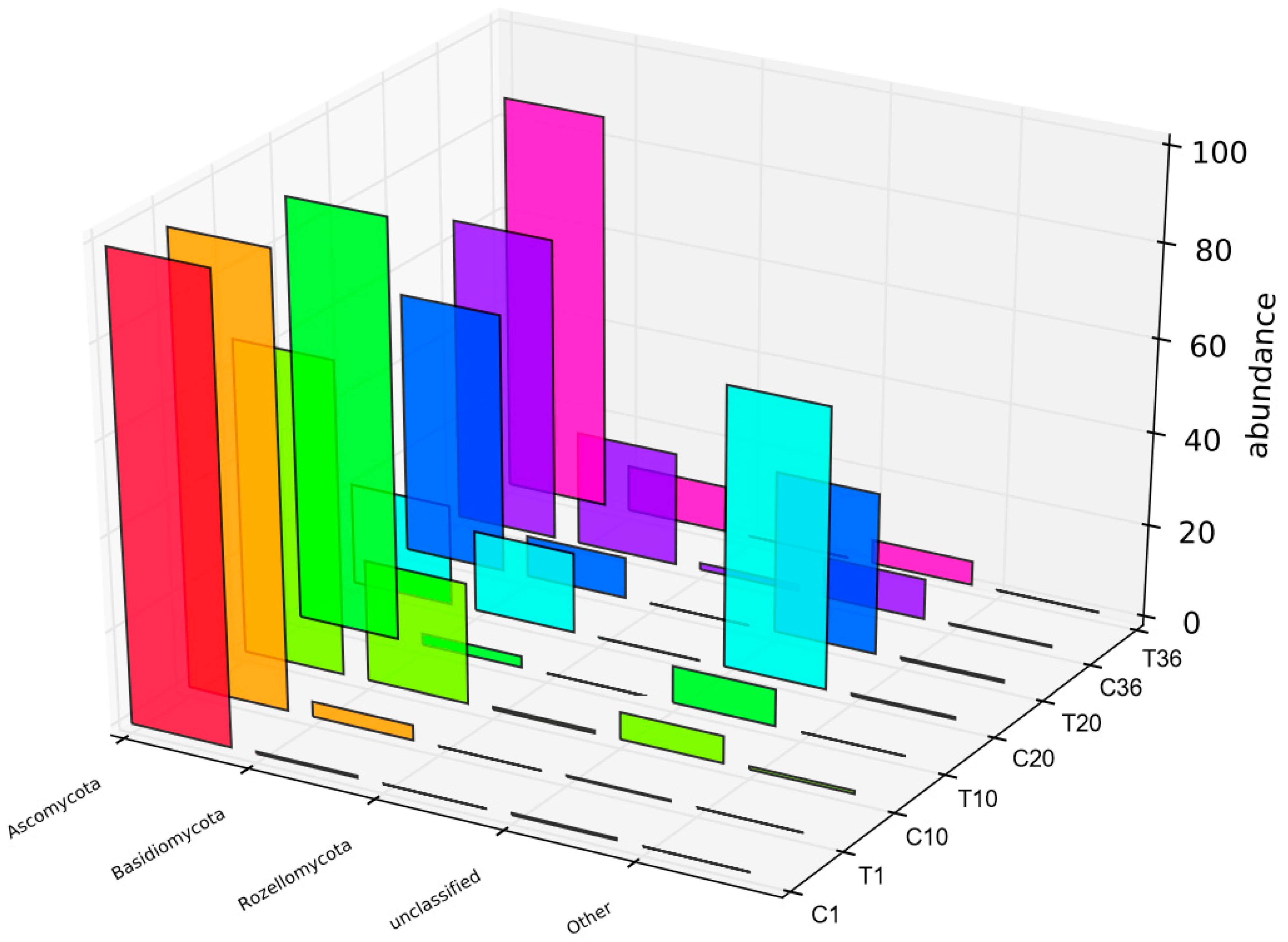
3.5. Community Dynamics of Activated Sludge
3.6. Environmental and Economic Benefits
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviation
| SMS | spent mushroom substrates |
| C/N | carbon–nitrogen ratio |
| EPS | extracellular polymeric substance |
| SBR | sequencing batch reactor |
| COD | chemical oxygen demand |
| qPCR | quantitative polymerase chain reaction |
| PN | extracellular protein |
| PS | extracellular polysaccharides |
| UV-vis | ultraviolet–visible spectrophotometer |
| HPLC | high-performance liquid chromatography |
| LC-MS | liquid chromatography mass spectrometry |
| ICP-OES | inductively coupled plasma emission spectrometry |
References
- Othman, N.Z.; Sarjuni, M.N.H.; Rosli, M.A.; Nadri, M.H.; Yeng, L.H.; Ying, O.P.; Sarmidi, M.R. Spent mushroom substrate as biofertilizer for agriculture application. In Valorisation of Agro-Industrial Residues–Volume I: Biological Approaches; Springer: Cham, Switzerland, 2020; Volume 3, pp. 7–57. [Google Scholar]
- Foluke, A.; Olutayo, A.; Olufemi, A. Assessing spent mushroom substrate as a replacement to wheat bran in the diet of broilers. Am. Int. J. Contemp. Res. 2014, 4, 178–183. [Google Scholar]
- Da Silva Alves, L.; de Almeida Moreira, B.R.; da Silva Viana, R.; Pardo-Gimenez, A.; Dias, E.S.; Noble, R.; Zied, D.C. Recycling spent mushroom substrate into fuel pellets for low-emission bioenergy producing systems. J. Clean. Prod. 2021, 313, 127875. [Google Scholar] [CrossRef]
- Jin, Y.; Teng, C.; Yu, S.; Song, T.; Dong, L.; Liang, J.; Bai, X.; Liu, X.; Hu, X.; Qu, J. Batch and fixed-bed biosorption of Cd (II) from aqueous solution using immobilized Pleurotus ostreatus spent substrate. Chemosphere 2018, 191, 799–808. [Google Scholar] [CrossRef]
- Liu, X.; Bai, X.; Dong, L.; Liang, J.; Jin, Y.; Wei, Y.; Li, Y.; Huang, S.; Qu, J. Composting enhances the removal of lead ions in aqueous solution by spent mushroom substrate: Biosorption and precipitation. J. Clean. Prod. 2018, 200, 1–11. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, T.; Chen, C.; Feng, L.; Su, X.; Zhou, L.; Chen, Y.; Xia, A.; Wang, X. Spent substrate of Ganodorma lucidum as a new bio-adsorbent for adsorption of three typical dyes. Bioresour. Technol. 2018, 266, 134–138. [Google Scholar] [CrossRef]
- Liu, M.; Liu, X.; Wu, Z.; Zhang, Y.; Meng, Q.; Yan, L. Sulfur-modified Pleurotus ostreatus spent substrate biochar enhances the removal of cadmium in aqueous solution: Characterization, performance, mechanism. J. Environ. Manag. 2022, 322, 115900. [Google Scholar] [CrossRef]
- Liu, F.; Tian, Y.; Ding, Y.; Li, Z. The use of fermentation liquid of wastewater primary sedimentation sludge as supplemental carbon source for denitrification based on enhanced anaerobic fermentation. Bioresour. Technol. 2016, 219, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zuo, J.; Wang, Y.; Zhao, J.; Tang, L.; Li, Z. Tertiary nitrogen removal for municipal wastewater using a solid-phase denitrifying biofilter with polycaprolactone as the carbon source and filtration medium. Water Res. 2016, 93, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Cho, S.; Jung, J. Key operating parameters affecting nitrogen removal rate in single-stage deammonification. Chemosphere 2018, 207, 357–364. [Google Scholar] [CrossRef]
- Yu, G.; Peng, H.; Fu, Y.; Yan, X.; Du, C.; Chen, H. Enhanced nitrogen removal of low C/N wastewater in constructed wetlands with co-immobilizing solid carbon source and denitrifying bacteria. Bioresour. Technol. 2019, 280, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.C.; Bao, X.; Che, L.; Wu, Q.L. Enhance biological nitrogen and phosphorus removal in wastewater treatment process by adding food waste fermentation liquid as external carbon source. Biochem. Eng. J. 2021, 165, 107811. [Google Scholar] [CrossRef]
- Xiong, R.; Yu, X.; Yu, L.; Peng, Z.; Cheng, L.; Li, T.; Fan, P. Biological denitrification using polycaprolactone-peanut shell as slow-release carbon source treating drainage of municipal WWTP. Chemosphere 2019, 235, 434–439. [Google Scholar] [CrossRef]
- Fu, X.; Hou, R.; Yang, P.; Qian, S.; Feng, Z.; Chen, Z.; Wang, F.; Yuan, R.; Chen, H.; Zhou, B. Application of external carbon source in heterotrophic denitrification of domestic sewage: A review. Sci. Total Environ. 2022, 817, 153061. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Y.; Chen, Y. Recent advances in partial denitrification in biological nitrogen removal: From enrichment to application. Bioresour. Technol. 2020, 298, 122444. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Dai, X.; Chai, X. Effect of different carbon sources on denitrification performance, microbial community structure and denitrification genes. Sci. Total Environ. 2018, 634, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xu, S. Edible mushroom industry in China: Current state and perspectives. Appl. Microbiol. Biotechnol. 2022, 106, 3949–3955. [Google Scholar] [CrossRef]
- Mohd Hanafi, F.H.; Rezania, S.; Mat Taib, S.; Md Din, M.F.; Yamauchi, M.; Sakamoto, M.; Hara, H.; Park, J.; Ebrahimi, S.S. Environmentally sustainable applications of agro-based spent mushroom substrate (SMS): An overview. J. Mater. Cycles Waste Manag. 2018, 20, 1383–1396. [Google Scholar] [CrossRef]
- Moon, Y.H.; Shin, P.G.; Cho, S.J. Feeding value of spent mushroom (Pleurotus eryngii) substrate. J. Mushroom 2012, 10, 236–243. [Google Scholar]
- Yang, Y.; Tao, X.; Lin, E.; Hu, K. Enhanced nitrogen removal with spent mushroom compost in a sequencing batch reactor. Bioresour. Technol. 2017, 244, 897–904. [Google Scholar] [CrossRef]
- Hu, H.; Ma, S.; Zhang, X.; Ren, H. Characteristics of dissolved organic nitrogen in effluent from a biological nitrogen removal process using sludge alkaline fermentation liquid as an external carbon source. Water Res. 2020, 176, 115741. [Google Scholar] [CrossRef]
- Tang, J.; Wang, X.C.; Hu, Y.; Pu, Y.; Huang, J.; Ngo, H.H.; Zeng, Y.; Li, Y. Nutrients removal performance and sludge properties using anaerobic fermentation slurry from food waste as an external carbon source for wastewater treatment. Bioresour. Technol. 2019, 271, 125–135. [Google Scholar] [CrossRef] [PubMed]
- American Public Health Association. Standard Methods for the Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 1985. [Google Scholar]
- Jiang, F.; Feng, X.; Jiang, X.; Wang, P. Enhanced dewaterability of lake dredged sediments by electrochemical oxidation of peroxydisulfate on BDD anode. Chemosphere 2022, 307, 135832. [Google Scholar] [CrossRef]
- Suzuki, M.; Hirai, T.; Arai, H.; Ishii, M.; Igarashi, Y. Purification, characterization, and gene cloning of thermophilic cytochrome cd1 nitrite reductase from Hydrogenobacter thermophilus TK-6. J. Biosci. Bioeng. 2006, 101, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Pomowski, A.; Zumft, W.G.; Kroneck, P.M.H.; Einsle, O. N2O binding at a [4Cu: 2S] copper–sulphur cluster in nitrous oxide reductase. Nature 2011, 477, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Tocheva, E.I.; Rosell, F.I.; Mauk, A.G.; Murphy, M.E. Side-on copper-nitrosyl coordination by nitrite reductase. Science 2004, 304, 867–870. [Google Scholar] [CrossRef]
- Wang, W.; Xie, H.; Wang, H.; Xue, H.; Wang, J.; Zhou, M.; Dai, X.; Wang, Y. Organic compounds evolution and sludge properties variation along partial nitritation and subsequent anammox processes treating reject water. Water Res. 2020, 184, 116197. [Google Scholar] [CrossRef]
- Yu, D.; Feng, M.; Sun, J.; Xu, X.L.; Zhou, G.H. Protein degradation and peptide formation with antioxidant activity in pork protein extracts inoculated with Lactobacillus plantarum and Staphylococcus simulans. Meat Sci. 2020, 160, 107958. [Google Scholar] [CrossRef]
- Wang, Y.; Fan, L.; Huang, J.; Liang, J.; Wang, X.; Ren, Y.; Li, H.; Yue, T.; Gao, Z. Evaluation of chemical composition, antioxidant activity, and gut microbiota associated with pumpkin juice fermented by Rhodobacter sphaeroides. Food Chem. 2023, 401, 134122. [Google Scholar] [CrossRef]
- Kim, J.H.; Jang, Y.A.; Seong, S.B.; Jang, S.A.; Hong, S.H.; Song, J.K.; Eom, G.T. High-level production and high-yield recovery of lactobionic acid by the control of pH and temperature in fermentation of Pseudomonas taetrolens. Bioprocess Biosyst. Eng. 2020, 43, 937–944. [Google Scholar] [CrossRef]
- Shankar, K.; Kulkarni, N.S.; Jayalakshmi, S.K.; Sreeramulu, K. Saccharification of the pretreated husks of corn, peanut and coffee cherry by the lignocellulolytic enzymes secreted by Sphingobacterium sp. ksn for the production of bioethanol. Biomass Bioenergy 2019, 127, 105298. [Google Scholar] [CrossRef]
- Geyik, A.G.; Kılıç, B.; Çeçen, F. Extracellular polymeric substances (EPS) and surface properties of activated sludges: Effect of organic carbon sources. Environ. Sci. Pollut. Res. 2016, 23, 1653–1663. [Google Scholar] [CrossRef]
- Sponza, D.T. Extracellular polymer substances and physicochemical properties of flocs in steady and unsteady-state activated sludge systems. Process Biochem. 2002, 37, 983–998. [Google Scholar] [CrossRef]
- Miao, L.; Zhang, Q.; Wang, S.; Li, B.; Wang, Z.; Zhang, S.; Zhang, M.; Peng, Y. Characterization of EPS compositions and microbial community in an Anammox SBBR system treating landfill leachate. Bioresour. Technol. 2018, 249, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Yang, S.F. Influence of loosely bound extracellular polymeric substances (EPS) on the flocculation, sedimentation and dewaterability of activated sludge. Water Res. 2007, 41, 1022–1030. [Google Scholar] [CrossRef]
- Su, F.; Wang, Z.; Huang, T.; Zhang, H.; Zhang, H. Simultaneous removal of nitrate, phosphorous and cadmium using a novel multifunctional biomaterial immobilized aerobic strain Proteobacteria Cupriavidus H29. Bioresour. Technol. 2020, 307, 123196. [Google Scholar] [CrossRef]
- Li, X.; Lu, Y.; Luo, H.; Liu, G.; Zhang, R. Microbial stratification structure within cathodic biofilm of the microbial fuel cell using the freezing microtome method. Bioresour. Technol. 2017, 241, 384–390. [Google Scholar] [CrossRef]
- Hosseinzadeh, A.; Zhou, J.L.; Navidpour, A.H.; Altaee, A. Progress in osmotic membrane bioreactors research: Contaminant removal, microbial community and bioenergy production in wastewater. Bioresour. Technol. 2021, 330, 124998. [Google Scholar] [CrossRef]
- Ma, X.; Wang, X.; Liu, Y.; Gao, J.; Wang, Y. Variations in toxicity of semi-coking wastewater treatment processes and their toxicity prediction. Ecotoxicol. Environ. Saf. 2017, 138, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Hanada, A.; Kurogi, T.; Giang, N.M.; Yamada, T.; Kamimoto, Y.; Kiso, Y.; Hiraishi, A. Bacteria of the candidate phylum TM7 are prevalent in acidophilic nitrifying sequencing-batch reactors. Microbes Environ. 2014, 29, 353–362. [Google Scholar] [CrossRef]
- Zhao, J.; Li, Y.; Chen, X.; Li, Y. Effects of carbon sources on sludge performance and microbial community for 4-chlorophenol wastewater treatment in sequencing batch reactors. Bioresour. Technol. 2018, 255, 22–28. [Google Scholar] [CrossRef]
- Xing, W.; Li, D.; Li, J.; Hu, Q.; Deng, S. Nitrate removal and microbial analysis by combined micro-electrolysis and autotrophic denitrification. Bioresour. Technol. 2016, 211, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xing, D.; Ren, N. p-Nitrophenol degradation and microbial community structure in a biocathode bioelectrochemical system. RSC Adv. 2016, 6, 89821–89826. [Google Scholar] [CrossRef]
- Assress, H.A.; Selvarajan, R.; Nyoni, H.; Ntushelo, K.; Mamba, B.B.; Msagati, T.A. Diversity, co-occurrence and implications of fungal communities in wastewater treatment plants. Sci. Rep. 2019, 9, 14056. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Ma, X.; Yin, X.; Wu, C.; Wang, Z.; Wu, Y.; Zhao, Y.; Tian, Y. Difference and interplay of microbial communities, metabolic functions, trophic modes and influence factors between sludge and bulking agent in a composting matrix. Bioresour. Technol. 2021, 336, 125085. [Google Scholar] [CrossRef] [PubMed]




| Operation Time (d) | C/N Ratio | Aeration Rate (m3/h) | Aeration Time (h) | Settling Time (min) | Fermentation Liquid Amount |
|---|---|---|---|---|---|
| 1–5 | 13 | 20 | 6.5 | 30 | 5% |
| 6–10 | 6.5 | 20 | 7 | 20 | 10% |
| 11–15 | 6.5 | 20 | 7 | 10 | 15% |
| 16–20 | 6.5 | 20 | 7 | 10 | 20% |
| 21–25 | 6.5 | 20 | 7 | 10 | 25% |
| 26–30 | 6.5 | 20 | 7 | 10 | 30% |
| Sequencing Type | Forward Primer | Reverse Primer | ||
|---|---|---|---|---|
| 16S rDNA V3-V4 | 341F | ACTCCTACGGGAGGCAGCAG | 805R | GACTACHVGGGTATCTAATCC |
| ITS ITS1-ITS2 | ITS1 | CTTGGTCATTTAGAGGAAGTAA | ITS2 | GCTGCGTTCTTCATCGATGC |
| Index | Method/Instrument | Value |
|---|---|---|
| COD | Potassium dichromate method | 20,000 mg/L |
| pH | pH meter | 6.19 |
| Water | Karl Fischer | 96.5–98.5% |
| Total protein | UV-vis | 0.4–0.7% |
| Total sugar | UV-vis | 0.5–0.8% |
| Acetic acid | HPLC | 0.33 mg/kg |
| Lactate | HPLC | 0.07 mg/kg |
| Glycollic acid | HPLC | 0.16 mg/kg |
| l-alanine | LC-MS | 205.1 mg/kg |
| l-leucine | LC-MS | 186.2 mg/kg |
| l-valine | LC-MS | 117.8 mg/kg |
| l-isoleucine | LC-MS | 116.3 mg/kg |
| l-threonine | LC-MS | 79.4 mg/kg |
| l-proline | LC-MS | 31.8 mg/kg |
| l-phenylalanine | LC-MS | 73.5 mg/kg |
| l-methionine | LC-MS | 25.5 mg/kg |
| l-aspartate | LC-MS | 21.5 mg/kg |
| l-glycine | LC-MS | 13.7 mg/kg |
| l-tryptophan | LC-MS | 12.3 mg/kg |
| K | ICP-OES | 1356.0 mg/kg |
| Ca | ICP-OES | 974.0 mg/kg |
| Mg | ICP-OES | 541.7 mg/kg |
| Na | ICP-OES | 66.7 mg/kg |
| Si | ICP-OES | 43.8 mg/kg |
| Cu | ICP-OES | 21.9 mg/kg |
| Fe | ICP-OES | 16.2 mg/kg |
| Al | ICP-OES | 7.2 mg/kg |
| Treatment | NirS (Average Copies) | NirK (Average Copies) |
|---|---|---|
| Control | 26,316.472 | 15,039.141 |
| Fermentation broth | 6724.9544 | 3912.6366 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, R.; Zhang, W.; Bi, X.; Jin, Y.; Yang, Y. Microbiological Mechanisms for Nitrogen Removal Using Anaerobic Fermentation Liquid from Spent Mushroom Substrates as a Carbon Source. Water 2023, 15, 3530. https://doi.org/10.3390/w15203530
Chen R, Zhang W, Bi X, Jin Y, Yang Y. Microbiological Mechanisms for Nitrogen Removal Using Anaerobic Fermentation Liquid from Spent Mushroom Substrates as a Carbon Source. Water. 2023; 15(20):3530. https://doi.org/10.3390/w15203530
Chicago/Turabian StyleChen, Ruihuan, Weihong Zhang, Xiaohui Bi, Yan Jin, and Yunlong Yang. 2023. "Microbiological Mechanisms for Nitrogen Removal Using Anaerobic Fermentation Liquid from Spent Mushroom Substrates as a Carbon Source" Water 15, no. 20: 3530. https://doi.org/10.3390/w15203530
APA StyleChen, R., Zhang, W., Bi, X., Jin, Y., & Yang, Y. (2023). Microbiological Mechanisms for Nitrogen Removal Using Anaerobic Fermentation Liquid from Spent Mushroom Substrates as a Carbon Source. Water, 15(20), 3530. https://doi.org/10.3390/w15203530