Abstract
Ag nanoclusters (AgNCs) have gained widespread applications in recent years due to their excellent antimicrobial efficacy and distinctive molecule-like characteristics. However, concerns about their potential effects on environmental and human health have been raised. Despite the fact that abundant research has been carried out to examine the possible ecotoxicology of AgNCs in a variety of living organisms, these studies have mostly concentrated on the toxicology of individual organisms and only a few have attempted to look into the impact of AgNCs across the aquatic food chain. This work evaluated the transcriptome level genotoxicity of AgNCs and their degraded Ag ions in two model species food chains: the aquatic green algae Scenedesmus obliquus and the invertebrate Daphnia magna. Daphnia magna’s digestive system and glycerophospholipid metabolism were hindered after feeding on Ag-containing algae as a result of down-regulation of the crucial gene PLA2G(SPLA2) that codes for secretory phospholipase A2. Our research also showed that the genotoxicity of AgNCs to Daphnia magna was mediated by a synergic interaction between the particulate form of AgNCs and their degraded Ag ions. The current work offers a fresh viewpoint on the mechanisms underlying AgNCs’ harmful effects and the possible ecological concern that metal-based nanoparticles provide to aquatic life.
1. Introduction
When compared to their larger counterparts, silver nanoparticles (AgNPs, >2 nm), silver nanoclusters (denoted AgNCs) with particle sizes below 2 nm, typically composed of several to one hundred silver atoms, exhibit obviously different physical and chemical properties [1,2,3]. Due to the significant quantum size confinement of free electrons in this sub-2 nm size range, AgNCs possess discrete electronic states and display distinctive molecule-like properties, such as intensive luminescence, well-defined molecular structure, quantized charge and HOMO–LUMO transitions [2,4,5,6,7,8]. In particular, the unique properties of AgNCs coupled with their ultra-small size and good biocompatibility have enabled them to emerge as promising functional materials for a wide variety of biomedical applications, especially as effective antimicrobial agents and as optical probes for bioimaging and biolabeling applications [2,6,9,10,11,12]. AgNCs will unavoidably be released into the environment as a result of their extensive use, especially in the aquatic environment. Given the growing public concern over the possible threats they pose to human health and living things, it is therefore worthwhile to investigate the potential ecotoxicology of AgNCs.
Although numerous studies have been conducted to explore the potential ecotoxicology of large AgNPs in a variety of microorganisms [12,13], algae [14], plants [15], invertebrates [16,17] and vertebrates [18,19], these studies have largely concentrated on the effect on individual organisms, and only a few have described the toxicology of ultra-small AgNCs. The trophic toxicology of AgNCs to the aquatic food chain is a significant issue that warrants further research because several previous studies have shown that nanoparticles, including AgNPs, could potentially accumulate in aquatic ecosystems and eventually cause harm to the “top-level” species [20,21,22]. As is generally known, freshwater algae, a great example of a unicellular organism, play a significant role in both photosynthesis and the cycle of carbon dioxide on a global scale [23,24]. Daphnia magna (D. magna) represents a model zooplankton in the aquatic environment with which to illuminate the toxic mechanisms of AgNPs in environmental assessments [17,25]. According to a previous study [22], freshwater algae have the capacity to internalize AgNPs, which may then be transmitted to higher tropic-level organisms like Daphnia magna. Therefore, we believe that through the food chain, algal-borne AgNCs may ultimately be hazardous to Daphnia magna.
In the past few years, many acute toxicity tests (short-term exposure and at high Ag concentrations) have been used to investigate the effects of AgNPs on freshwater organisms; however, environmental exposures to AgNPs are more likely to be long-term and at low concentrations [26,27,28,29]. In other words, testing for chronic toxicity is more relevant than testing for acute toxicity when evaluating the environmental consequences of AgNPs. Environmental conditions are also dynamic and complicated, and they are not easily reversible [26,27]. As a result, it is challenging to conduct an experiment in the lab to examine the impacts of AgNPs on the environment. If research on the long-term impacts of exposure to AgNPs could be done at the molecular level (for example, utilizing transcriptomic approaches), then the results should be able to provide useful information [30]. The biomolecular impacts of AgNPs and the released Ag ions on aquatic organisms have not received much attention until now. Transcriptomic analysis has recently been used to study, in great detail, small variations in gene expression in aquatic organisms after exposure to AgNPs. These investigations have provided the foundation for a better understanding of the molecular mechanisms behind the toxicity caused by AgNCs [14,31,32].
The goal of the current work is to use cutting-edge transcriptome technology to examine the molecular harmful effects of AgNCs carried by algae on the aquatic invertebrate Daphnia magna. Our results showed that Daphnia magna exposed to AgNCs carried by algae showed blatant changes in gene expression across the food chain. We suggest that the current study offers a fresh viewpoint on how to identify AgNCs-mediated toxicity as well as an early warning system for environmental risk brought on by the accumulation of other metal nanoparticles in aquatic life.
2. Materials and Methods
2.1. Materials
Prior to all experiments, all glassware and pipet tips were soaked in 0.05 M nitric acid for 24 h and then rinsed with Milli-Q water (18.2 MΩ). Silver nitrate (99.9999%), sodium borohydride (NaBH4), L-glutathione reduced (GSH) and L-cysteine (≥99%) were purchased from Sigma-Aldrich. The other reagents (analytical or ultrapure grades) were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). All solutions used in this study were prepared in Milli-Q water. Ultra-small AgNCs were prepared according to the method of Yuan [9,33]. The AgNC solutions were subsequently stored at 4 °C for later use. Further details on the characterization of AgNCs and their dissolution in exposure water are available in our previous work [34,35].
2.2. Algae Culture and Nanosilver Exposure Conditions
Scenedesmus obliquus (No. FACHB-417), was obtained from the Institute of Hydrobiology of the Chinese Academy of Sciences (Wuhan, China) and subsequently cultured in SE medium (Wuhan, China, Supplementary Materials Table S1) under controlled conditions (12:12 h light/dark photoperiod; 25 °C; illumination 2500~3000 lx; artificial shaking three times every day) in SE. To avoid biological contaminations, all glassware and algae culture media were sterilized in a SANYO sterilizing oven (Osaka, Japan) (121 °C, 30 min).
For algae exposure experiments, in consideration of the concentration level of AgNPs (0.1~146 μg/L) in the aquatic environment [28], algae in the log-phase of growth (5~10 × 105 cells/mL) were exposed to AgNC concentrations (according to the concentration of Ag atoms) of 0.00, 135 and 135 μg/L (containing 0.5 mM L-cysteine) and silver ions (5.0, 10.0 and 20.0 μg/L), respectively. The addition of L-cysteine, at a concentration of 0.5 mM, was sufficient to inhibit the dissolution of Ag+ into SE medium. According to our previous study, the concentration level of Ag+ released from AgNCs (μg/L) is almost equal to that of silver ions (5.0, 10.0 and 20.0 μg/L) dissolved from silver nitrate [34]. The control experiments were exposed to SE medium (without silver). The exposure experiments were carried out in triplicate.
2.3. Daphnia Magna Culture and Ag Exposure
Adult Daphnia magna were obtained commercially (Institute of Hydrobiology of Chinese Academy of Sciences) and grown in standard dilution water medium (SDW medium: 294 mg/L CaCl2·2H2O, 123 mg/L MgSO4·7H2O, 65 mg/L NaHCO3, 6.3 mg/L KCl). The growth conditions were the same as those of Scenedesmus obliquus mentioned above. Prior to the silver exposure experiments, the Daphnia magna were fed with SE medium-grown Scenedesmus obliquus (containing no AgNCs or AgNO3).
After exposure to Ag for 96 h, Scenedesmus obliquus was harvested on a 0.45 μm filter membrane for feeding to Daphnia magna. The algae cells were washed with Milli-Q water three times to remove remanent AgNCs or AgNO3 on the cell surface. The cells were then resuspended in SDW medium to reach a cell density of ~5 × 105 cells/mL. Finally, 100 mL of SDW medium containing algae cells (~5 × 105 cells/mL) was introduced into eighteen beakers (Figure S1, one control and five AgNCs or AgNO3 treatments). Algae cells were quickly consumed by Daphnia magna within 30 min. Considering the extremely small volume of Daphnia magna, each beaker contained ~100 Daphnia magna in order to ensure adequate RNA samples. The volume of SDW medium in each breaker remained relatively constant (1000 mL) through the addition of fresh medium. The experiment was done in triplicate.
2.4. RNA Sequencing
According to our previous studies using confocal microscopic images and TEM images, AgNCs are indeed internalized by the algae cells [34,35]. At the end of 48 h of exposure to algae cells grown with or without AgNCs/AgNO3, Daphnia magna fed with green algae but without silver were used as controls (sample A). Daphnia magna fed with green algae treated with 135 μg/L r-AgNCs but without 0.5 mM L-cysteine (sample B), and fed with green algae treated with the 135 μg/L r-AgNCs (with 0.5 mM L-cysteine) (sample C) and those fed with green algae treated with 10 μg/L silver ions (sample D) were collected and washed three times with Milli-Q water. To obtain sufficient RNA, the samples of each treatment were mixed and then immediately frozen in liquid nitrogen. Total RNA extraction of Daphnia magna was performed according to the manufacturer’s instructions for trizol reagent (Invitrogen, Carlsbad, CA, USA). The concentration and integrity of RNA was detected by a Nanodrop 2000 Spectrophotometer and a 2100 Bioanalyser (Agilent, Palo Alto, CA, USA), respectively. After the cDNA library of each treatment was prepared by polymerase chain reaction, the sequencing libraries were gradually diluted and quantified to 4–5 pM and sequenced on the Illumina NextSeq 500 platform.
2.5. Transcriptomic Analysis
A pair-end sequencing strategy was employed to generate RNA-Seq raw reads with an average length of 150 bp. In order to better conduct de novo transcriptome assembly, the raw reads were filtered to obtain high-quality reads by removing the adapter sequences and low-quality bases at the 3′ end (Q < 20) using the FastQC program(version: 0.11.2). Quality reads were converted into contigs, established de Bruijn graphs and optimized de Bruijn graphs to obtain the final transcripts, using the Trinity programme (http://trinityrnaseq.sf.net) (version: r20140717, accessed on 10 July 2023). The transcripts were subsequently subjected to the NCBI basic local alignment search tool (BLAST) (http://www.ncbi.nlm.nih.gov/) (version: 2.2.30+, accessed on 15 July 2023) against the NCBI non-redundant protein database (NR) (http://www.ncbi.nlm.nih.gov/) (version: GRCh36, accessed on 12 July 2023) (cut-off E-value < 10−5). The top-hit transcripts were identified as unigenes. For functional annotation of all the unigenes, we searched them against the public databases using NCBI BLAST, including the NCBI non-redundant protein database, the Gene Ontology (GO) database (http://geneontology.org/) (version: Conesa A and Gotz, accessed on 15 July 2023), Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg) (version: 81.0, accessed on 10 July 2023) and the Evolutionary genealogy of genes: non-supervised Orthologous Groups (eggNOG) database (http://eggnog.embl.de/) (version: 4.0.beta/, accessed on 12 July 2023).
To analyse the differentially expressed unigenes derived from Daphnia magna after exposure to algal-borne AgNCs and Ag NO3 compared to the controls, the number of reads per kilobase of the exon model per million mapped reads value (RPKM) was used to normalize the gene expression at the transcriptional level. Differentially expressed unigenes were identified by using the DESeq program (http://www-huber.embl.de/users/anders/DESeq) (version: 1.18.0, accessed on 10 July 2023). The false discovery rate (FRD, <0.05) method was introduced to determine the threshold of the p-value in multiple tests and further judge the significance of the difference in gene expression. When greater than a two-fold change (absolute value of log2 fold change > 1) and p-value < 0.05 in gene expression were observed in our study, unigenes were considered to have significant differential expression. KEGG Orthology (KO) metabolic pathways analysis of the differentially expressed unigenes was carried out on the public website (http://www.genome.jp/kegg/tool/map_pathway2.html) (version: 2015/05/22, accessed on 18 July 2023). GO and KEGG enrichment analysis of differentially expressed unigenes were conducted using the Goatools software (https://github.com/tanghaibao/GOatools) (version: v1.2.3, accessed on 10 July 2023) and the KOBAS software (http://kobas.cbi.pku.edu.cn/home.do) (version: 2.0, accessed on 15 July 2023), respectively. Cluster analysis of differential unigenes was performed by means of Cluster3.0/TreeView programme (http://bonsai.hgc.jp/~mdehoon/software/cluster/manual/index.html) (version: v1.2.0, accessed on 10 July 2023).
3. Results and Discussion
3.1. Sequencing and De Novo Transcriptome Assembly
Before cDNA libraries were constructed in our study, the concentration and integrity of total RNA were firstly examined by a UV-vis spectrophotometer (Nanodrop 2000, Waltham, MA, USA) and agarose gel electrophoresis, respectively (Table S2 and Figure S2). To obtain the Daphnia magna transcriptome expression profile after exposure to algal-borne AgNCs and Ag NO3, four independent cDNA libraries from various Daphnia magna treatments (samples A, B, C and D) were sequenced using the Illumina NextSeq 500 platform. After sequencing and data filtering, the raw reads and high-quality reads of various samples are presented in Table S3. Subsequently, transcript de novo assembly for quality reads was carried out by Trinity software(version: r20140717). A summary of all contigs, transcripts and unigene assemblies is shown in Table S3. A total of 32,009 unigenes were obtained with a maximum length of 237,122 bp, an average length of 923 bp and an N50 of 1517 bp, and these unigenes were further used to perform the analysis of differentially expressed genes at the transcriptional level. The length distribution of unigenes is displayed in Figure S3. The lengths of unigenes were ranged from 201 bp to 23,712 bp, and the most abundant unigenes were clustered in a group 200–299 bp in length. For further functional prediction and classifications, all unigenes were searched against several public databases, including the NCBI NR database, the GO database, eggNOG database and KEGG database, among others. The results of the annotation of the Daphnia magna transcriptome are summarized in Table S4, Figures S4–S6. Among the 32,009 unigenes, 32,009, 29,260 and 26,943 had significant hits in the NR, eggNOG and SwissProt databases, respectively.
3.2. Differential Expression Gene Analysis
A statistical analysis of differentially expressed genes between the three silver treatments and the controls was conducted using the gene expression data. Volcano plots showed the distribution trends of differential expression genes (red spots) and genes with no differential expression (blue spots) (Figure S7). It is quite clear that the expression levels of many differentially expressed genes derived from Daphnia magna after feeding with green algae treated with three silver (r-AgNCs, r-AgNCs containing L-cysteine and silver ions) were up- and down-regulated compared to the control treatment, which further demonstrates that r-AgNCs and their dissolved silver ions cause a toxic effect on the gene expression of Daphnia magna. The number of up- and down-regulated differentially expressed genes obtained per treatment group was coincident with the overlap between the differentially expressed genes for each silver treatment (Figure 1a,b). Figure 1c illustrates the heatmap of differentially expressed genes between the various algal-borne AgNC treatments and the control. The gene expression levels are visualised by using a plot colour ranging from green (low expression) to red (high expression), which demonstrates the significant differences in the level of gene expression for differentially expressed genes between each algal-borne AgNCs treatment and the control. The condition tree also shows that the gene expression profiles for differentially expressed genes were consistent across particle-specific AgNCs and their dissolved silver ions.
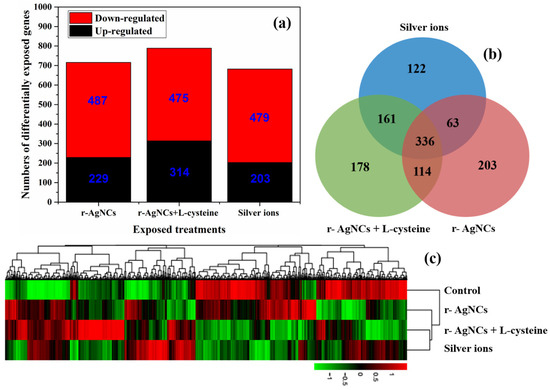
Figure 1.
Differentially expressed genes of Daphnia magna resulting from a diet of green algae exposed to three silver treatments (r-AgNCs, r-AgNCs + L-cysteine and silver ions) compared to the controls (Daphnia magna on a diet of green algae only). (a) Colour bars indicate the number of (red) up- and (blue) down-regulated genes derived from Daphnia magna (|fold change| > 2 and p-value < 0.05). (b) Venn diagram showing numbers of differentially expressed genes from Daphnia magna after the diet of green algae (|fold change| > 2 and p-value < 0.05). (c) Heatmap of differentially expressed genes (|fold change| > 2 and p-value < 0.05) derived from Daphnia magna as a result of the diet of green algae. Colour bars indicate the gene expression levels: (red) up-regulated genes and (green) down-regulated genes.
3.3. Enrichment Analysis of the Differential Expression Genes
The biological significance of the changes in gene expression of differentially expressed genes in Daphnia magna fed on green algae treated with silver has been investigated by using enrichment analysis in GO terms (denoted as GOSlim in our study, Figure S8) and the KEGG metabolic pathway (Figure 2). Gene Ontology (GO) is a standardized gene functional classification system including the terms for biological processes, cellular components and molecular functions. The KEGG metabolic pathway, which represents our knowledge of the molecular interaction and reaction networks of Daphnia magna, can help further our understanding of the biological functions of differentially expressed genes and how these genes interact. Enrichment analysis of differentially expressed genes (up- and down-regulated unigenes) was conducted to gain an overall perspective of the biological effects of each exposure to the green algae diet. The most significantly typical GO terms (structural molecule activity and external encapsulating structure) and KEGG pathways (lipid metabolism and digestive system) remained common across all three different algae diets, suggesting that there was a very significant overlap in the gene expression of differentially expressed unigenes in response to the three kinds of silver treatments. Based on this overlap, it is likely that the toxicity of AgNCs to Daphnia magna was predominantly due to the combined action between the AgNCs and their released silver ions.
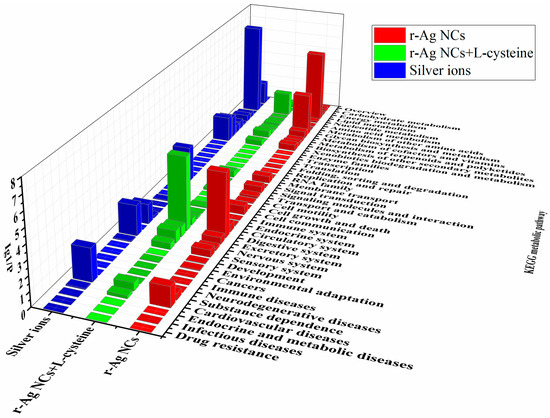
Figure 2.
KEGG enrichment analysis of differentially expressed genes of Daphnia magna from the diet of green algae treated with silver (AgNCs, AgNCs with L-cysteine and Ag ions). The enrichment analysis was conducted between silver treatments and controls (p values < 0.05) after 48 h of exposure, using all expressed unigenes as a background.
Scenedesmus obliquus, a freshwater green algae, is a well-known primary producer in the aquatic ecosystem and a source of food for primary or higher trophic-level consumers, including the freshwater cladoceran Daphnia magna. As is shown in Figure 2, the enrichment of differentially expressed unigenes in the KEGG metabolic pathway (lipid metabolism and digestive system) was more significant than other metabolic pathways (p-value < 0.05), which demonstrates that the processes of metabolism of Daphnia magna were inhibited at the molecular level when fed with the three different diets of green algae. Furthermore, Ig(1/p) of the terms of external encapsulating structure and structural molecular activity were larger than that of other terms of GO enrichment (Figure S8, adjusted p < 0.05), indicating that the differentially expressed genes coding the cellular components (e.g., cell membrane) and the biological macromolecules in the body (e.g., metabolic enzymes) were regulated after exposure to algal-borne AgNCs and Ag ions, and it is anticipated that this regulation was also associated with the process of metabolism in Daphnia magna. As a result, the AgNC-produced Ag ions and the diets of green algae that the Daphnia were treated with had a greater effect on their metabolism.
3.4. KO Metabolic Pathway Analysis of the Differential Expression Genes
As shown in Figure S1, after Daphnia being fed the diet of algae treated with AgNCs and Ag ions for 48 h, the content of Scenedesmus obliquus in the beaker decreased obviously, indicating that Daphnia magna can feed on Scenedesmus obliquus. Therefore, it is essential to investigate the effects of the diet of algae on the digestive system of Daphnia magna, especially with respect to the subtle differences in gene expression. Differentially expressed genes in the digestive system of Daphnia magna after being fed a diet of algae treated with silver for 48 h are listed in Table S5. The gene expression analysis demonstrated that many of the unigenes encoding for key enzymes and protein subunits of the digestive system were up- and down-regulated compared to the controls. Among all these genes, PLA2G and SPLA2 (gene ID: c75537_g1_i1) were especially sensitive to AgNC and Ag+ ion exposure. These genes encode for secretory phospholipase A2, a key enzyme for fat digestion and absorption, and they were down-regulated at the transcriptome level. Interestingly, PLA2G and SPLA2 (gene ID: c22033_g1_i2) also encode for secretory phospholipase A2 and they were also down-regulated as a result of exposure to AgNCs and Ag+ ions, except for AgNCs + L-cysteine, which suggests that a protein, such as secretory phospholipase A2, can be coded by different genes. Since the digestive system of Daphnia magna relies heavily on the lipase secreted by the hepatopancreas (Figure 3a), a down-regulation of the genes PLA2G and SPLA2, which encode phospholipase A2, suggests that a diet of algae treated with AgNCs and the subsequently released Ag ions may have an impact on how fats (such as glycerophospholipid) are metabolised.
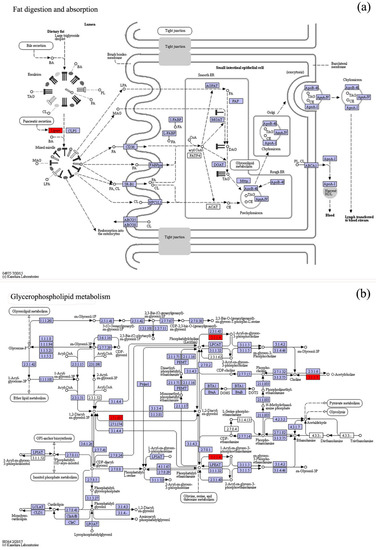
Figure 3.
Effects of the diets of algae treated with AgNCs and silver ions on differentially expressed genes related to glycerophospholipid metabolism and fat digestion and absorption of Daphnia magna. Shades of red, light blue and white represent down-regulation of target unigenes, no differential expression of unigenes (background genes) and unigene products not involved in Daphnia magna, respectively. (a) Effects of the diets of algae treated with AgNCs (without L-cysteine) for 48 h on fat digestion and absorption of Daphnia magna. (b) Effects of the diets of algae treated with AgNCs (without L-cysteine) for 48 h on glycerophospholipid metabolism of Daphnia magna. Based on the principles of classification of the Enzyme Commission, the identifier of the enzyme involved in the pathway was determined by the number in the boxes. Illustrations of the fat digestion and absorption and glycerophospholipid metabolism of Daphnia magna as a result of the diets of algae treated with AgNCs (containing L-cysteine) and silver ion are available in Table S5. Metabolic pathways were obtained through the KEGG annotation of differentially expressed unigenes.
Apart from the effect on the digestive system of Daphnia magna, the diet of algae treated with AgNCs and their released Ag ions can also lead to changes in glycerophospholipid metabolism and showed similarly significant alterations in the expression levels of differentially expressed genes (Table S5, gene ID: c75537_g1_i1). Glycerophospholipid is one of the most important membrane lipids and can be hydrolyzed to produce aliphatic acid and glycerol by the catalysis of secretory phospholipase A2 (Figure 3b). Aliphatic acid will further produce acetyl coenzyme A by β-oxidation and finally enter the tricarboxylic acid cycle (TCA cycle). Furthermore, intermediate products of β-oxidation (e.g., arachidonic acid metabolism, linoleic acid and alpha-linolenic acid) were also regulated by secretory phospholipase A2 (Table S5). The down-regulation of key enzymes, such as secretory phospholipase A2 coded by gene PLA2G (SPLA2), indicates that glycerophospholipid metabolism was probably also regulated as a result of the diet of algae treated with AgNCs and their released Ag ions, resulting in a decrease in the lipid metabolic capacity of Daphnia magna. As a model zooplankton in an aquatic ecosystem, after exposure to algae-born silver, Daphnia magna can adjust its metabolic processes to adapt to the external environmental conditions. However, metabolic processes were regulated by enzyme activity and gene expression in the ordinary course of events. From the analyses, it can be concluded that the gene PLA2G (SPLA2), coding for the key enzyme secretory phospholipase A2, can not only be regarded as a biomarker but also be applied to the assessment of the aquatic food chain (e.g., Scenedesmus obliquus and Daphnia magna) exposed to metal-based nanoparticles.
Although the released Ag ions from AgNCs help to explain some of their toxicity, it is still not obvious whether AgNCs itself is the root of the problem. For example, Yuan et al. reported that AgNCs possessed superior antimicrobial properties against pathogenic bacteria (for example, P. aeruginosa), via the generation of intracellular reactive oxygen species (ROS) [33]. However, our previous work demonstrated that no significant ROS were induced in Scenedesmus obliquus treated with various concentrations of AgNCs and Ag ions for 96 h compared to the controls, suggesting that the AgNC- or Ag-ion-mediated toxicity to algae cells could not be attributable to ROS accumulation [34,35]. Another study reported that AgNCs had much higher antimicrobial activities towards both Gram-positive bacteria (B. subtilis, and S. aureus) and Gram-negative bacteria (P. aeruginosa, and E. coli) owing to abundantly released Ag ions on the surface [36]. With regard to the effect of their larger counterpart, AgNPs, Helen et al. reported that a 15k oligonucleotide microarray for Daphnia magna was used to differentiate between particle-specific and ionic silver toxicity [32]. AgNPs and AgNO3 were found to have different gene expression profiles, which may indicate their different modes of toxicity. AgNPs and AgNO3 can both affect key biological processes, including protein metabolism and signal transduction, as well as developmental processes (such as sensory development). A previous investigation of the toxicity of AgNPs in comparison with AgNO3 in two aquatic organisms, green algae Chlamydomonas reinhardtii and the invertebrate Daphnia magna, revealed that Daphnia magna could equally accumulate AgNPs and AgNO3 after feeding on Ag-containing algae, and the finding suggested that AgNO3 was more toxic than nano-Ag [22]. Based on these previous studies and the significant changes in the expression levels of differentially expressed genes in Daphnia magna after exposure for 48 h to Ag-containing algae, we can confirm that the AgNCs and their released ionic-form silver have a synergistic effect on the invertebrate organism Daphnia magna. Whether the AgNCs entering the Daphnia magna cells will gradually degrade into silver ions and further cause its toxicity deserves further investigation. However, Chen and colleagues revealed that the toxicity of AgNPs on human monocytes (THP-1) is largely due to the chemical transformation of particulate silver from elemental silver (Ag0)n to Ag+ ions with the help of synchrotron radiation beam transmission X-ray microscopy (SR-TXM) and SR-X-ray absorption near edge structure (SR-XANES) spectroscopy, suggesting that this could be a potential mechanism [37].
4. Conclusions
In conclusion, we showed that Daphnia magna fed with Scenedesmus obliquus contained Ag-experienced transcriptome alterations as a result of exposure to AgNCs. AgNCs in their particulate form and the degraded Ag ions that they produce worked in concert to modify Daphnia magna’ transcriptiome. Because the gene PLA2G (SPLA2), which codes for secretory phospholipase A2, was downregulated, the digestive process and glycerophospholipid metabolism were hindered. Additionally, it needs to be thoroughly researched in the future if the AgNCs that enter Daphnia magna cells gradually break down into Ag ions and further mediate AgNCs toxicity. The current study not only provides a different method for identifying AgNCs-mediated toxicity, but it also adds to our knowledge of early aquatic ecological risk mediated by other metal nanoparticles.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/w15183172/s1. Figure S1: The toxicity test of Daphnia magna through the diet of Scenedesmus obliquus treated with silver nanoparticles, and two photos represented the beginning (a) and end (b) of the toxicity test. Figure S2: The total RNA checked by agarose gel electrophoresis. Figure S3: Histogram of length distribution of contiges (a), transcripts (b) and unigenes (c). Figure S4: The Gene Ontology (GO) annotations of unigenes. Figure S5: The evolutionary genealogy of genes: non-supervised orthologous groups (eggNOG) annotations of unigenes. Figure S6: The Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations of unigenes. Figure S7: The “volcano plot” pictures with log2(fold change) plotted versus −log10(p-value) of differentially expressed genes derived from Daphnia magna. Figure S8: GO enrichment analysis of differentially expressed genes of Daphnia magna from the diet of green algae. Table S1: The composition of SE culture medium. Table S2: The total RNA was checked using NanoDrop 2000 UV-Vis Spectrophotometer. Table S3: Illumina de novo assembly statistics of transcriptomic profiles of Daphnia magna from green algae diet. Table S4: Annotation of unigenes of Daphnia magna transcriptome. Table S5: List of differentially expressed genes for the digestive system and lipid metabolism of Daphnia magna after the diets of algae treated with silver for 48 h.
Author Contributions
Conceptualization, L.Z. and H.T.; methodology, H.T.; validation, L.Z. and H.T.; resources, H.T.; writing—original draft preparation, L.Z.; writing—review and editing, L.Z. and H.T.; visualization, L.Z.; funding acquisition, L.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Water Pollution Control and Treatment Projects of the Ministry of Science and Technology of China (2014ZX07206001), the National Natural Science fund of China (41272381) and the Campus for Research Excellence and Technological Enterprise (CREATE) programme between Singapore and Shanghai Jiao Tong University.
Data Availability Statement
Data are available from the corresponding author on request.
Acknowledgments
We appreciate Personal Biotechnology Co., Ltd. (Shanghai, China) for their supports in Transcriptome Sequencing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lu, Y.Z.; Chen, W. Sub-nanometre sized metal clusters: From synthetic challenges to the unique property discoveries. Chem. Soc. Rev. 2012, 41, 3594–3623. [Google Scholar] [CrossRef] [PubMed]
- Diez, I.; Ras, R.H. Fluorescent silver nanoclusters. Nanoscale 2011, 3, 1963–1970. [Google Scholar] [CrossRef] [PubMed]
- Shang, L.; Dong, S.J.; Nienhaus, G.U. Ultra-small fluorescent metal nanoclusters: Synthesis and biological applications. Nano Today 2011, 6, 401–418. [Google Scholar] [CrossRef]
- Xu, H.X.; Suslick, K.S. Water-soluble fluorescent silver nanoclusters. Adv. Mater. 2010, 22, 1078–1082. [Google Scholar] [CrossRef] [PubMed]
- Guidez, E.B.; Aikens, C.M. Theoretical analysis of the optical excitation spectra of silver and gold nanowires. Nanoscale 2012, 4, 4190–4198. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Yeow, T.J.; Zhang, Q.; Lee, J.Y.; Xie, J.P. Highly luminescent Ag+ nanoclusters for Hg2+ ion detection. Nanoscale 2012, 4, 1968–1971. [Google Scholar] [CrossRef]
- Zheng, K.; Yuan, X.; Kuah, K.; Luo, Z.; Yao, Q.; Zhang, Q.; Xie, J.P. Boiling water synthesis of ultrastable thiolated silver nanoclusters with aggregation-induced emission. Chem. Commun. 2015, 51, 15165–15168. [Google Scholar] [CrossRef]
- Murray, R.W. Nanoelectrochemistry: Metal Nanoparticles, Nanoelectrodes, and Nanopores. Chem. Rev. 2008, 108, 2688–2720. [Google Scholar] [CrossRef]
- Yuan, X.; Tay, Y.; Dou, X.; Luo, Z.; Leong, D.T.; Xie, J.P. Glutathione-protected silver nanoclusters as cysteine-selective fluorometric and colorimetric probe. Anal. Chem. 2013, 85, 1913–1919. [Google Scholar] [CrossRef]
- Guo, W.W.; Yuan, J.P.; Dong, Q.Z.; Wang, E. Highly sequence-dependent formation of fluorescent silver nanoclusters in hybridized DNA duplexes for single nucleotide mutation identification. J. Am. Chem. Soc. 2010, 132, 932–934. [Google Scholar] [CrossRef]
- Yeh, H.C.; Sharma, J.; Han, J.J.; Martinez, J.S.; Werner, J.H. A DNA–silver nanocluster probe that fluoresces upon hybridization. Nano Lett. 2010, 10, 3106–3110. [Google Scholar] [CrossRef]
- Zheng, C.R.; Li, S.; Ye, C.; Li, X.; Zhang, C.; Yu, X. Particulate respirators functionalized with silver nanoparticles showed excellent real-time antimicrobial effects against pathogens. Environ. Sci. Technol. 2016, 50, 7144–7151. [Google Scholar] [CrossRef]
- Xiu, Z.M.; Zhang, Q.B.; Puppala, H.L.; Colvin, V.L.; Alvarez, P.J. Negligible particle-specific antibacterial activity of silver nanoparticles. Nano Lett. 2012, 12, 4271–4275. [Google Scholar] [CrossRef]
- Leclerc, S.; Wilkinson, K.J. Bioaccumulation of nanosilver by Chlamydomonas reinhardtii-nanoparticle or the free ion? Environ. Sci. Technol. 2014, 48, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Kaveh, R.; Li, Y.S.; Ranjbar, S.; Tehrani, R.; Brueck, C.L.; Van Aken, B. Changes in Arabidopsis thaliana gene expression in response to silver nanoparticles and silver ions. Environ. Sci. Technol. 2013, 47, 10637–10644. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.S.; Guagliardo, P.; Jiang, H.B.; Wang, W.X. Intra- and intercellular silver nanoparticle translocation and transformation in Oyster gill filaments: Coupling nanoscale secondary ion mass spectrometry and dual stable isotope tracing study. Environ. Sci. Technol. 2021, 55, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Gray, E.P.; Coleman, J.G.; Bednar, A.J.; Kennedy, A.J.; Ranville, J.F.; Higgins, C.P. 2013. Extraction and analysis of silver and gold nanoparticles from biological tissues using single particle inductively coupled plasma mass spectrometry. Environ. Sci. Technol. 2013, 47, 14315–14323. [Google Scholar] [CrossRef]
- Liu, N.; Li, Y.; Liu, L.H.; Liu, X.L.; Yin, Y.G.; Qu, G.B.; Shi, J.B.; Song, M.Y.; He, B.; Hu, L.G.; et al. Administration of silver nasal spray leads to nanoparticle accumulation in rat brain tissues. Environ. Sci. Technol. 2022, 56, 403–413. [Google Scholar] [CrossRef]
- Wang, M.Y.; Wang, W.X. Nanoscale whole-body expansion microscopy revealed the early skeletal developmental malformation induced by silver nanoparticles. Environ. Sci. Technol. Lett. 2023, 10, 471–477. [Google Scholar] [CrossRef]
- Farre, M.; Gajda-Schrantz, K.; Kantiani, L.; Barcelo, D. Ecotoxicity and analysis of nanomaterials in the aquatic environment. Anal. Bioanal. Chem. 2009, 393, 81–95. [Google Scholar] [CrossRef]
- Gupta, G.S.; Kumar, A.; Senapati, V.A.; Pandey, A.K.; Shanker, R.; Dhawan, A. Laboratory scale microbial food chain to study bioaccumulation, biomagnification, and ecotoxicity of cadmium telluride quantum dots. Environ. Sci. Technol. 2017, 51, 1695–1706. [Google Scholar] [CrossRef] [PubMed]
- McTeer, J.; Dean, A.P.; White, K.N.; Pittman, J.K. Bioaccumulation of silver nanoparticles into Daphnia magna from a freshwater algal diet and the impact of phosphate availability. Nanotoxicology 2014, 8, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Quigg, A.; Chin, W.-C.; Chen, C.-S.; Zhang, S.; Jiang, Y.; Miao, A.J.; Schwehr, K.A.; Xu, C.; Santschi, P.H. Direct and Indirect Toxic Effects of Engineered Nanoparticles on Algae: Role of Natural Organic Matter. ACS Sustain. Chem. Eng. 2013, 1, 686–702. [Google Scholar] [CrossRef]
- Chisholm, S.W. Stirring times in the Southern Ocean. Nature 2000, 407, 685–687. [Google Scholar] [CrossRef]
- Yan, N.; Tang, B.Z.; Wang, W.X. In vivo bioimaging of silver nanoparticle dissolution in the gut environment of zooplankton. ACS Nano 2018, 12, 12212–12223. [Google Scholar] [CrossRef]
- Levard, C.; Hotze, E.M.; Lowry, G.V.; Brown, G.E., Jr. Environmental transformations of silver nanoparticles: Impact on stability and toxicity. Environ. Sci. Technol. 2012, 46, 6900–6914. [Google Scholar] [CrossRef]
- Lowry, G.V.; Gregory, K.B.; Apte, S.C.; Lead, J.R. Transformations of nanomaterials in the environment. Environ. Sci. Technol. 2012, 46, 6893–6899. [Google Scholar] [CrossRef]
- Yu, S.J.; Yin, Y.G.; Liu, J.F. Silver nanoparticles in the environment. Environ. Sci. Process. Impacts 2013, 15, 78–92. [Google Scholar] [CrossRef]
- Gil-Allue, C.; Schirmer, K.; Tlili, A.; Gessner, M.O.; Behra, R. Silver nanoparticle effects on stream periphyton during short-term exposures. Environ. Sci. Technol. 2015, 49, 1165–1172. [Google Scholar] [CrossRef]
- Ankley, G.T.; Daston, G.P.; Degitz, S.J. Toxicogenomics in Regulatory Ecotoxicology. Environ. Sci. Technol. 2006, 40, 4055–4065. [Google Scholar] [CrossRef]
- Chen, C.; Unrine, J.M.; Judy, J.D.; Lewis, R.W.; Guo, J.; McNear, D.H.; Tsyusko, O.V. Toxicogenomic responses of the model legume medicago truncatula to aged biosolids containing a mixture of nanomaterials (TiO2, Ag, and ZnO) from a pilot wastewater treatment plant. Environ. Sci. Technol. 2015, 49, 8759–8768. [Google Scholar] [CrossRef] [PubMed]
- Poynton, H.C.; Lazorchak, J.M.; Impellitteri, C.A.; Blalock, B.J.; Rogers, K.; Allen, H.J.; Loguinov, A.; Heckman, J.L.; Govindasmawy, S. Toxicogenomic responses of nanotoxicity in Daphnia magna exposed to silver nitrate and coated silver nanoparticles. Environ. Sci. Technol. 2012, 46, 6288–6296. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Setyawati, M.I.; Tan, A.S.; Ong, C.N.; Leong, D.T.; Xie, J.P. Highly luminescent silver nanoclusters with tunable emissions: Cyclic reduction–decomposition synthesis and antimicrobial properties. NPG Asia Mater. 2013, 5, e39. [Google Scholar] [CrossRef]
- Zhang, L.; He, Y.L.; Goswami, N.; Xie, J.P.; Zhang, B.; Tao, X.J. Uptake and effect of highly fluorescent silver nanoclusters on Scenedesmus obliquus. Chemosphere 2016, 153, 322–331. [Google Scholar] [CrossRef]
- Zhang, L.; Goswami, N.; Xie, J.P.; Zhang, B.; He, Y.L. Unraveling the molecular mechanism of photosynthetic toxicity of highly fuorescent silver nanoclusters to Scenedesmus obliquus. Sci. Rep. 2017, 7, 16432. [Google Scholar] [CrossRef]
- Yuan, X.; Setyawati, M.I.; Leong, D.T.; Xie, J.P. Ultrasmall Ag+-rich nanoclusters as highly efficient nanoreservoirs for bacterial killing. Nano Res. 2013, 7, 301–307. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, T.; Li, P.; Huang, W.; Tang, J.; Wang, P.; Liu, J.; Yuan, Q.; Bai, R.; Li, B.; et al. Use of Synchrotron radiation-analytical techniques to reveal chemical origin of silver-nanoparticle cytotoxicity. ACS Nano 2015, 9, 6532–6547. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).