Recent Advances in Bacterial Degradation of Hydrocarbons
Abstract
:1. Introduction
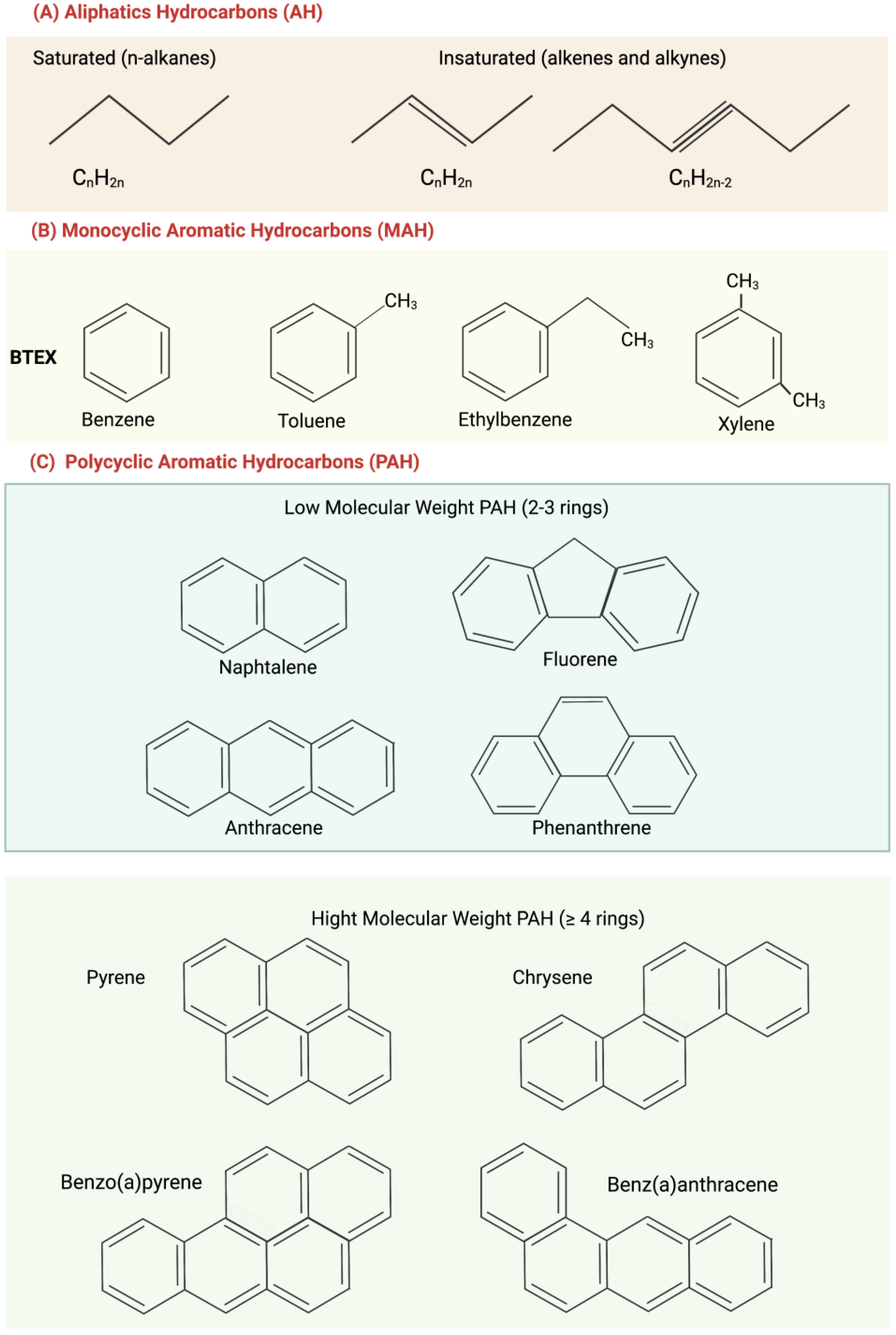
2. Aliphatic Hydrocarbons and Polycyclic Aromatic Hydrocarbons
3. Environmental Fate and Toxic Effects
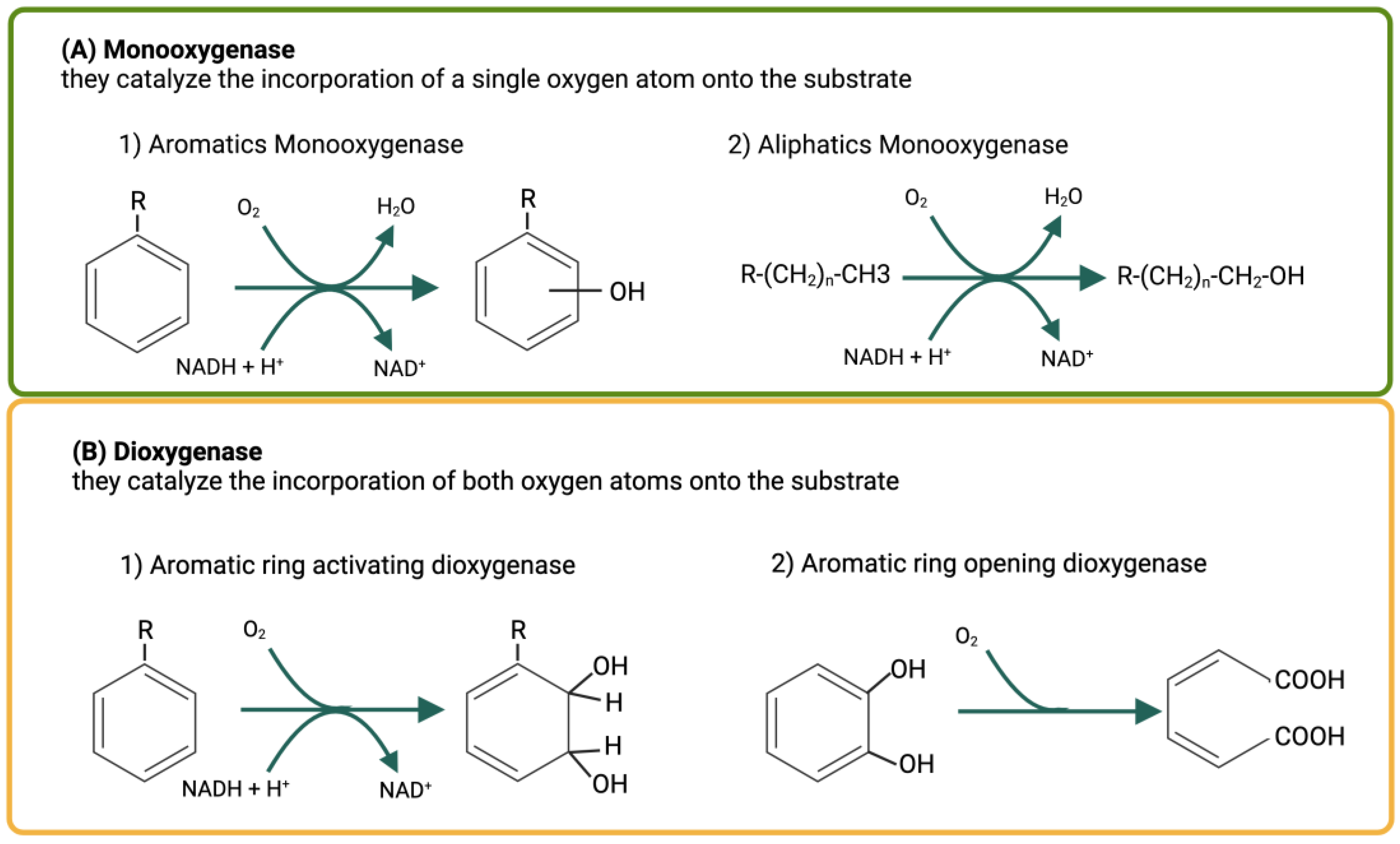
4. Microbial Degradation of Hydrocarbons
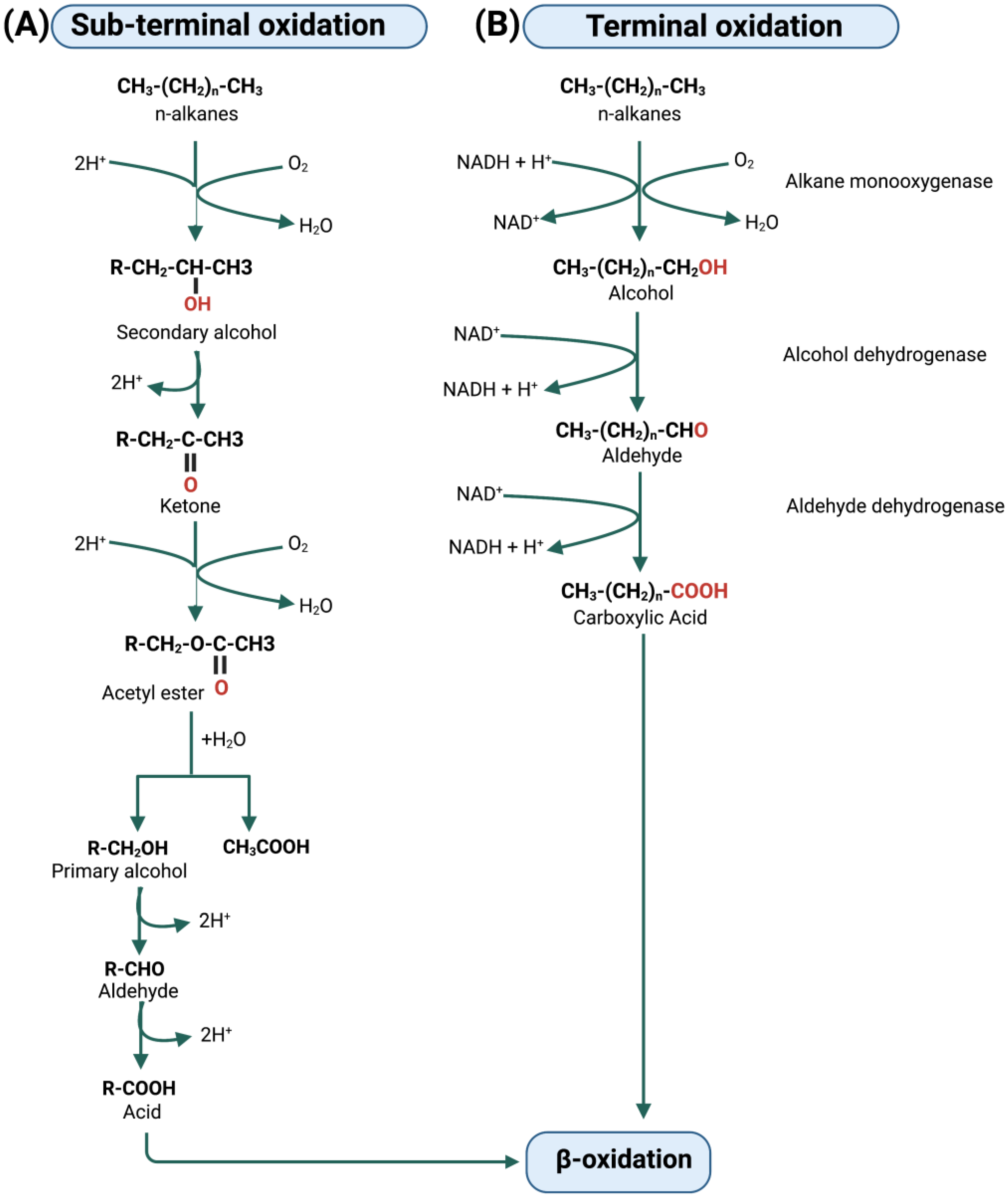
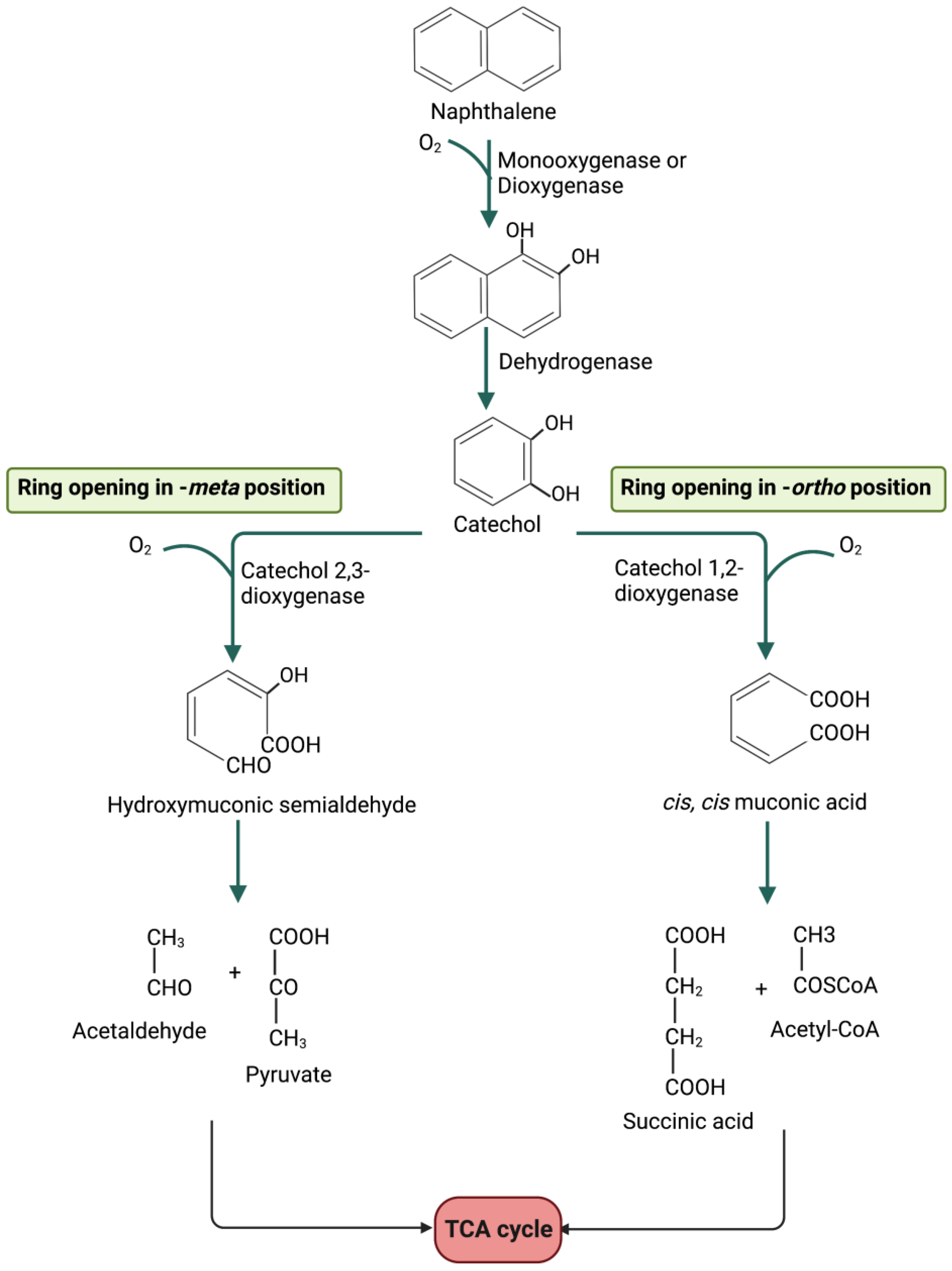
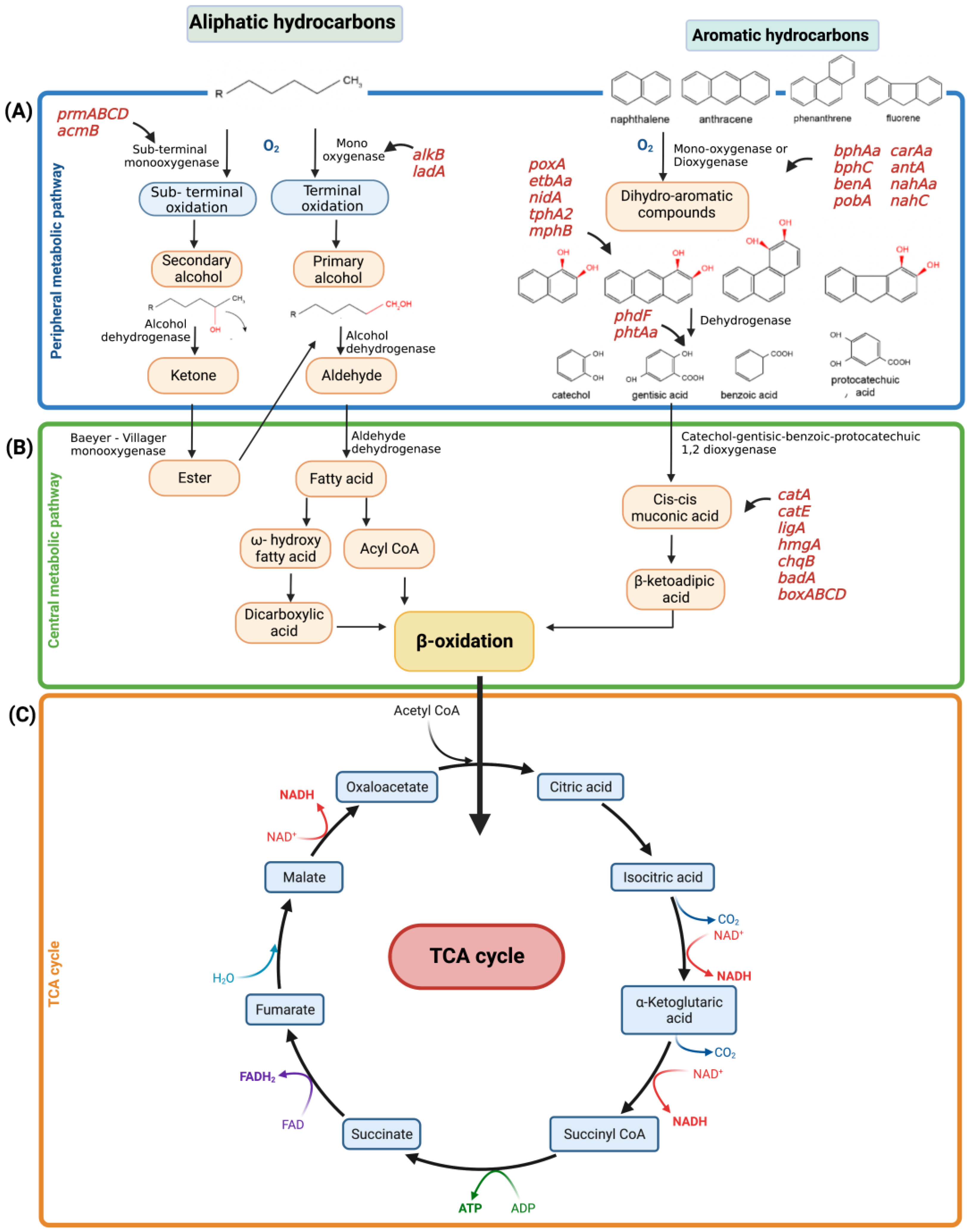
5. Metabolic Degradation Pathways
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Giri, K.; Rai, J.P.N. Bacterial Metabolism of Petroleum Hydrocarbons. Biotechnology 2014, 11, 73–93. [Google Scholar]
- Giraldez, P.; Aboal, J.R.; Fernandez, A.; Di Guardo, A.; Terzaghi, E. Plant-air partition coefficients for thirteen urban conifer tree species: Estimating the best gas and particulate matter associated PAH removers. Environ. Pollut. 2022, 315, 120409. [Google Scholar] [CrossRef] [PubMed]
- Devatha, C.P.; Vishnu Vishal, A.; Purna Chandra Rao, J. Investigation of physical and chemical characteristics on soil due to crude oil contamination and its remediation. Appl. Water Sci. 2019, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Garrido-Sanz, D.; Sansegundo-Lobato, P.; Redondo-Nieto, M.; Conlon, R.; Martin, M.; Mali, R.; Liu, X.; Dowling, D.N.; Rivilla, R.; et al. Soil Microbiome Structure and Function in Ecopiles Used to Remediate Petroleum-Contaminated Soil. Front. Environ. Sci. 2021, 9, 39. [Google Scholar] [CrossRef]
- Nagkirti, P.; Shaikh, A.; Vasudevan, G.; Paliwal, V.; Dhakephalkar, P. Bioremediation of terrestrial oil spills. Feasibility Assessment. In Optimization and Applicability of Bioprocess; Springer: Singapore, 2017; pp. 141–173. [Google Scholar]
- Abdullah, S.R.S.; Al-Baldawi, I.A.; Almansoory, A.F.; Purwanti, I.F.; Al-Sbani, N.H.; Sharuddin, S.S.N. Plant-assisted remediation of hydrocarbons in water and soil: Application, mechanisms, challenges and opportunities. Chemosphere 2020, 247, 125932. [Google Scholar] [CrossRef] [PubMed]
- Logeshwaran, P.; Megharaj, M.; Chadalavada, S.; Bowman, M.; Naidu, R. Petroleum hydrocarbons (PH) in groundwater aquifers; An overview of environmental fate, toxicity, microbial degradation and risk-based remediation approaches. Environ. Technol. Innov. 2018, 10, 175–193. [Google Scholar] [CrossRef]
- Barbieri, P.; Bestetti, G.; Galli, E.; Zannoni, D. Microbiologia Ambientale ed Elementi di Biologia Microbica; Ambrosiana: Milano, Italy, 2012; pp. 218–388. [Google Scholar]
- Varjani, S.J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2017, 223, 277–286. [Google Scholar] [CrossRef]
- Huang, L.; Batterman, S.A. Multimedia model for polycyclic aromatic hydrocarbons (PAHs) and nitro-PAHs in Lake Michigan. Environ. Sci. Technol. 2014, 48, 13817–13825. [Google Scholar] [CrossRef] [Green Version]
- Ghosal, D.; Ghosh, S.; Dutta, T.K.; Ahn, Y. Current State of Knowledge in Microbial Degradation of Polycyclic Aromatic Hydrocarbons (PAHs): A Review. Front. Microbiol. 2016, 7, 1369. [Google Scholar] [CrossRef] [Green Version]
- Lebov, J.; Grieger, K.; Womack, D.; Zaccaro, D.; Whitehead, N.; Kowalcyk, B.; MacDonald, P.D.M. A framework for One Health research. One Health 2017, 3, 44–50. [Google Scholar] [CrossRef]
- Nzila, A. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons under anaerobic conditions: Overview of studies, proposed pathways and future perspectives. Environ. Pollut. 2018, 239, 788–802. [Google Scholar] [CrossRef] [PubMed]
- Sudip, K.S.; Om, V.S.; Rakesh, K.J. Polycyclic aromatic hydrocarbons: Environmental pollution and bioremediation. Trends Biotechnol. 2002, 20, 243–248. [Google Scholar]
- Boström, C.-E.; Gerde, P.; Hanberg, A.; Jernström, B.; Johansson, C.; Kyrklund, T.; Rannug, A.; Törnqvist, M.; Victorin, K.; Westerholm, R.; et al. Cancer risk assessment, indicators, and guidelines for polycyclic aromatic hydrocarbons in the ambient air. Environ. Health Perspect. 2002, 110, 451–488. [Google Scholar]
- Kuppusamy, S.; Maddela, N.R.; Megharaj, M.; Venkateswarlu, K. Total Petroleum Hydrocarbons: Environmental Fate, Toxicity and Remediation; Springer: Cham, Switzerland, 2020. [Google Scholar]
- Truskewycz, A.; Gundry, T.D.; Khudur, L.S.; Kolobaric, A.; Taha, M.; Alburto-Medina, A.; Ball, A.S.; Shahsavari, E. Petroleum Hydrocarbons Contamination in Terrestrial Ecosystem- Fate and Microbial Responses. Molecules 2019, 24, 3400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, X.; Zheng, B.; Li, X.; Zhao, X.; Dionysiou, D.D.; Liu, Y. Influencing factors and health risk assessment of polycyclic aromatic hydrocarbons in groundwater in China. J. Hazard. Mater. 2021, 402, 123419. [Google Scholar] [CrossRef]
- Ruiz, O.N.; Radwan, O.; Striebich, R.C. GC–MS hydrocarbon degradation profile data of Pseudomonas frederiksbergensis SI8, a bacterium capable of degrading aromatics at low temperatures. Data Brief 2021, 35, 106864. [Google Scholar] [CrossRef]
- Directive 2013/39/EU of the European Parliament and of the Council of 12 August 2013 amending Directives 2000/60/EC and 2008/105/EC as regards priority substances in the field of water policy. Off. J. Eur. Union 2013, L226, 1–17.
- Dedysh, S.N.; Dunfield, P.F. Facultative methane oxidizers. In Taxonomy, Genomics and Ecophysiology of Hydrocarbon-Degrading Microbes; Springer: Cham, Switzerland, 2019; pp. 279–297. [Google Scholar]
- Gupta, G.; Kumar, V.; Pal, A.K. Microbial Degradation of High Molecular Weight Polycyclic Aromatic Hydrocarbons with Emphasis on Pyrene. Polycycl. Aromat. Compd. 2017, 39, 124–138. [Google Scholar] [CrossRef]
- Heitkamp, M.A.; Franklin, W.; Cerniglia, C.E. Microbial metabolism of polycyclic aromatic hydrocarbons: Isolation and characterization of a pyrene-degrading bacterium. Appl. Environ. Microbiol. 1988, 54, 2549–2555. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.; Zhang, Y.; Su, J.; Qiu, Q.; Jia, Z.; Zhu, Y.G. Bacterial communities predominant in the degradation of 13C4-4, 5, 9, 10-pyrene during composting. Bioresour. Technol. 2013, 143, 608–614. [Google Scholar] [CrossRef]
- Madigan, M.T.; Martinko, J.M.; Bender, K.S.; Buckley, D.H.; Stahl, D.A. Brock Biologia dei Microrganismi. In Microbiologia Generale, Ambientale e Industriale; Pearson: Milano/Torino, Italy, 2016; pp. 698–699. [Google Scholar]
- Posada-Baquero, R.; Jiménez-Volkerink, S.N.; García, J.L.; Vila, J.; Cantos, M.; Grifoll, M.; Ortega-Calvo, J.J. Rhizosphere-enhanced biosurfactant action on slowly desorbing PAHs in contaminated soil. Sci. Total Environ. 2020, 720, 137608. [Google Scholar] [CrossRef] [PubMed]
- Chebbi, A.; Hentati, D.; Zaghden, H.; Baccar, N.; Rezgui, F.; Chalbi, M.; Sayadi, S.; Chamkha, M. Polycyclic aromatic hydrocarbon degradation and biosurfactant production by a newly isolated Pseudomonas sp. strain from used motor oil-contaminated soil. Int. Biodeterior. Biodegrad. 2017, 122, 128–140. [Google Scholar] [CrossRef]
- Medic, A.; Lješevic, M.; Inui, H.; Beškoski, V.; Kojic, I.; Stojanovic, K.; Karadžic, I. Efficient biodegradation of petroleum n-alkanes and polycyclic aromatic hydrocarbons by polyextremophilic Pseudomonas aeruginosa san ai with multidegradative capacity. R. Soc. Chem. 2019, 10, 14060–14070. [Google Scholar]
- Ahmed, A.W.; Alzubaidi, F.S.; Hamza, S.J. Biodegradation of crude oil in contaminated water by local isolates of Enterobacter cloacae. Iraqi J. Sci. 2014, 55, 1025–1033. [Google Scholar]
- Lenchi, N.; Kebbouche-Gana, S.; Servais, P.; Gana, M.L.; Llirós, M. Diesel biodegradation capacities and biosurfactant production in saline-alkaline conditions by Delftia sp NL1, isolated from an Algerian oilfield. Geomicrobiol. J. 2020, 37, 454–466. [Google Scholar] [CrossRef]
- Li, J.; Chen, W.; Zhou, W.; Wang, Y.; Deng, M.; Zhou, S. Synergistic degradation of pyrene by Pseudomonas aeruginosa PA06 and Achromobacter sp. AC15 with sodium citrate as the co- metabolic carbon source. Ecotoxicology 2020, 30, 1487–1498. [Google Scholar] [CrossRef] [PubMed]
- Czarny, J.; Staninska-Pięta, A.; Piotrowska-Cyplik, W.; Juzwa, A.; Wolniewicz, R.; Marecik, Ł.; Ławniczak, Ł. Acinetobacter sp. as the key player in diesel oil degrading community exposed to PAHs and heavy metals. J. Hazard. Mater. 2020, 383, 121168. [Google Scholar] [CrossRef]
- Van Hamme, J.D.; Singh, A.; Ward, O.P. Recent advances in petroleum microbiology. Microbiol. Mol. Biol. Rev. 2003, 67, 503–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thangaraj, K.; Kapley, A.; Purohit, H.J. Characterization of diverse Acinetobacter isolates for utilization of multiple aromatic compounds. Bioresour. Technol. 2008, 99, 2488–2494. [Google Scholar] [CrossRef]
- Ghosal, D.; Dutta, A.; Chakraborty, J.; Basu, S.; Dutta, T.K. Characterization of the metabolic pathway involved in assimilation of acenaphthene in Acinetobacter sp. strain AGAT-W. Res. Microbiol. 2013, 164, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Rolando, L.; Vila, J.; Baquero, R.P.; Castilla-Alcantara, J.C.; Barra Caracciolo, A.; Ortega-Calvo, J.-J. Impact of bacterial motility on biosorption and cometabolism of pyrene in a porous medium. Sci. Total Environ. 2020, 717, 137210. [Google Scholar] [CrossRef] [PubMed]
- Elufisan, T.O.; Rodríguez-Luna, I.C.; Oyedara, O.O.; Sánchez-Varela, A.; Hernández-Mendoza, A.; Dantán Gonzalez, E.; Paz-González, A.D.; Muhammad, K.; Rivera, G.; Villalobos-Lopez, M.A.; et al. The Polycyclic Aromatic Hydrocarbon (PAH) degradation activities and genome analysis of a novel strain Stenotrophomonas sp. Pemsol isolated from Mexico. Peer J. 2020, 8, e8102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, X.; Xu, Y.; Li, G.; Zhang, Y.; Huang, T.; Hu, Z. Isolation, characterization of Rhodococcus sp. P14 capable of degrading high-molecular-weight polycyclic aromatic hydrocarbons and aliphatic hydrocarbons. Mar. Pollut. Bull. 2011, 62, 2122–2128. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Sanz, D.; Sansegundo-Lobato, P.; Redondo-Nieto, M.; Suman, J.; Cajthaml, T.; Blanco-Romero, E.; Rivilla, R. Analysis of the biodegradative and adaptive potential of the novel polychlorinated biphenyl degrader Rhodococcus sp. WAY2 revealed by its complete genome sequence. Microb. Genom. 2020, 6, e000363. [Google Scholar] [CrossRef]
- Devarapalli, P.; Kumavath, R.N. Metagenomics-a technological drift in bioremediation. In Advances in Bioremediation of Wastewater and Polluted Soil; IntechOpen: London, UK, 2015; pp. 73–91. [Google Scholar]
- Quintella, C.M.; Mata, A.M.T.; Lima, L.C.P. Overview of bioremediation with technology assessment and emphasis on fungal bioremediation of oil contaminated soils. J. Environ. Manag. 2019, 241, 156–166. [Google Scholar] [CrossRef]
- Gallego, S.; Vila, J.; Tauler, M.; Nieto, J.M.; Breugelmans, P.; Springael, D.; Grifoll, M. Community structure and PAH ring-hydroxylating dioxygenase genes of a marine pyrene-degrading microbial consortium. Biodegradation 2014, 25, 543–556. [Google Scholar] [CrossRef]
- Vaidya, S.; Jain, K.; Madamwar, D. Metabolism of pyrene through phthalic acid pathway by enriched bacterial consortium composed of Pseudomonas, Burkholderia, and Rhodococcus (PBR). 3 Biotech 2017, 7, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Wanapaisan, P.; Laothamteep, N.; Vejarano, F.; Chakraborty, J.; Shintani, M.; Muangchinda, C.; Morita, T.; Suzuki-Minakuchi, C.; Inoue, K.; Nojiri, H.; et al. Synergistic degradation of pyrene by five culturable bacteria in a mangrove sediment-derived bacterial consortium. J. Hazard. Mater. 2018, 342, 561–570. [Google Scholar] [CrossRef]
- Ferraro, A.; Massini, G.; Mazzurco Miritana, V.; Panico, A.; Pontoni, L.; Race, M.; Rosa, S.; Signorini, A.; Fabbricino, M.; Pirozzi, F. Bioaugmentation strategy to enhance polycyclic aromatic hydrocarbons anaerobic biodegradation in contaminated soils. Chemosphere 2021, 275, 130091. [Google Scholar] [CrossRef]
- Phulpoto, I.A.; Hu, B.; Wang, Y.; Ndayisenga, F.; Li, J.; Yu, Z. Effect of natural microbiome and culturable biosurfactants-producing bacterial consortia of freshwater lake on petroleum-hydrocarbon degradation. Sci. Total Environ. 2021, 751, 141720. [Google Scholar] [CrossRef]
- Adebusoye, S.A.; Ilori, M.O.; Amund, O.O.; Teniola, O.D.; Olatope, S. Microbial degradation of petroleum hydrocarbons in a polluted tropical stream. World J. Microbiol. Biotechnol. 2007, 23, 1149–1159. [Google Scholar] [CrossRef]
- Mandri, T.; Lin, J. Isolation and characterization of engine oil degrading indigenous microorganisms in Kwazulu-Natal, South Africa. Afr. J. Biotechnol. 2007, 6, 23–27. [Google Scholar]
- Bacosa, H.; Suto, K.; Inoue, C. Preferential degradation of aromatic hydrocarbons in kerosene by a microbial consortium. Int. Biodeterior. Biodegrad. 2010, 64, 702–710. [Google Scholar] [CrossRef]
- Mamdoh, T.J. Enrichment of Potential Halophilic Marinobacter Consortium for Mineralization of Petroleum Hydrocarbons and Also as Oil Reservoir Indicator in Red Sea, Saudi Arabia. Polycycl. Aromat. Compd. 2020, 42, 400–411. [Google Scholar]
- Feng, L.; Jiang, X.; Huang, Y.; Wen, D.; Fu, T.; Fu, R. Petroleum hydrocarbon-contaminated soil bioremediation assisted by isolated bacterial consortium and sophorolipid. Environ. Pollut. 2021, 273, 116476. [Google Scholar] [CrossRef]
- Vasilyeva, G.; Kondrashina, V.; Strijakova, E.; Ortega-Calvo, J.-J. Adsorptive bioremediation of soil highly contaminated with crude oil. Sci. Total Environ. 2020, 706, 135739. [Google Scholar] [CrossRef]
- Fahid, M.; Arslan, M.; Shabir, G.; Younus, S.; Yasmeen, T.; Rizwan, M.; Siddique, K.; Ahmad, S.R.; Tahseen, R.; Iqbal, S.; et al. Phragmites australis in combination with hydrocarbons degrading bacteria is a suitable option for remediation of diesel-contaminated water in floating wetlands. Chemosphere 2019, 240, 124890. [Google Scholar] [CrossRef]
- Kuppusamy, S.; Thavamani, P.; Venkateswarlu, K.; Lee, Y.B.; Naidu, R.; Megharaj, M. Remediation approaches for polycyclic aromatic hydrocarbons (PAHs) contaminated soils: Technological constraints, emerging trends and future directions. Chemosphere 2017, 168, 944–968. [Google Scholar] [CrossRef]
- Koolivand, A.; Saeedi, R.; Coulon, F.; Kumar, V.; Villaseñor, J.; Asghari, F.; Hesampoor, F. Bioremediation of petroleum hydrocarbons by vermicomposting process bioaugmentated with indigenous bacterial consortium isolated from petroleum oily sludge. Ecotoxicol. Environ. Saf. 2020, 198, 110645. [Google Scholar] [CrossRef]
- Parthipan, P.; Cheng, L.; Dhandapani, P.; Elumalai, P.; Huang, M.; Rajasekar, A. Impact of biosurfactant and iron nanoparticles on biodegradation of polyaromatic hydrocarbons (PAHs). Environ. Pollut. 2022, 306, 119384. [Google Scholar] [CrossRef]
- Kumari, B.; Singh, D.P. A review on multifaceted application of nanoparticles in the field of bioremediation of petroleum hydrocarbons. Ecol. Eng. 2016, 97, 98–105. [Google Scholar] [CrossRef]
- Van Beilen, J.B.; Li, Z.; Duetz, W.A.; Smits, T.H.M.; Witholt, B. Diversity of alkane hydroxylase systems in the environment. Oil Gas Sci. Technol. 2003, 58, 427–440. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Liu, X.; Yang, W.; Xu, F.; Wang, W.; Feng, L.; Bartlam, M.; Wang, L.; Rao, Z. Crystal structure of long- chain alkane monooxygenase (LadA) in complex with coenzyme FMN: Unveiling the long-chain alkane hydroxylase. J. Mol. Biol. 2008, 376, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Mao, G.; Wang, Y.; Bartlam, M. Structural insights into diversity and n- alkane biodegradation mechanisms of alkane hydroxylases. Front. Microbiol. 2013, 4, 58. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.J.; Kweon, O.; Cerniglia, C.E. Proteomic applications to elucidate bacterial aromatic hydrocarbons metabolic pathways. Curr. Opin. Microbiol. 2009, 12, 301–309. [Google Scholar] [CrossRef]
| Substance Name | CAS Number | AA-EQS Surface Waters (μg/L) | MAC-EQS Inland Surface Waters (μg/L) | MAC-EQS Other Surface Waters (μg/L) | EQS Biota (μg/kg) |
|---|---|---|---|---|---|
| Anthracene | 120-12-7 | 0.1 | 0.1 | 0.1 | |
| benzo(a)pyrene | 50-32-8 | 1.7 × 10–4 | 0.27 | 0.027 | 5 |
| benzo(b)fluoranthene | 205-99-2 | * | 0.017 | 0.017 | * |
| benzo(g,h,i)perylene | 207-08-9 | * | 8.2 × 10–3 | 8.2 × 10–4 | * |
| benzo(k)fluoranthene | 191-24-2 | * | 0.017 | 0.017 | * |
| Fluoranthene | 206-44-0 | 0.0063 | 0.12 | 0.12 | 30 |
| indeno(1,2,3-cd)pyrene | 193-39-5 | * | - | - | * |
| Naphthalene | 91-20-3 | 2 | 130 | 130 |
| Genus/Species | Mechanism of Action/ Hydrocarbon | References | |
|---|---|---|---|
| Proteobacteria | Pseudomonas W10 | Biosurfactant production/ phenanthrene | [25] |
| Pseudomonas aeruginosa | Biosurfactant production/ n-alkanes (C16–C19), fluorene, phenanthrene, pyrene | [26] | |
| Enterobacter cloacae | Biosurfactant production/ crude oil | [27] | |
| Delftia sp. NL1 | Biosurfactant production/diesel | [28] | |
| Achromobacter (AC15) | Biosurfactant production/ Pyrene | [29] | |
| Acinetobacter | Biosurfactant production/ naphthalene, acenaphthene, acenaphthylene | [11,32,33] | |
| Pseudomonas putida | Chemotaxis/ Pyrene | [36] | |
| Stenotrophomonas sp. Pemsol | Horizontal gene transfer/ biphenyl, anthraquinone, phenanthrene, naphthalene, phenanthridine | [37] | |
| Actino bacteria | Rhodococcus sp. P14 | Change in fatty acid composition of cell membrane/biofilm formation/phenanthrene, pyrene, benzo(a)pyrene | [38] |
| Gene | Enzyme | Metabolism | |
|---|---|---|---|
| Aliphatics | alkB | Alkane 1-monooxygenase | Peripheral |
| ladA | Long chain alkane monooxygenase | Peripheral | |
| prmABCD | Propane monooxygenase | Peripheral | |
| acmAB | Bayer-Villiger monooxygenase and esterase | Peripheral | |
| Aromatics | benA | Benzoate 1,2 dioxygenase | Peripheral |
| pobA | Hydrobenzoate 3-monooxygenase | Peripheral | |
| bphAa | Biphenyl 2,3-dioxygenase | Peripheral | |
| bphC | Dihydroxyphenyl 2,3-dioxygenase | Peripheral | |
| carAa | Carbazole 1,9-dioxygenase | Peripheral | |
| antA | Anthranilate 1,2-dioxygenase | Peripheral | |
| nahAa | Naphthalene1,2-dioxygenase (α-subunit) | Peripheral | |
| nahAb | Naphthalene1,2-dioxygenase (β-subunit) | Peripheral | |
| nahAc | Naphthalene1,2-dioxygenase (ferrodoxin) | Peripheral | |
| nahC | 1,2-dihydroxy naphthalene dioxygenase | Peripheral | |
| nagG | Salicylate 5-hydroxylase | Peripheral | |
| poxA | Phenol hydroxylase | Peripheral | |
| etbAa | Ethylbenzene dioxygenase | Peripheral | |
| nidA | Phenanthrene dioxygenase | Peripheral | |
| tphA2 | Terephthalate 1,2-dioxygenase | Peripheral | |
| phdF | 3,4-dihydroxy-phenanthrene dioxygenase | Peripheral | |
| pht3 | Phthalate 4,5-dioxygenase | Peripheral | |
| phtAa | Phthalate 3,4-dioxygenase | Peripheral | |
| cmtAb | p-coumate dioxygenase | Peripheral | |
| cmtC | 2,3-dihydroxy-p-coumate 3,4-dioxygenase | Peripheral | |
| hcaE | 3-phenylpropanoate/trans-cinnamate dioxygenase | Peripheral | |
| mhpB | 2,3-dihydroxy phenylpropionate 1,2-dioxygenase | Peripheral | |
| catA | Catechol 1,2-dioxygenase | Central | |
| catE | Catechol 2,3-dioxygenase | Central | |
| pcaG | Protocateucate 3,4-dioxygenase | Central | |
| hmgA | Homogensite 1,2-dioxygenase | Central | |
| ligA | Protocateucate 4,5-dioxygenase | Central | |
| chqB | Hydroxyquinol 1,2-dioxygenase | Central | |
| badA | Benzoate-CoA ligase | Central | |
| boxA | Benzoyl-CoA 2,3-epoxidase (α-subunit) | Central | |
| boxB | Benzoyl-CoA 2,3-epoxidase (β-subunit) | Central | |
| boxC | Benzoyl-CoA dihydrodiol lyase | Central | |
| boxD | 3,4-dehydroadipyl-CoA-semialdehyde dehydrogenase | Central |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandolfo, E.; Barra Caracciolo, A.; Rolando, L. Recent Advances in Bacterial Degradation of Hydrocarbons. Water 2023, 15, 375. https://doi.org/10.3390/w15020375
Pandolfo E, Barra Caracciolo A, Rolando L. Recent Advances in Bacterial Degradation of Hydrocarbons. Water. 2023; 15(2):375. https://doi.org/10.3390/w15020375
Chicago/Turabian StylePandolfo, Emiliana, Anna Barra Caracciolo, and Ludovica Rolando. 2023. "Recent Advances in Bacterial Degradation of Hydrocarbons" Water 15, no. 2: 375. https://doi.org/10.3390/w15020375





