Effects of Bacillus pumilus BP-171 and Carbon Sources on the Growth Performance of Shrimp, Water Quality and Bacterial Community in Penaeus vannamei Culture System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Experimental Strain and Carbon Sources
2.3. Experimental Design
2.4. Experimental Management
2.5. Sample Collection and Measurement
2.5.1. Growth Performance of Shrimp
2.5.2. Water Quality Parameters
2.5.3. DNA Extraction, Amplification, Purification, and Sequencing
2.6. Statistical Analysis
3. Results
3.1. Growth Performance of Shrimp
3.2. Water Quality Parameters
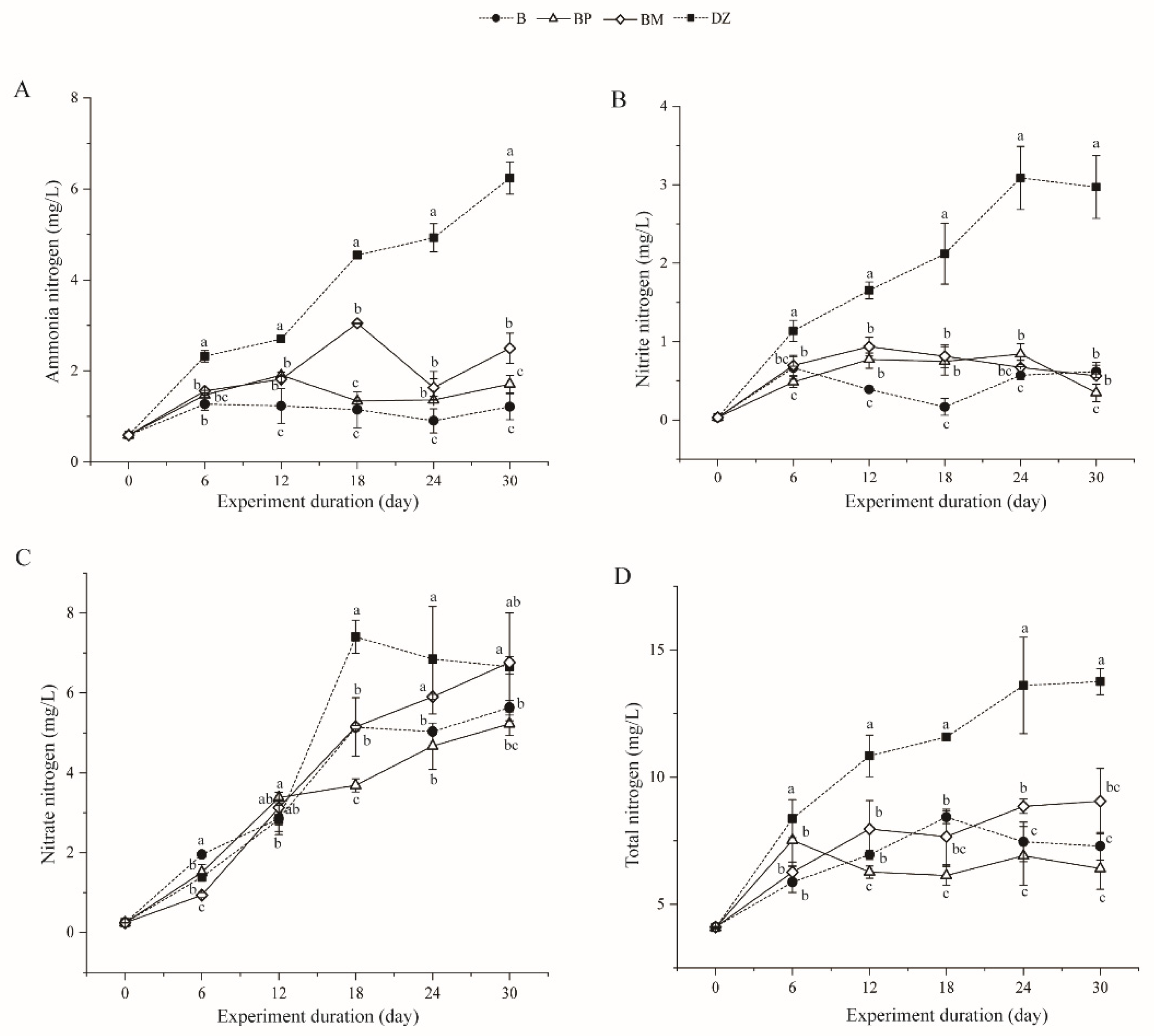
3.2.1. Changes in Ammonia, Nitrite, Nitrate, and Total Nitrogen
3.2.2. Changes in Soluble Reactive Phosphorus (SRP) and Total Phosphorus (TP)
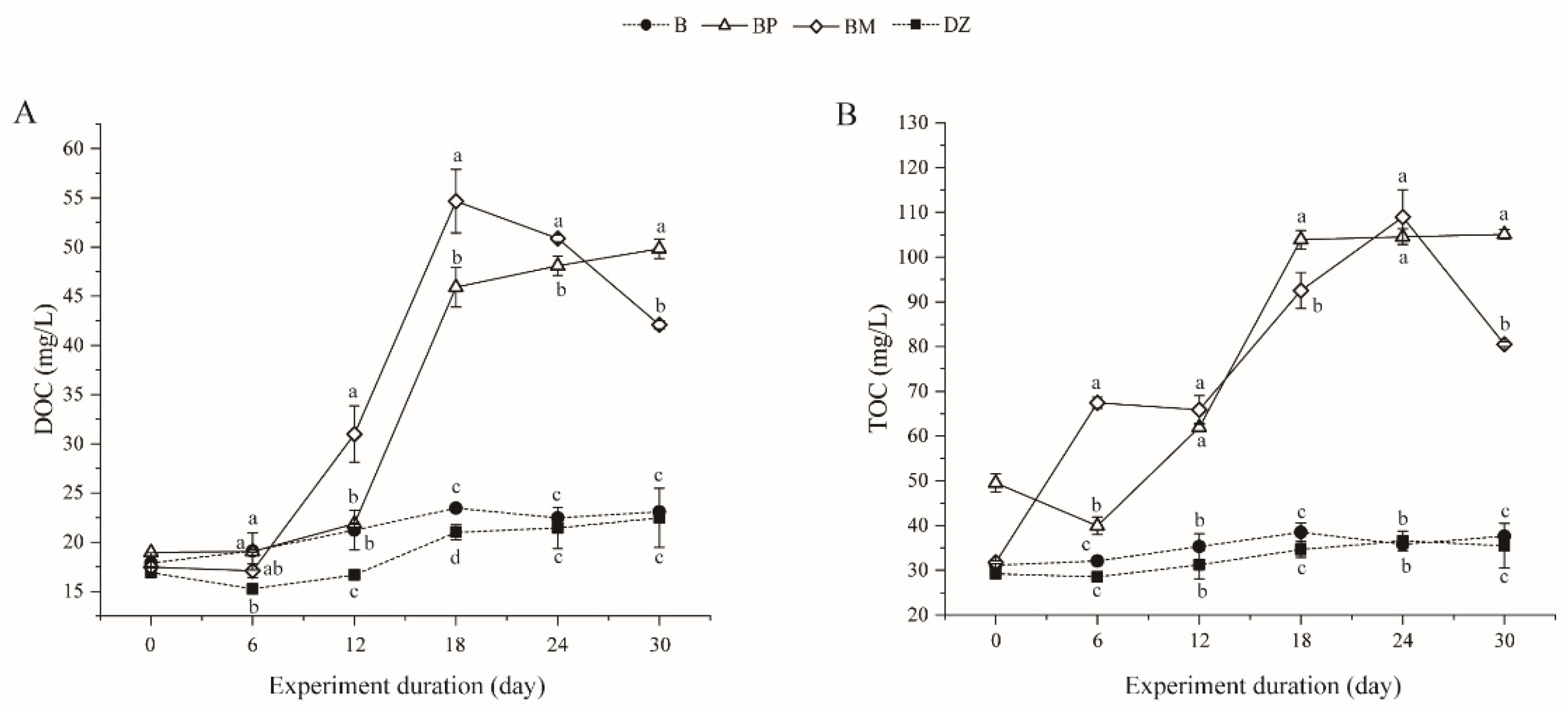
3.2.3. Changes of Dissolved Organic Carbon (DOC) and Total Organic Carbon (TOC)
3.3. The Microbial Diversity, Community Structure, and Function
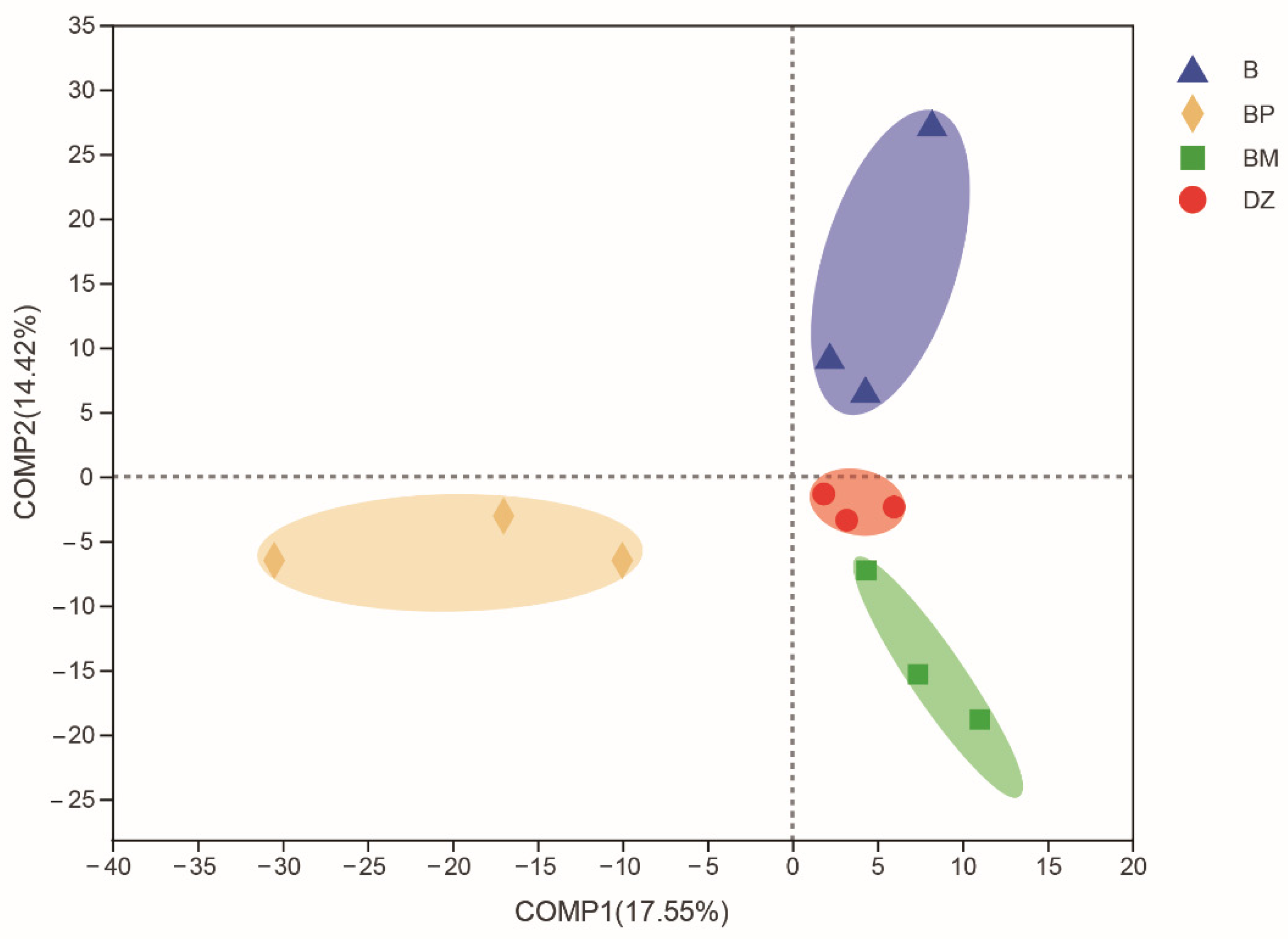
3.3.1. Microbial Diversity
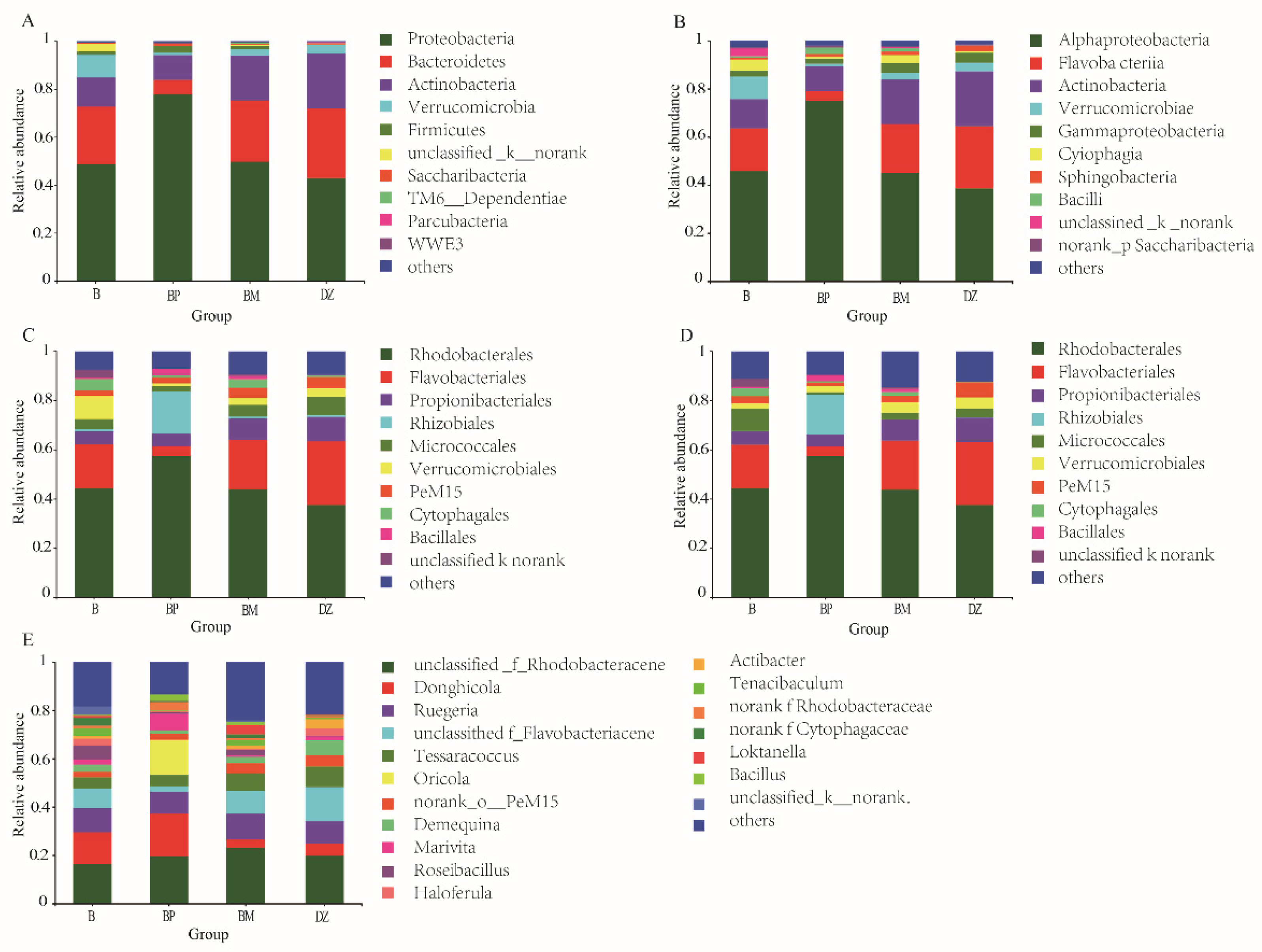
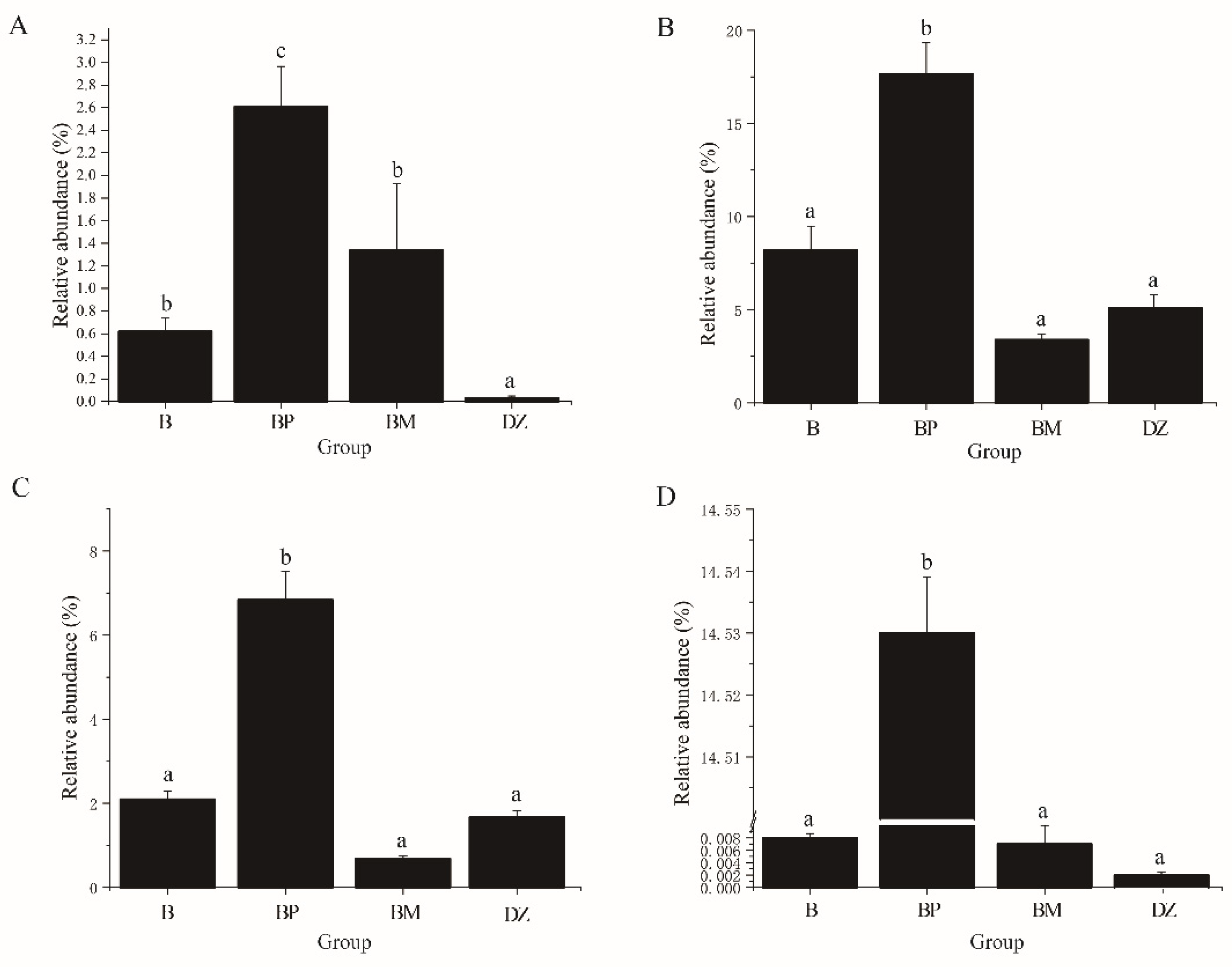
3.3.2. Bacterial Community Composition
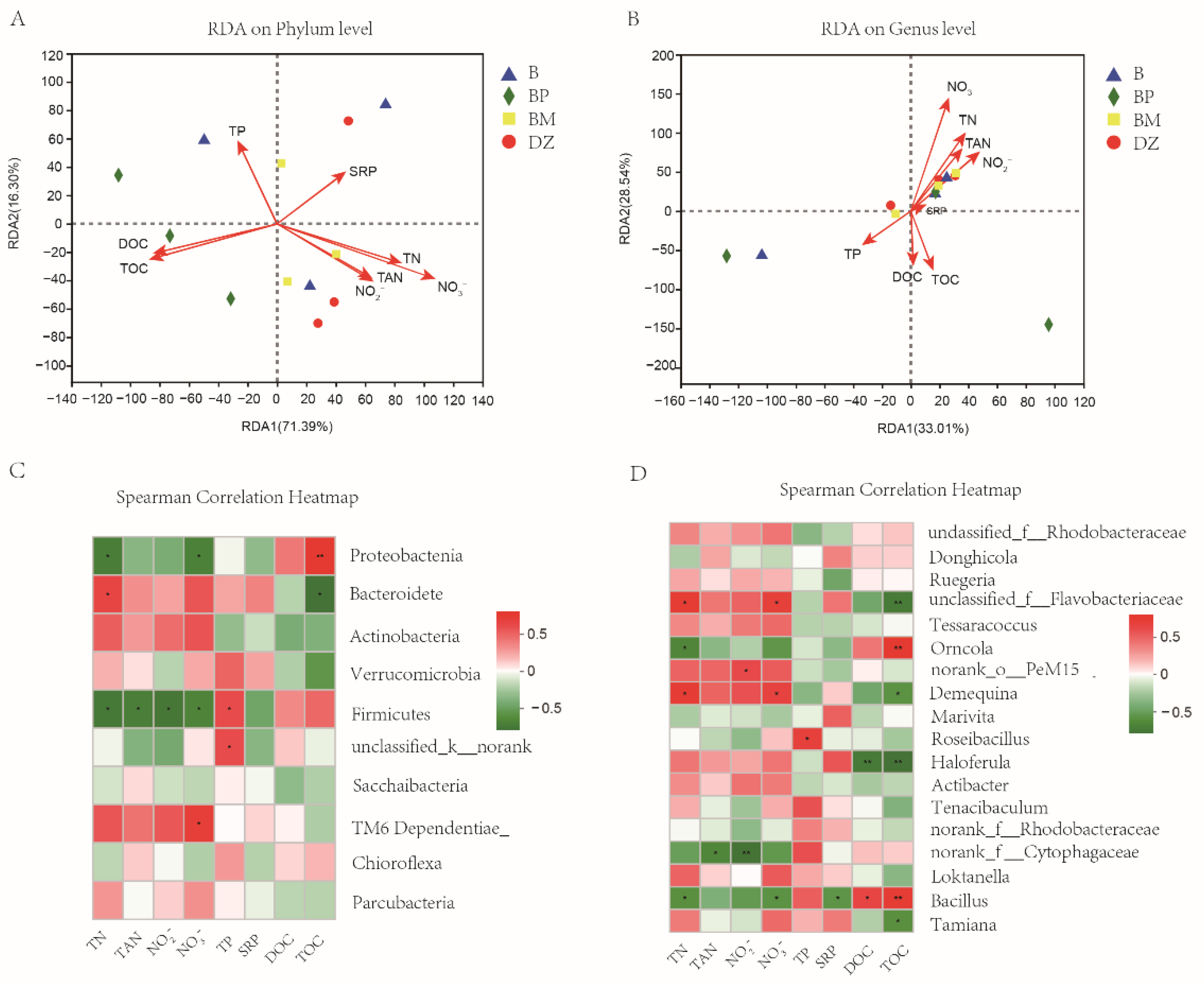
3.3.3. The Correlation of Water Quality Indexes and Microbial Community Structure
3.3.4. Ecological Network Analysis
3.3.5. Predictive Functions of Microbiota in the Water
4. Discussion
4.1. Effects of Addition of B. pumilus 171 and Carbon Sources on the Growth Performance of Shrimp
4.2. Effects of the Addition of B. pumilus 171 and Carbon Sources on the Water Quality
4.3. Effects of Addition of B. pumilus 171 and Carbon Sources on the Microbial Diversity and Microbiota Compositions in Water
4.4. Effects of Addition of B. pumilus 171 and Carbon Sources on the Ecological Network and Function of the Microbial Community
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goh, J.X.H.; Tan, L.T.; Law, J.W.; Ser, H.; Khaw, K.; Letchumanan, V.; Lee, L.; Goh, B. Harnessing the Potentialities of Probiotics, Prebiotics, Synbiotics, Paraprobiotics, and Postbiotics for Shrimp Farming. Rev. Aquac. 2022, 14, 1478–1557. [Google Scholar] [CrossRef]
- Robles-Porchas, G.R.; Gollas-Galván, T.; Martínez-Porchas, M.; Martínez-Cordova, L.R.; Miranda-Baeza, A.; Vargas-Albores, F. The Nitrification Process for Nitrogen Removal in Biofloc System Aquaculture. Rev. Aquac. 2020, 12, 2228–2249. [Google Scholar] [CrossRef]
- Kokkuar, N.; Li, L.; Srisapoome, P.; Dong, S.; Tian, X. Application of Biodegradable Polymers as Carbon Sources in Ex Situ Biofloc Systems: Water Quality and Shift of Microbial Community. Aquac. Res. 2021, 52, 3570–3579. [Google Scholar] [CrossRef]
- Hlordzi, V.; Kuebutornye, F.K.A.; Afriyie, G.; Abarike, E.D.; Lu, Y.; Chi, S.; Anokyewaa, M.A. The Use of Bacillus Species in Maintenance of Water Quality in Aquaculture: A Review. Aquac. Rep. 2020, 18, 100503. [Google Scholar] [CrossRef]
- Lieke, T.; Meinelt, T.; Hoseinifar, S.H.; Pan, B.; Straus, D.L.; Steinberg, C.E.W. Sustainable Aquaculture Requires Environmental-friendly Treatment Strategies for Fish Diseases. Rev. Aquac. 2020, 12, 943–965. [Google Scholar] [CrossRef] [Green Version]
- Zokaeifar, H.; Babaei, N.; Saad, C.R.; Kamarudin, M.S.; Sijam, K.; Balcazar, J.L. Administration of Bacillus subtilis Strains in the Rearing Water Enhances the Water Quality, Growth Performance, Immune Response, and Resistance against Vibrio Harveyi Infection in Juvenile White Shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 2014, 36, 68–74. [Google Scholar] [CrossRef]
- Devaraja, T.; Banerjee, S.; Yusoff, F.; Shariff, M.; Khatoon, H. A holistic approach for selection of Bacillus spp. as a bioremediator for shrimp postlarvae culture. Turk. J. Biol. 2013, 37, 92–100. [Google Scholar] [CrossRef]
- Kuebutornye, F.K.A.; Abarike, E.D.; Lu, Y. A Review on the Application of Bacillus as Probiotics in Aquaculture. Fish Shellfish Immunol. 2019, 87, 820–828. [Google Scholar] [CrossRef]
- Soltani, M.; Ghosh, K.; Hoseinifar, S.H.; Kumar, V.; Lymbery, A.J.; Roy, S.; Ringø, E. Genus Bacillus, Promising Probiotics in Aquaculture: Aquatic Animal Origin, Bio-Active Components, Bioremediation and Efficacy in Fish and Shellfish. Rev. Fish. Sci. Aquac. 2019, 27, 331–379. [Google Scholar] [CrossRef] [Green Version]
- Crab, R.; Defoirdt, T.; Bossier, P.; Verstraete, W. Biofloc Technology in Aquaculture: Beneficial Effects and Future Challenges. Aquaculture 2012, 356–357, 351–356. [Google Scholar] [CrossRef]
- Abbaszadeh, A.; Keyvanshokooh, S.; Yavari, V.; Naderi, M. Proteome Modifications of Pacific White Shrimp (Litopenaeus vannamei) Muscle under Biofloc System. Aquac. Nutr. 2019, 25, 358–366. [Google Scholar] [CrossRef]
- Gao, F.; Liao, S.; Liu, S.; Bai, H.; Wang, A.; Ye, J. The Combination Use of Candida tropicalis HH8 and Pseudomonas stutzeri LZX301 on Nitrogen Removal, Biofloc Formation and Microbial Communities in Aquaculture. Aquaculture 2019, 500, 50–56. [Google Scholar] [CrossRef]
- El-Saadony, M.T.; Alagawany, M.; Patra, A.K.; Kar, I.; Tiwari, R.; Dawood, M.A.O.; Dhama, K.; Abdel-Latif, H.M.R. The Functionality of Probiotics in Aquaculture: An Overview. Fish Shellfish Immunol. 2021, 117, 36–52. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Zhang, Y.; Dong, H.; Wang, Y.; Zhang, J. Effect of the Dietary Probiotic Clostridium Butyricum on Growth, Intestine Antioxidant Capacity and Resistance to High Temperature Stress in Kuruma Shrimp Marsupenaeus japonicus. J. Therm. Biol. 2017, 66, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Dash, P.; Tandel, R.S.; Bhat, R.A.H.; Mallik, S.; Pandey, N.N.; Singh, A.K.; Sarma, D. The Addition of Probiotic Bacteria to Microbial Floc: Water Quality, Growth, Non-Specific Immune Response and Disease Resistance of Cyprinus Carpio in Mid-Himalayan Altitude. Aquaculture 2018, 495, 961–969. [Google Scholar] [CrossRef]
- Dauda, A.B. Biofloc Technology: A Review on the Microbial Interactions, Operational Parameters and Implications to Disease and Health Management of Cultured Aquatic Animals. Rev. Aquac. 2020, 12, 1193–1210. [Google Scholar] [CrossRef]
- Ray, A.J.; Lotz, J.M. Comparing a Chemoautotrophic-Based Biofloc System and Three Heterotrophic-Based Systems Receiving Different Carbohydrate Sources. Aquac. Eng. 2014, 63, 54–61. [Google Scholar] [CrossRef]
- Pérez-Fuentes, J.A.; Hernández-Vergara, M.P.; Pérez-Rostro, C.I.; Fogel, I. C: N Ratios Affect Nitrogen Removal and Production of Nile Tilapia Oreochromis niloticus Raised in a Biofloc System under High Density Cultivation. Aquaculture 2016, 452, 247–251. [Google Scholar] [CrossRef]
- De Schryver, P.; Crab, R.; Defoirdt, T.; Boon, N.; Verstraete, W. The Basics of Bio-Flocs Technology: The Added Value for Aquaculture. Aquaculture 2008, 277, 125–137. [Google Scholar] [CrossRef]
- Li, Y.; Li, T.; Tian, X.; Luo, K.; Wang, L.; Zhang, S.; Wei, C.; Liu, Y. Effects of probiotics and polyhydroxybutyrate on growth and non-specific immunity of Litopenaeus vannamei and its culturing water quality. Period. Ocean Univ. China 2021, 51, 22–31. [Google Scholar] [CrossRef]
- Xu, Z.; Song, L.; Dai, X.; Chai, X. PHBV Polymer Supported Denitrification System Efficiently Treated High Nitrate Concentration Wastewater: Denitrification Performance, Microbial Community Structure Evolution and Key Denitrifying Bacteria. Chemosphere 2018, 197, 96–104. [Google Scholar] [CrossRef]
- Xu, Z.; Dai, X.; Chai, X. Biological Denitrification Using PHBV Polymer as Solid Carbon Source and Biofilm Carrier. Biochem. Eng. J. 2019, 146, 186–193. [Google Scholar] [CrossRef]
- Barman, P.; Kati, A.; Mandal, A.K.; Bandyopadhyay, P.; Mohapatra, P.K.D. Biopotentiality of Bacillus cereus PB45 for Nitrogenous Waste Detoxification in Ex Situ Model. Aquac. Int. 2017, 25, 1167–1183. [Google Scholar] [CrossRef]
- Sun, X.; Li, Q.; Zhang, Y.; Liu, H.; Zhao, J.; Qu, K. Phylogenetic analysis and nitrogen removal characteristics of a heterotrophic nitrifying-aerobic denitrifying bacteria strain from marine environment. Acta Microbiol. Sin. 2012, 52, 687–695. [Google Scholar] [CrossRef]
- Zhao, K.; Tian, X.; Li, H.; Dong, S.; Jiang, W. Characterization of a Novel Marine Origin Aerobic Nitrifying–Denitrifying Bacterium Isolated from Shrimp Culture Ponds. Aquac. Res. 2019, 50, 1770–1781. [Google Scholar] [CrossRef]
- Xie, F.; Thiri, M.; Wang, H. Simultaneous Heterotrophic Nitrification and Aerobic Denitrification by a Novel Isolated Pseudomonas mendocina X49. Bioresour. Technol. 2021, 319, 124198. [Google Scholar] [CrossRef]
- Lei, X.; Jia, Y.; Chen, Y.; Hu, Y. Simultaneous Nitrification and Denitrification without Nitrite Accumulation by a Novel Isolated Ochrobactrum Anthropic LJ81. Bioresour. Technol. 2019, 272, 442–450. [Google Scholar] [CrossRef]
- Chen, P.; Li, J.; Li, Q.X.; Wang, Y.; Li, S.; Ren, T.; Wang, L. Simultaneous Heterotrophic Nitrification and Aerobic Denitrification by Bacterium Rhodococcus Sp. CPZ24. Bioresour. Technol. 2012, 116, 266–270. [Google Scholar] [CrossRef]
- Song, J.; Zhao, K.; Tian, X.; Xie, Y.; He, Y.; Dong, S. Effects of different environmental factors and carbon and nitrogen sources on the nitrogen removing performance of Bacillus pumilus BP-171. Period. Ocean. Univ. China 2019, 49, 34–42. [Google Scholar] [CrossRef]
- Avnimelech, Y. Carbon/Nitrogen Ratio as a Control Element in Aquaculture Systems. Aquaculture 1999, 176, 227–235. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly Accurate OTU Sequences from Microbial Amplicon Reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.X.; Zhao, S.M.; Peng, N.; Xu, C.P.; Wang, J.; Liang, Y.X. Effects of a Probiotic (Bacillus subtilis FY99-01) on the Bacterial Community Structure and Composition of Shrimp (Litopenaeus vannamei, Boone) Culture Water Assessed by Denaturing Gradient Gel Electrophoresis and High-Throughput Sequencing. Aquac. Res. 2016, 47, 857–869. [Google Scholar] [CrossRef]
- Moriarty, D.J.W. Control of Luminous Vibrio Species in Penaeid Aquaculture Ponds. Aquaculture 1998, 164, 351–358. [Google Scholar] [CrossRef]
- Azim, M.E.; Little, D.C. The Biofloc Technology (BFT) in Indoor Tanks: Water Quality, Biofloc Composition, and Growth and Welfare of Nile Tilapia (Oreochromis Niloticus). Aquaculture 2008, 283, 29–35. [Google Scholar] [CrossRef]
- Megahed, M.E.; Mohamed, K. Sustainable Growth of Shrimp Aquaculture Through Biofloc Production as Alternative to Fishmeal in Shrimp Feeds. J. Agric. Sci. 2014, 6, p176. [Google Scholar] [CrossRef] [Green Version]
- Khanjani, M.H.; Sajjadi, M.M.; Alizadeh, M.; Sourinejad, I. Nursery Performance of Pacific White Shrimp (Litopenaeus vannamei Boone, 1931) Cultivated in a Biofloc System: The Effect of Adding Different Carbon Sources. Aquac. Res. 2017, 48, 1491–1501. [Google Scholar] [CrossRef]
- Crab, R.; Avnimelech, Y.; Defoirdt, T.; Bossier, P.; Verstraete, W. Nitrogen Removal Techniques in Aquaculture for a Sustainable Production. Aquaculture 2007, 270, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Emerenciano, M.; Cuzon, G.; Arévalo, M.; Miquelajauregui, M.M.; Gaxiola, G. Effect of Short-Term Fresh Food Supplementation on Reproductive Performance, Biochemical Composition, and Fatty Acid Profile of Litopenaeus vannamei (Boone) Reared under Biofloc Conditions. Aquac. Int. 2013, 21, 987–1007. [Google Scholar] [CrossRef]
- Ahmad, I.; Babitha Rani, A.M.; Verma, A.K.; Maqsood, M. Biofloc Technology: An Emerging Avenue in Aquatic Animal Healthcare and Nutrition. Aquac. Int. 2017, 25, 1215–1226. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Kim, S.; Shin, J.; Kim, M.-G.; Gunathilaka, B.E.; Kim, S.H.; Kim, J.E.; Ji, S.-C.; Han, J.E.; Lee, K.-J. Dietary Supplementations of Bacillus Probiotic Improve Digestibility, Growth Performance, Innate Immunity, and Water Ammonia Level for Pacific White Shrimp, Litopenaeus vannamei. Aquac. Int. 2021, 29, 2463–2475. [Google Scholar] [CrossRef]
- Abdel-Latif, H.M.R.; Yilmaz, E.; Dawood, M.A.O.; Ringø, E.; Ahmadifar, E.; Yilmaz, S. Shrimp Vibriosis and Possible Control Measures Using Probiotics, Postbiotics, Prebiotics, and Synbiotics: A Review. Aquaculture 2022, 551, 737951. [Google Scholar] [CrossRef]
- Op den Camp, H.J.M.; Islam, T.; Stott, M.B.; Harhangi, H.R.; Hynes, A.; Schouten, S.; Jetten, M.S.M.; Birkeland, N.-K.; Pol, A.; Dunfield, P.F. Environmental, Genomic and Taxonomic Perspectives on Methanotrophic Verrucomicrobia: Perspectives on Methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 2009, 1, 293–306. [Google Scholar] [CrossRef]
- Yoon, J.; Matsuo, Y.; Adachi, K.; Nozawa, M.; Matsuda, S.; Kasai, H.; Yokota, A. Description of Persicirhabdus sediminis Gen. Nov., Sp. Nov., Roseibacillus ishigakijimensis Gen. Nov., Sp. Nov., Roseibacillus ponti Sp. Nov., Roseibacillus persicicus Sp. Nov., Luteolibacter pohnpeiensis Gen. Nov., Sp. Nov. and Luteolibacter algae Sp. Nov., Six Marine Members of the Phylum “Verrucomicrobia”, and Emended Descriptions of the Class Verrucomicrobiae, the Order Verrucomicrobiales and the Family Verrucomicrobiaceae. Int. J. Syst. Evol. Microbiol. 2008, 58, 998–1007. [Google Scholar] [CrossRef] [PubMed]
- Szuróczki, S.; Abbaszade, G.; Szabó, A.; Bóka, K.; Schumann, P.; Tóth, E. Phragmitibacter flavus Gen. Nov., Sp. Nov. a New Member of the Family Verrucomicrobiaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2108–2114. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, R.A.; Peeters, S.H.; Versantvoort, W.; Picone, N.; Pol, A.; Jetten, M.S.M.; Op den Camp, H.J.M. Verrucomicrobial Methanotrophs: Ecophysiology of Metabolically Versatile Acidophiles. FEMS Microbiol. Rev. 2021, 45, fuab007. [Google Scholar] [CrossRef] [PubMed]
- Arrigo, K.R. Marine Microorganisms and Global Nutrient Cycles. Nature 2005, 437, 349–355. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, X.; Li, J.; Lee, C.T.; Ong, P.Y.; Zhang, Z.; Li, C. Effect of Aquaculture Salinity on Nitrification and Microbial Community in Moving Bed Bioreactors with Immobilized Microbial Granules. Bioresour. Technol. 2020, 297, 122427. [Google Scholar] [CrossRef]
- Wyman, M.; Hodgson, S.; Bird, C. Denitrifying Alphaproteobacteria from the Arabian Sea That Express NosZ, the Gene Encoding Nitrous Oxide Reductase, in Oxic and Suboxic Waters. Appl. Environ. Microbiol. 2013, 79, 2670–2681. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Qi, L.; Huang, R.; Wang, F.; Wang, Z.; Gao, M. Characterization of Denitrifying Community for Application in Reducing Nitrogen: A Comparison of NirK and NirS Gene Diversity and Abundance. Appl. Biochem. Biotechnol. 2020, 192, 22–41. [Google Scholar] [CrossRef]
- Hu, D.; Wang, L.; Zhao, R.; Zeng, J.; Shao, Z. Core Microbiome Involved in Nitrite Removal in Shrimp Culture Ponds. Aquac. Res. 2022, 53, 1663–1675. [Google Scholar] [CrossRef]
- Hameed, A.; Shahina, M.; Lin, S.-Y.; Nakayan, P.; Liu, Y.-C.; Lai, W.-A.; Hsu, Y.-H. Youngimonas vesicularis Gen. Nov., Sp. Nov., of the Family Rhodobacteraceae, Isolated from Surface Seawater, Reclassification of Donghicola xiamenensis Tan et al. 2009 as Pseudodonghicola xiamenensis Gen. Nov., Comb. Nov. and Emended Description of the Genus Donghicola yoon et al. 2007. Int. J. Syst. Evol. Microbiol. 2014, 64, 2729–2737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam Chowdhury, M.M.; Nakhla, G. Anammox Enrichment: Impact of Sludge Retention Time on Nitrogen Removal. Environ. Technol. 2021, 43, 4426–4437. [Google Scholar] [CrossRef] [PubMed]
- Brailo, M.; Schreier, H.J.; McDonald, R.; Maršić-Lučić, J.; Gavrilović, A.; Pećarević, M.; Jug-Dujaković, J. Bacterial Community Analysis of Marine Recirculating Aquaculture System Bioreactors for Complete Nitrogen Removal Established from a Commercial Inoculum. Aquaculture 2019, 503, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Mergaert, J.; Swings, J. Phyllobacterium. In Bergey’s Manual of Systematics of Archaea and Bacteria; Whitman, W.B., Rainey, F., Kämpfer, P., Trujillo, M., Chun, J., DeVos, P., Hedlund, B., Dedysh, S., Eds.; Wiley: Hoboken, NJ, USA, 2015; pp. 1–7. ISBN 978-1-118-96060-8. [Google Scholar]
- Zheng, Q.; Wang, Y.; Lu, J.; Lin, W.; Chen, F.; Jiao, N. Metagenomic and Metaproteomic Insights into Photoautotrophic and Heterotrophic Interactions in a Synechococcus Culture. mBio 2020, 11, e03261-19. [Google Scholar] [CrossRef] [Green Version]
- Rout, P.R.; Bhunia, P.; Dash, R.R. Simultaneous Removal of Nitrogen and Phosphorous from Domestic Wastewater Using Bacillus cereus GS-5 Strain Exhibiting Heterotrophic Nitrification, Aerobic Denitrification and Denitrifying Phosphorous Removal. Bioresour. Technol. 2017, 244, 484–495. [Google Scholar] [CrossRef]
- Verbaendert, I.; Boon, N.; De Vos, P.; Heylen, K. Denitrification Is a Common Feature among Members of the Genus Bacillus. Syst. Appl. Microbiol. 2011, 34, 385–391. [Google Scholar] [CrossRef]
- Deng, Y.; Jiang, Y.-H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular Ecological Network Analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef] [Green Version]
- Venturelli, O.S.; Carr, A.V.; Fisher, G.; Hsu, R.H.; Lau, R.; Bowen, B.P.; Hromada, S.; Northen, T.; Arkin, A.P. Deciphering Microbial Interactions in Synthetic Human Gut Microbiome Communities. Mol. Syst. Biol. 2018, 14, e8157. [Google Scholar] [CrossRef]
- Coyte, K.Z.; Schluter, J.; Foster, K.R. The Ecology of the Microbiome: Networks, Competition, and Stability. Science 2015, 350, 663–666. [Google Scholar] [CrossRef]
- Liu, L.; Wang, M.; Wei, C.; Liu, Y.; Pan, M.; Wang, S.; Cui, L.; Tian, X. Effects of Dietary Poly-β-Hydroxybutyrate Supplementation on the Growth, Non-Specific Immunity, and Intestinal Microbiota of the Sea Cucumber Apostichopus japonicus. Front. Mar. Sci. 2022, 9, 855938. [Google Scholar] [CrossRef]
- Kanehisa, M. Enzyme Annotation and Metabolic Reconstruction Using KEGG. In Protein Function Prediction; Kihara, D., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; Volume 1611, pp. 135–145. ISBN 978-1-4939-7013-1. [Google Scholar]
- Peh, E.; Kittler, S.; Reich, F.; Kehrenberg, C. Antimicrobial Activity of Organic Acids against Campylobacter Spp. and Development of Combinations—A Synergistic Effect? PLoS ONE 2020, 15, e0239312. [Google Scholar] [CrossRef]
- Yu, H.; Huang, G.H.; Zhang, X.D.; Li, Y. Inhibitory Effects of Organic Acids on Bacteria Growth During Food Waste Composting. Compos. Sci. Util. 2010, 18, 55–63. [Google Scholar] [CrossRef]
- Nieto-Peñalver, C.G.; Savino, M.J.; Bertini, E.V.; Sánchez, L.A.; de Figueroa, L.I.C. Gluconic Acid Produced by Gluconacetobacter Diazotrophicus Pal5 Possesses Antimicrobial Properties. Res. Microbiol. 2014, 165, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Defoirdt, T.; Mai Anh, N.T.; De Schryver, P. Virulence-Inhibitory Activity of the Degradation Product 3-Hydroxybutyrate Explains the Protective Effect of Poly-β-Hydroxybutyrate against the Major Aquaculture Pathogen Vibrio campbellii. Sci. Rep. 2018, 8, 7245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukami, K.; Takagi, F.; Sonoda, K.; Okamoto, H.; Kaneno, D.; Horikawa, T.; Takita, M. Effects of the Monomeric Components of Poly-Hydroxybutyrate-Co-Hydroxyhexanoate on the Growth of Vibrio penaeicida In Vitro and on the Survival of Infected Kuruma Shrimp (Marsupenaeus japonicus). Animals 2021, 11, 567. [Google Scholar] [CrossRef]
- Cho, B.; Park, M.; Shim, J.; Azam, F. Significance of Bacteria in Urea Dynamics in Coastal Surface Waters. Mar. Ecol. Prog. Ser. 1996, 142, 19–26. [Google Scholar] [CrossRef] [Green Version]
- Jørgensen, N. Uptake of Urea by Estuarine Bacteria. Aquat. Microb. Ecol. 2006, 42, 227–242. [Google Scholar] [CrossRef]
- Siebers, D. Bacterial—Invertebrate Interactions in Uptake of Dissolved Organic Matter. Am Zool 1982, 22, 723–733. [Google Scholar] [CrossRef] [Green Version]
- Preston, R.L. Transport of Amino Acids by Marine Invertebrates. J. Exp. Zool. 1993, 265, 410–421. [Google Scholar] [CrossRef]
- Moss, S.M.; Pruder, G.D. Characterization of Organic Particles Associated with Rapid Growth in Juvenile White Shrimp, Penaeus vannamei Boone, Reared under Intensive Culture Conditions. J. Exp. Mar. Biol. Ecol. 1995, 187, 175–191. [Google Scholar] [CrossRef]
- Vogt, G. Synthesis of Digestive Enzymes, Food Processing, and Nutrient Absorption in Decapod Crustaceans: A Comparison to the Mammalian Model of Digestion. Zoology 2021, 147, 125945. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Han, T.; Zheng, S.; Wu, G. Nutrition and Functions of Amino Acids in Aquatic Crustaceans. Amino Acids Nutr. Health 2021, 1285, 169–198. [Google Scholar] [CrossRef]




| Treatment | B | BP | BM | DZ |
|---|---|---|---|---|
| Initial body weight (g) | 6.05 ± 0.05 | 6.07 ± 0.07 | 6.03 ± 0.06 | 6.12 ± 0.01 |
| Final body weight (g) | 13.64 ± 0.63 a | 17.23 ± 0.18 c | 15.12 ± 0.13 b | 12.92 ± 0.53 a |
| Gross weight (g) | 504.23 ± 7.74 b | 637.78 ± 14.89 c | 526.13 ± 16.88 b | 478.15 ± 12.56 a |
| Survival rate (%) | 70.00 ± 0.06 b | 88.57 ± 0.01 b | 82.74 ± 0.03 b | 64.31 ± 0.03 a |
| SGR (%) | 2.70 ± 0.13 a | 3.47 ± 0.07 c | 3.07 ± 0.06 b | 2.48 ± 0.14 a |
| FER (%) | 51.36 ± 6.96 a | 70.27 ± 2.65 b | 55.77 ± 3.50 a | 45.34 ± 1.94 a |
| Groups | α-Diversity Index | Coverage | |||
|---|---|---|---|---|---|
| Shannon | Simpson | Ace | Chao | ||
| B | 3.23 ± 0.69 | 0.10 ± 0.07 | 438.93 ± 97.23 | 440.16 ± 87.69 | 0.998 |
| BP | 2.74 ± 0.69 | 0.18 ± 0.11 | 443.16 ± 134.35 | 435.90 ± 122.11 | 0.997 |
| BM | 3.56 ± 0.46 | 0.06 ± 0.02 | 459.62 ± 30.45 | 447.41 ± 31.13 | 0.998 |
| DZ | 3.36 ± 0.08 | 0.07 ± 0.01 | 411.01 ± 29.19 | 370.11 ± 19.18 | 0.998 |
| B | BP | BM | DZ | |
|---|---|---|---|---|
| Nodes | 328 | 323 | 334 | 285 |
| Edges | 1270 | 1056 | 1256 | 1223 |
| Positive relationship | 800 | 616 | 753 | 733 |
| Negative relationship | 470 | 440 | 503 | 490 |
| negative interactions/positive interaction ratio | 58.75% | 71.43% | 66.80% | 66.85% |
| Module number | 7 | 26 | 16 | 7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Liu, Y.; Luo, K.; Li, T.; Liu, Q.; Tian, X. Effects of Bacillus pumilus BP-171 and Carbon Sources on the Growth Performance of Shrimp, Water Quality and Bacterial Community in Penaeus vannamei Culture System. Water 2022, 14, 4037. https://doi.org/10.3390/w14244037
Wang M, Liu Y, Luo K, Li T, Liu Q, Tian X. Effects of Bacillus pumilus BP-171 and Carbon Sources on the Growth Performance of Shrimp, Water Quality and Bacterial Community in Penaeus vannamei Culture System. Water. 2022; 14(24):4037. https://doi.org/10.3390/w14244037
Chicago/Turabian StyleWang, Mingyang, Yang Liu, Kai Luo, Tengfei Li, Qingbing Liu, and Xiangli Tian. 2022. "Effects of Bacillus pumilus BP-171 and Carbon Sources on the Growth Performance of Shrimp, Water Quality and Bacterial Community in Penaeus vannamei Culture System" Water 14, no. 24: 4037. https://doi.org/10.3390/w14244037
APA StyleWang, M., Liu, Y., Luo, K., Li, T., Liu, Q., & Tian, X. (2022). Effects of Bacillus pumilus BP-171 and Carbon Sources on the Growth Performance of Shrimp, Water Quality and Bacterial Community in Penaeus vannamei Culture System. Water, 14(24), 4037. https://doi.org/10.3390/w14244037






