HCH Removal in a Biochar-Amended Biofilter
Abstract
:1. Introduction
2. Material and Methods
2.1. Biochar Characteristics
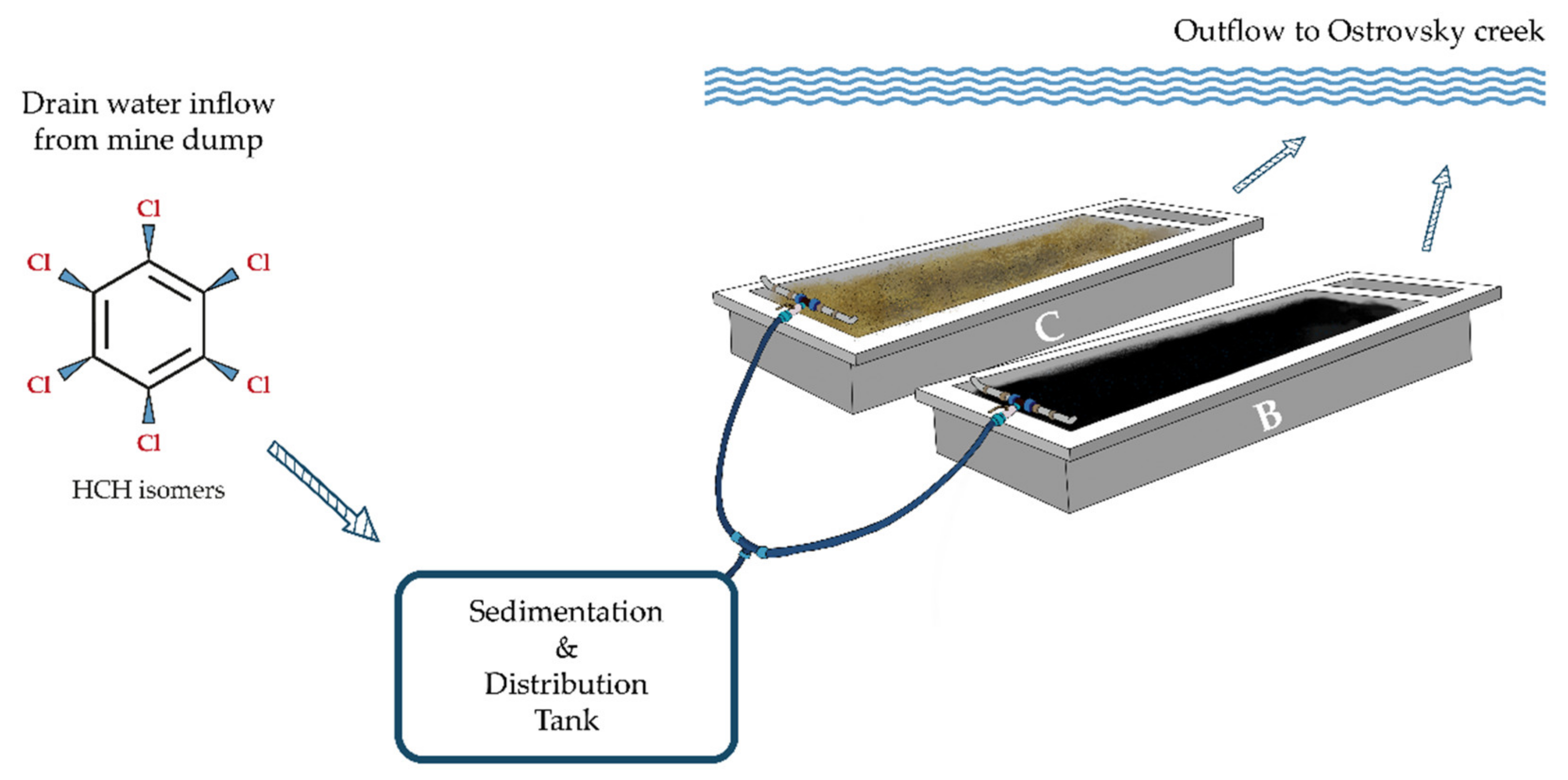
2.2. Site Description and Experimental Design
2.3. Sampling
2.4. Chemical Analysis
2.5. Molecular Biology Analysis
2.5.1. Real-Time Quantitative PCR
2.5.2. Amplicon 16S rRNA Sequencing
2.6. Statistical Analysis
3. Results and Discussion
3.1. General Characteristics of Inflow Water
3.2. Pollutant Removal Efficiency
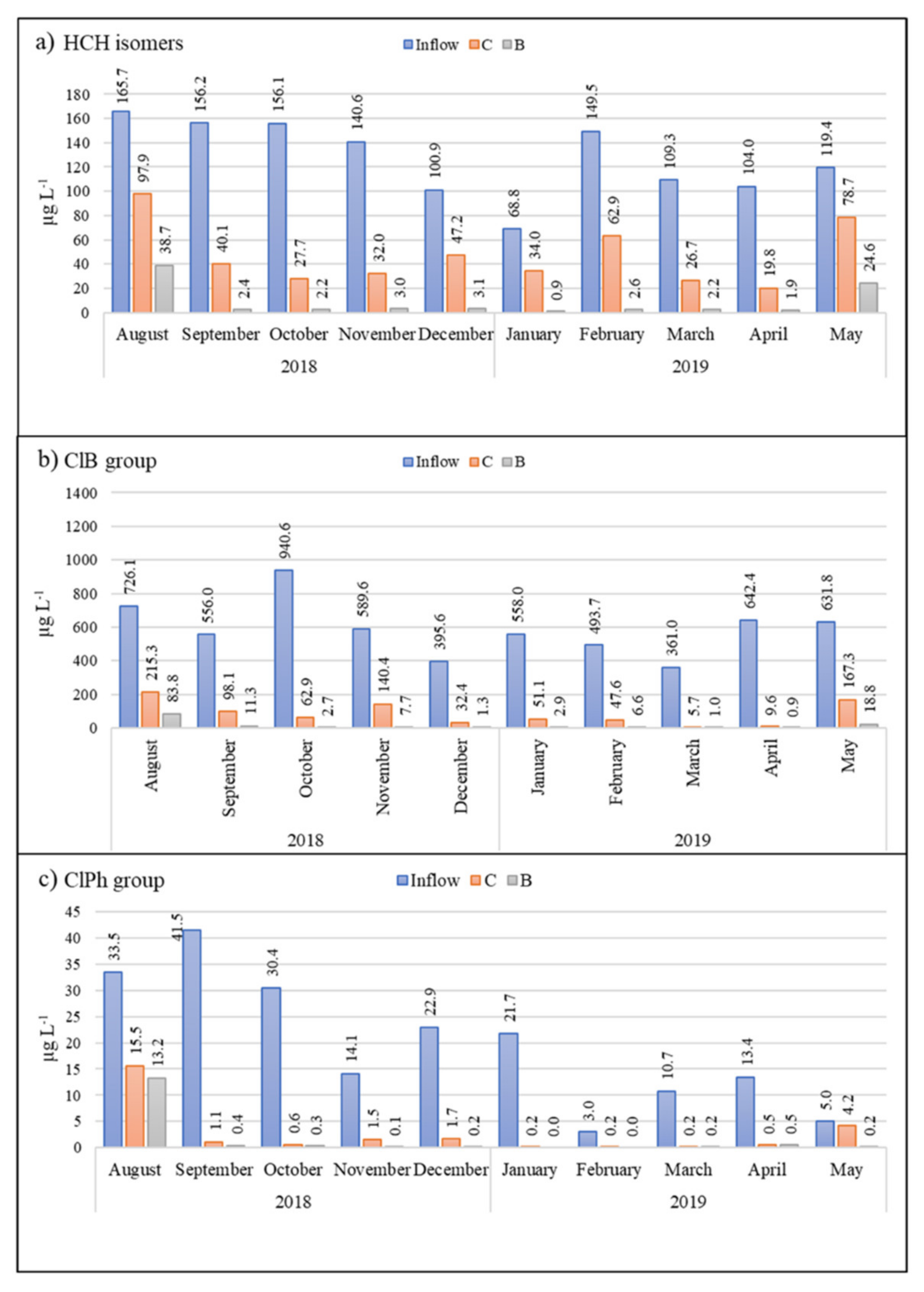
3.2.1. HCH Isomers
3.2.2. Chlorobenzenes
- From August to November, the decrease was oscillatory (58.2%, 28%, 57.4%, and 41%, respectively);
- From December (8.3%), its removal consistently increased and in April reached 84.4%.
3.2.3. Chlorophenols
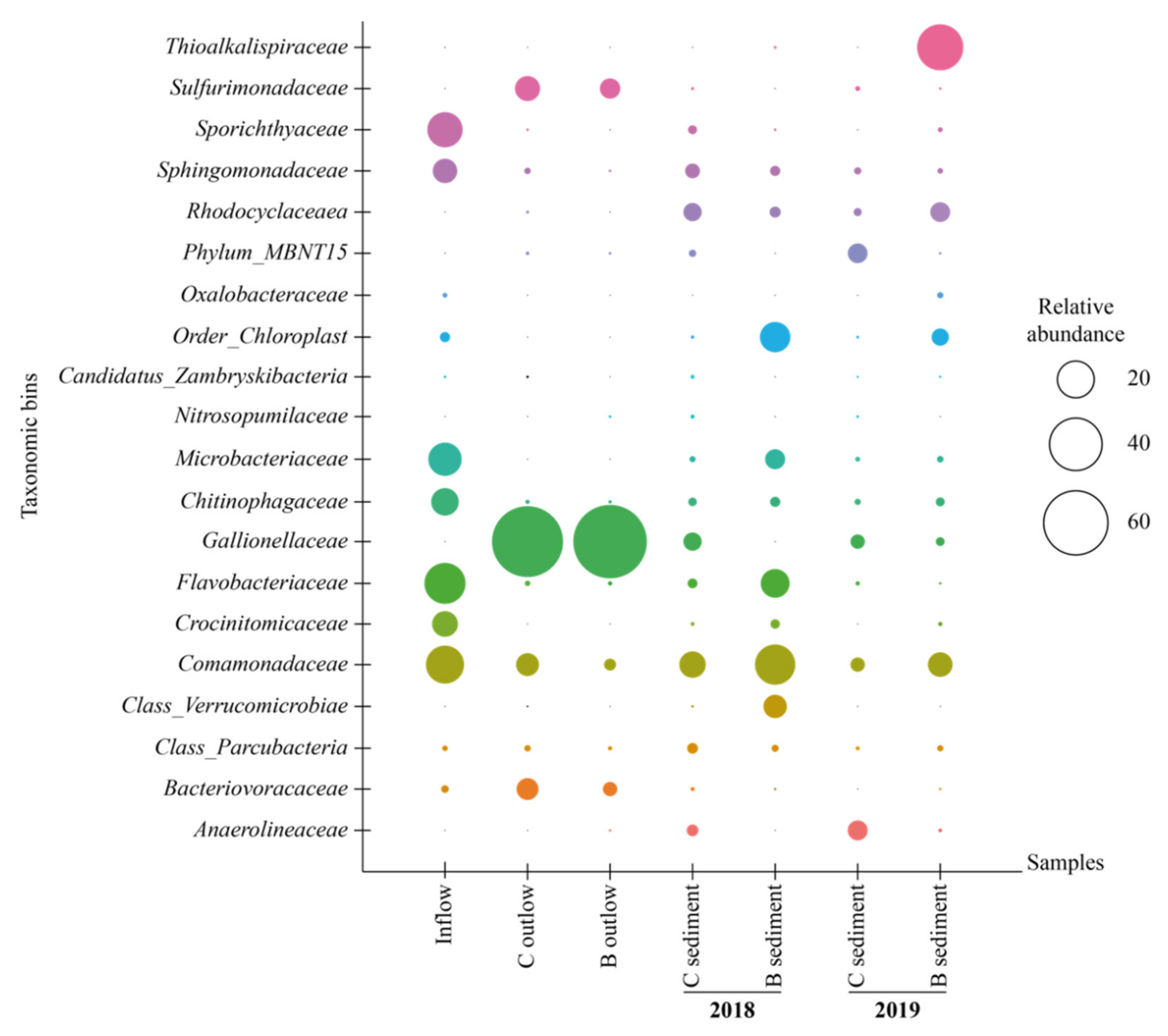
3.3. Microbial Community Structure and Function
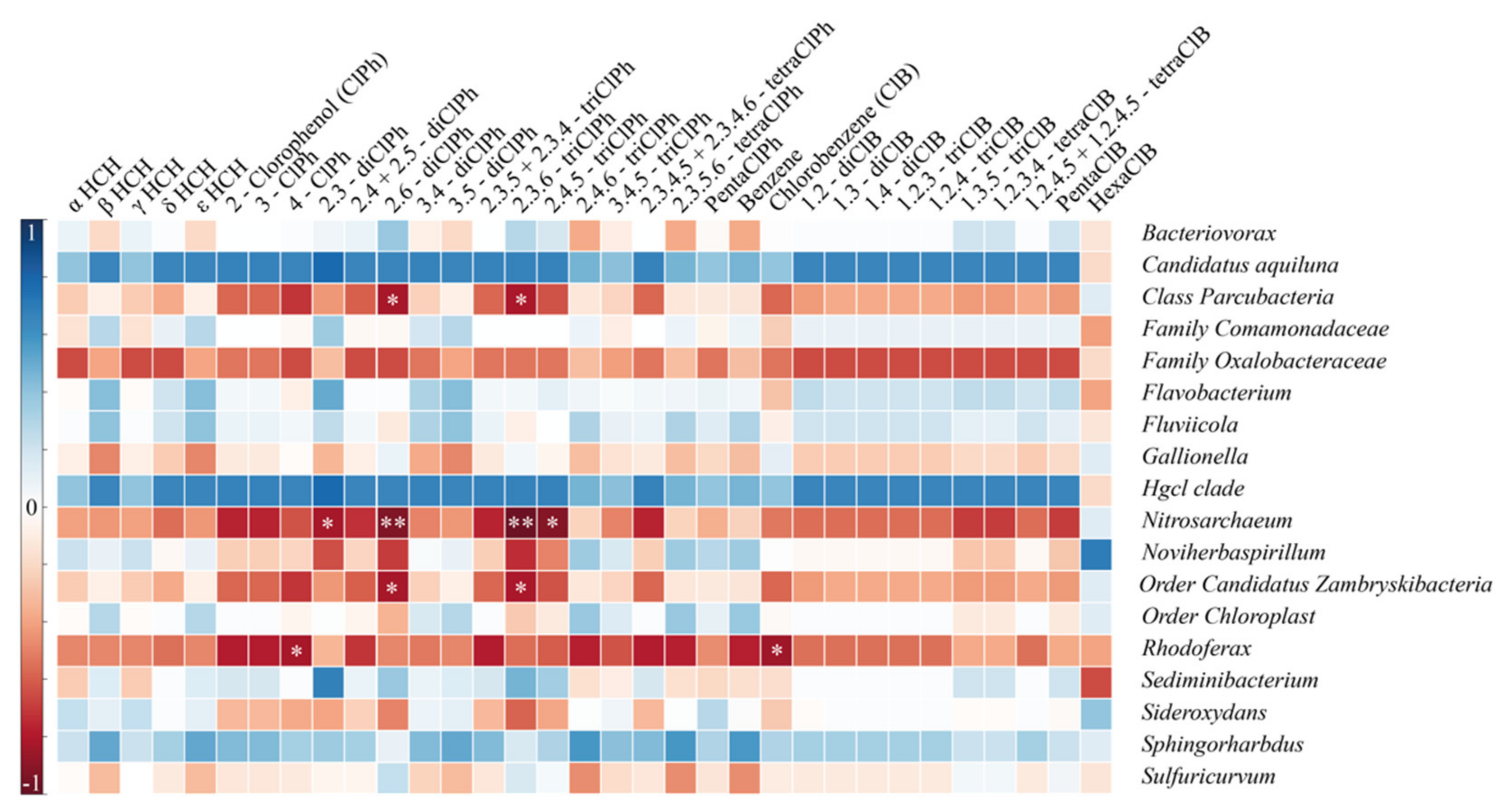
3.4. Functional Genes Involved in HCH Biodegradation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stockholm Convention—An Overview ScienceDirect Topics. Available online: https://www.sciencedirect.com/topics/earth-and-planetary-sciences/stockholm-convention (accessed on 21 June 2021).
- Bajpai, A.; Shukla, P.; Dixit, B.S.; Banerji, R. Concentrations of organochlorine insecticides in edible oils from different regions of India. Chemosphere 2007, 67, 1403–1407. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, P.; Zhang, G.; Li, J.; Sampathkumar, P.; Balasubramanian, T.; Kathiresan, K.; Shin Takahashi, S.; Subramanian, A.; Tanabe, S.; Jones, C.K. Seasonal variation of atmospheric organochlorine pesticides and polybrominated diphenyl ethers in Parangipettai, Tamil Nadu, India: Implication for atmospheric transport. Sci. Total Environ. 2019, 649, 1653–1660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erkmen, B.; Kolankaya, D. Determination of organochlorine pesticide residues in water, sediment, and fish samples from the Meriç Delta, Turkey. Int. J. Environ. Anal. Chem. 2006, 86, 161–169. [Google Scholar] [CrossRef]
- Rigét, F.; Bignert, A.; Braune, B.; Dam, M.; Dietz, R.; Evans, M.; Green, N.; Gunnlaugsdottir, H.; Hoydal, K.S.; Kucklick, J.; et al. Temporal trends of persistent organic pollutants in Arctic marine and freshwater biota. Sci. Total Environ. 2019, 649, 99–110. [Google Scholar] [CrossRef]
- Zhao, G.; Xu, Y.; Li, W.; Han, G.; Ling, B. PCBs and OCPs in human milk and selected foods from Luqiao and Pingqiao in Zhejiang, China. Sci. Total Environ. 2007, 378, 281–292. [Google Scholar] [CrossRef] [PubMed]
- ERA-Consult Madrid, Directorate-General for Internal Policies of the Union (European Parliament), M. Vega, D. Romano, and E. Uotila. Lindane (Persistant Organic Pollutant) in the EU. 30 January 2017. Available online: http://op.europa.eu/en/publication-detail/-/publication/8e37ab69-bd13-11e6-a237-01aa75ed71a1 (accessed on 24 May 2021).
- Breivik, K.; Pacyna, J.M.; Münch, J. Use of α-, β- and γ-hexachlorocyclohexane in Europe, 1970–1996. Sci. Total Environ. 1999, 239, 151–163. [Google Scholar] [CrossRef]
- Dominguez, C.M.; Parchão, J.; Rodriguez, S.; Lorenzo, D.; Romero, A.; Santos, A. Kinetics of Lindane Dechlorination by Zerovalent Iron Microparticles: Effect of Different Salts and Stability Study. Ind. Eng. Chem. Res. 2016, 55, 12776–12785. [Google Scholar] [CrossRef]
- Homolková, M.; Hrabák, P.; Kolář, M.; Černík, M. Degradability of hexachlorocyclohexanes in water using ferrate (VI). Water Sci. Technol. 2015, 71, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Joo, S.H.; Zhao, D. Destruction of lindane and atrazine using stabilized iron nanoparticles under aerobic and anaerobic conditions: Effects of catalyst and stabilizer. Chemosphere 2008, 70, 418–425. [Google Scholar] [CrossRef]
- Merz, J.P.; Gamoke, B.C.; Foley, M.P.; Raghavachari, K.; Peters, D.G. Electrochemical reduction of (1R,2r,3S,4R,5r,6S)-hexachlorocyclohexane (Lindane) at carbon cathodes in dimethylformamide. J. Electroanal. Chem. 2011, 660, 121–126. [Google Scholar] [CrossRef]
- Wacławek, S.; Silvestri, D.; Hrabák, P.; Padil, V.V.; Torres-Mendieta, R.; Wacławek, M.; Černík, M.; Dionysiou, D. Chemical oxidation and reduction of hexachlorocyclohexanes: A review. Water Res. 2019, 162, 302–319. [Google Scholar] [CrossRef] [PubMed]
- Beesley, L.; Moreno-Jiménez, E.; Gomez-Eyles, J.L.; Harris, E.; Robinson, B.; Sizmur, T. A review of biochars’ potential role in the remediation, revegetation and restoration of contaminated soils. Environ. Pollut. 2011, 159, 3269–3282. [Google Scholar] [CrossRef] [PubMed]
- Denyes, M.J.; Langlois, V.S.; Rutter, A.; Zeeb, B.A. The use of biochar to reduce soil PCB bioavailability to Cucurbita pepo and Eisenia fetida. Sci. Total Environ. 2012, 437, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Ghosh, C.; Boettinger, W.J. Diffusion Parameters and Growth Mechanism of Phases in the Cu-Sn System. Metall. Mater. Trans. A 2011, 42, 952–963. [Google Scholar] [CrossRef]
- Mohan, D.; Sarswat, A.; Ok, Y.S.; Pittman, C.U. Organic and inorganic contaminants removal from water with biochar, a renewable, low cost and sustainable adsorbent—A critical review. Bioresour. Technol. 2014, 160, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Eyles, J.L.; Sizmur, T.; Collins, C.D.; Hodson, M.E. Effects of biochar and the earthworm Eisenia fetida on the bioavailability of polycyclic aromatic hydrocarbons and potentially toxic elements. Environ. Pollut. 2011, 159, 616–622. [Google Scholar] [CrossRef]
- Silvani, L.; Cornelissen, G.; Hale, S.E. Sorption of α-, β-, γ- and δ-hexachlorocyclohexane isomers to three widely different biochars: Sorption mechanisms and application. Chemosphere 2019, 219, 1044–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Shi, Z.; Li, D.; Rey, A.; Ruan, H.; Craine, J.M.; Liang, J.; Zhou, J.; Luo, Y. Soil properties control decomposition of soil organic carbon: Results from data-assimilation analysis. Geoderma 2016, 262, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Zimmerman, A.R.; Gao, B.; Ahn, M.-Y. Positive and negative carbon mineralization priming effects among a variety of biochar-amended soils. Soil Biol. Biochem. 2011, 43, 1169–1179. [Google Scholar] [CrossRef]
- Ali, I.; Asim, M.; Khan, T.A. Low cost adsorbents for the removal of organic pollutants from wastewater. J. Environ. Manag. 2012, 113, 170–183. [Google Scholar] [CrossRef]
- Ahmad, M.; Rajapaksha, A.U.; Lim, J.E.; Zhang, M.; Bolan, N.; Mohan, D.; Vithanage, M.; Lee, S.S.; Ok, Y.S. Biochar as a sorbent for contaminant management in soil and water: A review. Chemosphere 2014, 99, 19–33. [Google Scholar] [CrossRef]
- Grgić, M.; Maletić, S.; Beljin, J.; Isakovski, M.K.; Rončević, S.; Tubić, A.; Agbaba, J. Lindane and hexachlorobenzene sequestration and detoxification in contaminated sediment amended with carbon-rich sorbents. Chemosphere 2019, 220, 1033–1040. Available online: https://agris.fao.org/agris-search/search.do?recordID=US201900062586 (accessed on 1 May 2020). [CrossRef]
- Lal, R.; Pandey, G.; Sharma, P.; Kumari, K.; Malhotra, S.; Pandey, R.; Raina, V.; Kohler, H.-P.E.; Holliger, C.; Jackson, C.; et al. Biochemistry of Microbial Degradation of Hexachlorocyclohexane and Prospects for Bioremediation. Microbiol. Mol. Biol. Rev. MMBR 2010, 74, 58–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bala, K.; Sharma, P.; Lal, R. Sphingobium quisquiliarum sp. nov., a hexachlorocyclohexane (HCH)-degrading bacterium isolated from an HCH-contaminated soil. Int. J. Syst. Evol. Microbiol. 2010, 60, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Böltner, D.; Moreno-Morillas, S.; Ramos, J.-L. 16S rDNA phylogeny and distribution of lin genes in novel hexachlorocyclohexane-degrading Sphingomonas strains. Environ. Microbiol. 2005, 7, 1329–1338. [Google Scholar] [CrossRef]
- Mohn, W.W.; Mertens, B.; Neufeld, J.D.; Verstraete, W.; Lorenzo, V.D. Distribution and phylogeny of hexachlorocyclohexane-degrading bacteria in soils from Spain. Environ. Microbiol. 2006, 8, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Saxena, A.; Anand, S.; Dua, A.; Sangwan, N.; Khan, F.; Lal, R. Novosphingobium lindaniclasticum sp. nov., a hexachlorocyclohexane (HCH)-degrading bacterium isolated from an HCH dumpsite. Int. J. Syst. Evol. Microbiol. 2013, 63, 2160–2167. [Google Scholar] [CrossRef] [PubMed]
- Nagata, Y.; Miyauchi, K.; Takagi, M. Complete analysis of genes and enzymes for γ-hexachlorocyclohexane degradation in Sphingomonas paucimobilis UT26. J. Ind. Microbiol. Biotechnol. 1999, 23, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Oakley, A.J.; Klvaňa, M.; Otyepka, M.; Nagata, Y.; Wilce, M.C.J.; Damborský, J. Crystal Structure of Haloalkane Dehalogenase LinB from Sphingomonas paucimobilis UT26 at 0.95 Å Resolution: Dynamics of Catalytic Residues. Biochemistry 2004, 43, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Okai, M.; Kubota, K.; Fukuda, M.; Nagata, Y.; Nagata, K.; Tanokura, M. Crystal Structure of γ-Hexachlorocyclohexane Dehydrochlorinase LinA from Sphingobium japonicum UT26. J. Mol. Biol. 2010, 403, 260–269. [Google Scholar] [CrossRef]
- Brittain, D.R.B.; Pandey, R.; Kumari, K.; Sharma, P.; Pandey, G.; Lal, R.; Coote, M.L.; Oakeshott, J.G.; Jackson, C.J. Competing SN2 and E2 reaction pathways for hexachlorocyclohexane degradation in the gas phase, solution and enzymes. Chem. Commun. 2010, 47, 976–978. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, T.; Chang, S.X.; Jiang, X.; Song, Y. Biochar increases soil microbial biomass but has variable effects on microbial diversity: A meta-analysis. Sci. Total Environ. 2020, 749, 141593. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Yin, C.; Chen, T.; Quan, G.; Ippolito, J.A.; Liu, B.; Yan, J.; Ding, C.; Hussain, Q.; Umer, M. Remediation of organic halogen- contaminated wetland soils using biochar. Sci. Total Environ. 2019, 696, 134087. [Google Scholar] [CrossRef]
- Odedishemi, F.O.; Wang, H.-C.; Guadie, A.; Ajibade, T.F.; Fang, Y.-K.; Sharif, H.M.A.; Liu, W.-Z.; Wang, A.-J. Total nitrogen removal in biochar amended non-aerated vertical flow constructed wetlands for secondary wastewater effluent with low C/N ratio: Microbial community structure and dissolved organic carbon release conditions. Bioresour. Technol. 2021, 322, 124430. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Huang, L.; Liang, Y.; Xiang, H.; Jiang, J.; Wang, Q.; Hou, J.; Chen, Y. Response of microbes to biochar strengthen nitrogen removal in subsurface flow constructed wetlands: Microbial community structure and metabolite characteristics. Sci. Total Environ. 2019, 694, 133687. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Lal, D.; Lata, P.; Sangwan, N.; Garg, N.; Holliger, C.; Lal, R. Changes in the bacterial community and lin genes diversity during biostimulation of indigenous bacterial community of hexachlorocyclohexane (HCH) dumpsite soil. Microbiology 2013, 82, 234–240. [Google Scholar] [CrossRef]
- Suar, M.; van der Meer, J.R.; Lawlor, K.; Holliger, C.; Lal, R. Dynamics of Multiple lin Gene Expression in Sphingomonas paucimobilis B90A in Response to Different Hexachlorocyclohexane Isomers. Appl. Environ. Microbiol. 2004, 70, 6650–6656. [Google Scholar] [CrossRef] [Green Version]
- Clifford, R.J.; Milillo, M.; Prestwood, J.; Quintero, R.; Zurawski, D.V.; Kwak, Y.I.; Waterman, P.E.; Lesho, E.P.; Mc Gann, P. Detection of Bacterial 16S rRNA and Identification of Four Clinically Important Bacteria by Real-Time PCR. PLoS ONE 2012, 7, e48558. [Google Scholar] [CrossRef]
- Nechanická, M.D.L.; Dolinová, I. Use of Nanofiber Carriers for Monitoring of Microbial Biomass. In Topical Issues of Rational Use of Natural Resources: Proceedings of the International Forum-Contest of Young Researchers, St. Petersburg, Russia, 18–20 April 2018; CRC Press: Boca Raton, FL, USA, 2018; p. 361. Available online: https://www.routledge.com/Topical-Issues-of-Rational-Use-of-Natural-Resources-Proceedings-of-the/Litvinenko/p/book/9780367027438 (accessed on 18 November 2020).
- Claesson, M.J.; Wang, Q.; O’Sullivan, O.; Greene-Diniz, R.; Cole, J.R.; Ross, R.; O’Toole, P. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010, 38, e200. [Google Scholar] [CrossRef]
- Dowd, S.E.; Callaway, T.R.; Wolcott, R.D.; Sun, Y.; McKeehan, T.; Hagevoort, R.G.; Edrington, T.S. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 2008, 8, 125. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ssekagiri, A.; Sloan, W.T.; Ijaz, U.Z. microbiomeSeq: An R package for analysis of microbial communities in an environmental context. In Proceedings of the ISCB Africa ASBCB Conference, Kumasi, Ghana, 10–12 October 2017. [Google Scholar] [CrossRef]
- Torondel, B.; Ensink, J.H.; Gundogdu, O.; Ijaz, U.Z.; Parkhill, J.; Abdelahi, F.; Nguyen, V.-A.; Sudgen, S.; Gibson, W.; Walker, A.W.; et al. Assessment of the influence of intrinsic environmental and geographical factors on the bacterial ecology of pit latrines. Microb. Biotechnol. 2016, 9, 209–223. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, B.; Simpson, G.L.; Solymos, P.; Stevens, H.; Henry, M.; Wagner, H. The Vegan Package; R Core Team: Vienna, Austria, 2009. [Google Scholar]
- Buser, H.-R.; Mueller, M.D. Isomer and Enantioselective Degradation of Hexachlorocyclohexane Isomers in Sewage Sludge under Anaerobic Conditions. Environ. Sci. Technol. 1995, 29, 664–672. [Google Scholar] [CrossRef]
- Wypych, G. Handbook of Solvents; ChemTec Publishing: Scarborough, ON, Canada, 2001. [Google Scholar]
- Arey, J.S.; Green, W.H.; Gschwend, P.M. The Electrostatic Origin of Abraham’s Solute Polarity Parameter. J. Phys. Chem. B 2005, 109, 7564–7573. [Google Scholar] [CrossRef]
- Lamarche, O.; Platts, J.A.; Hersey, A. Theoretical prediction of the polarity/polarizability parameter π2H. Phys. Chem. Chem. Phys. 2001, 3, 2747–2753. [Google Scholar] [CrossRef]
- Chen, W.; Wei, R.; Ni, J.; Yang, L.; Qian, W.; Yang, Y. Sorption of chlorinated hydrocarbons to biochars in aqueous environment: Effects of the amorphous carbon structure of biochars and the molecular properties of adsorbates. Chemosphere 2018, 210, 753–761. [Google Scholar] [CrossRef]
- Marcus, Y. Linear solvation energy relationships. Correlation and prediction of the distribution of organic solutes between water and immiscible organic solvents. J. Phys. Chem. 1991, 95, 8886–8891. [Google Scholar] [CrossRef]
- Yang, K.; Wu, W.; Jing, Q.; Zhu, L. Aqueous Adsorption of Aniline, Phenol, and their Substitutes by Multi-Walled Carbon Nanotubes. Environ. Sci. Technol. 2008, 42, 7931–7936. [Google Scholar] [CrossRef]
- Murthy, H.M.R.; Manonmani, H.K. Aerobic degradation of technical hexachlorocyclohexane by a defined microbial consortium. J. Hazard. Mater. 2007, 149, 18–25. [Google Scholar] [CrossRef]
- Liu, X.; Peng, P.; Fu, J.; Huang, W. Effects of FeS on the Transformation Kinetics of γ-Hexachlorocyclohexane. Environ. Sci. Technol. 2003, 37, 1822–1828. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Z.; Belchik, S.M.; Edwards, M.J.; Liu, C.; Kennedy, D.W.; Merkley, E.D.; Lipton, M.S.; Butt, J.N.; Richardson, D.J.; et al. Identification and Characterization of MtoA: A Decaheme c-Type Cytochrome of the Neutrophilic Fe(II)-Oxidizing Bacterium Sideroxydans lithotrophicus ES-1. Front. Microbiol. 2012, 3, 37. [Google Scholar] [CrossRef] [Green Version]
- Dadhwal, M.; Singh, A.; Prakash, O.; Gupta, S.; Kumari, K.; Sharma, P.; Jit, S.; Verma, M.; Holliger, C.; Lal, R. Proposal of biostimulation for hexachlorocyclohexane (HCH)-decontamination and characterization of culturable bacterial community from high-dose point HCH-contaminated soils. J. Appl. Microbiol. 2009, 106, 381–392. [Google Scholar] [CrossRef]
- Leoni, C.; Volpicella, M.; Fosso, B.; Manzari, C.; Piancone, E.; Dileo, M.C.G.; Arcadi, E.; Yakimov, M.; Pesole, G.; Ceci, L.R. A Differential Metabarcoding Approach to Describe Taxonomy Profiles of Bacteria and Archaea in the Saltern of Margherita di Savoia (Italy). Microorganisms 2020, 8, 936. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, K.; Suh, S.-K.; Nagata, Y.; Takagi, M. Cloning and Sequencing of a 2,5-Dichlorohydroquinone Reductive Dehalogenase Gene Whose Product Is Involved in Degradation of γ-Hexachlorocyclohexane by Sphingomonas paucimobilis. J. Bacteriol. 1998, 180, 1354–1359. [Google Scholar] [CrossRef] [Green Version]
- Nagata, Y.; Nariya, T.; Ohtomo, R.; Fukuda, M.; Yano, K.; Takagi, M. Cloning and sequencing of a dehalogenase gene encoding an enzyme with hydrolase activity involved in the degradation of gamma-hexachlorocyclohexane in Pseudomonas paucimobilis. J. Bacteriol. 1993, 175, 6403–6410. [Google Scholar] [CrossRef] [Green Version]
| Gene | ||||||
|---|---|---|---|---|---|---|
| Biofilter | U16SRT | linA | linB | linB-RT | linD | |
| 2018 | C | +++ | +++ | ++ | +++ | +++ |
| B | + | ND | ND | ND | ND | |
| 2019 | C | +++ | ++ | ++ | ++ | ++ |
| B | +++ | +++ | +++ | +++ | +++ | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amirbekov, A.; Mamirova, A.; Sevcu, A.; Spanek, R.; Hrabak, P. HCH Removal in a Biochar-Amended Biofilter. Water 2021, 13, 3396. https://doi.org/10.3390/w13233396
Amirbekov A, Mamirova A, Sevcu A, Spanek R, Hrabak P. HCH Removal in a Biochar-Amended Biofilter. Water. 2021; 13(23):3396. https://doi.org/10.3390/w13233396
Chicago/Turabian StyleAmirbekov, Aday, Aigerim Mamirova, Alena Sevcu, Roman Spanek, and Pavel Hrabak. 2021. "HCH Removal in a Biochar-Amended Biofilter" Water 13, no. 23: 3396. https://doi.org/10.3390/w13233396
APA StyleAmirbekov, A., Mamirova, A., Sevcu, A., Spanek, R., & Hrabak, P. (2021). HCH Removal in a Biochar-Amended Biofilter. Water, 13(23), 3396. https://doi.org/10.3390/w13233396






