Abstract
Cork matrices biofunctionalized with Moringa oleifera seed extracts (MoSe) have potential for use as a biofilter with antibacterial properties to reduce waterborne pathogens. The aim of this study was to evaluate the effect of cork biofunctionalized with active antimicrobial compounds of MoSe (f-cork) on the inhibition of Escherichia coli (InhEc). The LacZ gene from a strain of E. coli was used as the target sequence using viability quantification Polymerase Chain Reaction (qPCR) and differentiation of viable and dead bacteria through selective cell viability PMA staining. To perform this, a 27−4 fractional factorial design and a biofiltration system were used to evaluate the effect of the active protein in MoSe immobilized in granulated cork on InhEc. We found that the potential for antimicrobial activity increased with f-cork for an effective maximal bacterial reduction (99.99%; p < 0.05). The effect of f-cork functionalized with MoSe on E. coli viability was of 0.024% and 0.005% for the cells exposed to PMA, respectively, being the relevant conditions in treatment 2: (0 L/min) without aeration, (5%) MoSe and (5 mm) cork particle. In conclusion, the f-cork functionalized with MoSe presented biosorbent and antibacterial properties that effectively reduced the E. coli growth.
1. Introduction
Pathogenic bacteria from expanding human settlements can be an important cause of disease, giving rise to global health concerns, such as diarrhoea, episodes associated with a lack of access to safe drinking water and inadequate sanitation, which result in millions of deaths annually [1]. Escherichia coli and intestinal Enterococci are among the main bacteria used as faecal indicators and can be found frequently in irrigation and drinking water systems, groundwater, water reservoirs and rivers [2,3,4]. These water supplies, found to be contaminated with faecal bacteria due to untreated or inadequately treated water and thus considered to be unfit for human consumption, are of rising concern [5,6].
For human settlements, water pollution control, other nature-based solutions (NbS) and green infrastructure can be effectively used in combination with biofiltration systems for water’s reuse and its sustainable sanitation [7]. Many biotechnological alternatives have been proposed to treat wastewater, such as the application of natural coagulants, such as plant seed extracts, which has been shown to reduce and/or neutralize colloid loads and to remove pathogens [8]. However, one of the major limitations to the purification of water with natural coagulants is the release of organic matter [9,10]. Thus, a new strategy has been established with a functionalization process to retain the active protein adsorbed on a porous material that maintains the antimicrobial and clarification capacity of the seed extracts of Moringa oleifera, Mangifera indica and Prunus armeniaca, among others [8,11].
Some studies reported that the major antimicrobial activity of seed extracts has been attributed to active ingredients of the positively charged protein chain and their hydrophobic sections, which penetrates and disturbs the bacterial cell wall. Likewise, phytochemical analysis showed the presence of highly effective bioactive compounds, such as alkaloids, polyphenols, tannins, flavonoids and saponins, that presented antibacterial properties by inhibiting extracellular microbial enzymes, by depriving the substrates required for microbial growth and by avoiding oxidative phosphorylation [11,12,13]. Additionally, the biofiltration technique has been used as a water treatment technology to remove pathogens, the first biofilters being developed using rock or slag as a filter medium [14]. A large number of alternatives have been utilized to design biofilters, such as peat, different types of coal, tea leaves, coconut waste, shellfish, banana and orange peel, pine needles, bamboo leaves and grass roots, that eliminate heavy metals and microorganisms such as bacteria, fungi and different types of algae [15].
In this line, cork has been presented as an environmentally friendly material and sustainable resource with the potential to remove emergent pollutants and waterborne pathogens [16,17,18]. Some studies have shown that cork residues can be modified to increase their effectiveness as possible biosorbents of pollutants in wastewater treatment [19,20]. The chemical composition of cork depends on its origin, containing 40–60% suberin, 10–50% lignin, 5–15% polysaccharides and 10–25% of extractable components that include waxes and tannins [21,22]. It has been demonstrated that enzymatic coupling of the phenolic residues in cork matrices with functionalized biopolymers and nanoparticles reduces the growth of E. coli efficiently [23]. For this reason, the aim of this study was to evaluate the effect of cork functionalized with M. oleifera seed extract (MoSe), which as far as we know has not yet been elucidated, since studies with this material have been mainly focused on its use as a biocatalytic functional matrix with silver nanoparticles that efficiently reduced the growth of E. coli [23]. However, to confirm the antimicrobial effect of MoSe, it is necessary to apply faster methods than conventional ones to demonstrate the membrane cell disturbance caused by the bioactive compounds present in the seed extract [24,25]. Studies have reported that among these could be phytochemical compounds with antimicrobial properties that can reduce high cellular concentrations of pathogenic bacteria such as E. coli, Salmonella sp. and Staphylococcus sp. [26].
One of the alternatives for confirming the inhibitory effect of cork biofunctionalized with seed extract on bacteria is the differentiation of viable and dead bacterial cells using selective nucleic acid intercalating dyes, such as ethidium monoazide (EMA) and propidium monoazide (PMA), and the subsequent detection and quantification of viable cells by quantitative PCR [27]. These compounds can penetrate only into dead cells to compromise membrane integrity and to intercalate with DNA, via their photoinducible azide groups, in turn inhibiting DNA amplification during PCR [28]. Using this approach, the present study evaluated the cell viability of E. coli as a representative indicator of faecal contamination of water, using viability quantitative PCR (vqPCR) with PMA dye to evaluate the bacterial inhibitory effect of cork biofunctionalized with MoSe as a potential disinfection bioremediation technology for water contaminated with E. coli.
2. Materials and Methods
2.1. Bacterial Growth Kinetic and Selection of the Reporter Gene
Escherichia coli NBRC 102203 was identified by the Microbial Culture Collection, CINVESTAV, Mexico City. Cells were used to carry out the viability test through amplification of the LacZ gene. The bacterium was isolated from wastewater a Xochimilco canal, Mexico City and grown in tryptic soy broth (TSB) medium (Sigma-Aldrich, St. Louis, MO, USA) in an orbital shaker (New Brunswick Scientific, NJ, USA) at 37 °C, 120 rpm for 12 h. Then, cells were sown in tryptic soy agar (TSA) (Sigma-Aldrich, St. Louis, MO, USA) plates and Chromocult coliform medium (Merck, Darmstadt, Germany) using the micro-drop technique with serial dilution in 0.85% sterile saline solution (NaCl). Next, cell growth was monitored by optical density at 600 nm to evaluate the inhibitory effect of cork functionalized with MoSe and to carry out the viability test (vqPCR) [28,29].
Escherichia DH5α cells were used as hosts for preserving and propagating all recombinant DNA constructs. Escherichia DH5α cells carrying LacZ inserts in a pJET1.2/blunt cloning vector (Thermo Fisher Scientific, MA, USA) were grown in LB (Luria–Bertani) liquid medium supplemented with 30 μg/mL ampicillin (Sigma-Aldrich, St. Louis, MO, USA).
As it is constantly expressed in the DNA of the bacterium, LacZ gene (beta-d-galactosidase), used in axenic culture at the laboratory level, was selected to carry out the vqPCR tests, following the procedure described by Foulds et al. [30].
2.2. Genomic DNA Extraction
E. coli cells were processed, and genomic DNA was extracted by using an E.Z.N.A.® Tissue DNA Kit ( Omega Bio-Tek, Norcross, GA, USA). Likewise, all the samples generated during this study were centrifuged, pellets were washed three times with PBS (phosphate buffer solution) and DNA samples were analysed in a Nanodrop 1000 Spectrophotometer (Thermo Scientific, Wilmington, MA, USA) in order to determine their concentration and purity.
2.3. Real-Time PCR for Detection of E. coli
2.3.1. LacZ Gene Amplification
To verify the DNA quality of some of the samples, the LacZ gene was amplified by PCR with the conditions already established by Foulds et al. [30] and Ferreira et al. [31]. The PCR product was subjected to 1% agarose gel electrophoresis and stained with SYBRTM Safe DNA Gel Stain (Thermo Fisher Scientific, MA, USA) (0.5 µg/mL). Table S1 shows the sequences of the primers used for amplification of the LacZ gene and the size of the PCR product. Furthermore, the PCR product was cloned into the vector pJET1.2 using the manufacturer’s protocol of the CloneJET PCR cloning kit (Thermo Fisher Scientific, MA, USA). According to the protocol of Ausubel et al. [32], to verify that the amplicon corresponds to the LacZ gene of E. coli NBRC 102203, competent chemical cells of E. coli DH5-alpha were transformed with the LacZ/pJET1.2 construct. Finally, two clones previously identified by colony PCR were selected for DNA sequencing analysis using the kit primers. The sequencing analysis of the LacZ gene in the LacZ/pJET1.2 construct was carried out by the sequencing service of the Laboratorio Nacional de Servicios Experimentales (LaNSE) CINVESTAV-IPN, (Mexico City, Mexico).
2.3.2. LacZ Gene Standard Curves
A standard curve of the LacZ gene was obtained to estimate the cell concentration present in the dilutions [28]. The different dilutions of a DNA solution of known concentration allow the acquisition of a line that relates the concentration to the cycle threshold (CT) value.
2.3.3. Quantitative Real-Time PCR
Experiments were performed using 5× HOT FIREPol EvaGreen qPCR Mix (Capillary). The reaction mixture consisted of 5 µL of genomic DNA template, 4 µL of master mix and 0.3 µL of each primer and PCR water to achieve a final volume of 20 µL. A LightCycler 1.5 thermal cycler (Roche, Basel, Switzerland) was used. The thermal cycler program was set at 95 °C for 15 min, followed by 40 cycles of 95 °C for 30 s, 52 °C for 1 min and 72 °C for 20 s. Positive and negative controls were included for all qPCR reactions. The negative control consisted of adding 5 μL of PCR water to the capillary instead of DNA template. The positive control was one of the dilutions used for the performance of standard curves and data were expressed as absolute abundance (gene copy number/mL of sample) [33].
2.4. Sample Concentration, PMA Treatment and Relevant Test Controls
Samples of the biofilter system effluent (see Section 2.7) were centrifuged in an Eppendorf Microcentrifuge 5424 (Hamburg, Germany) 10,000× g for 5 min and washed three times with PBS. Then, 10 µL of PMA (2.5 mM) was added, following the Blue-V PMA Viability Kit protocol (Qiagen, Hilden, Germany). Dead and live cell samples were confirmed by growth on Chromocult coliform agar plates. The corresponding samples with PMA were irradiated for 10 min (after incubation in the dark for 10 min) using a PhotoActivation System for Tubes-PhAST Blue (GenIUL, Terrassa, Spain).
Viability Test (vqPCR) with PMA Treatment
A bacterial suspension of E. coli NBRC 102203 was adjusted with PBS to a cell concentration of 108 cells/mL at a wavelength of 600 nm in a final volume of 50 mL. This bacterial suspension was added to the biofiltration system of cork functionalized with MoSe. The viability test was carried out at two times (0 and 6 h). For each test time, the samples were divided into those without treatment (absence of PMA) and with treatment (presence of PMA). Test samples are described in Table S2.
2.5. Live/Dead Cell Viability Assays
Samples of the biofilter system effluent were stained with a BacLight double staining kit for bacteria (Thermo Fisher Scientific, Spain) and visualized in a Zeiss epifluorescence microscope. This kit uses two nucleic acid stains: green, fluorescent staining of SYTOTM 9 and red fluorescent staining of propidium iodide, which differ in their ability to penetrate bacterial cells. SYTOTM 9 dye can penetrate all bacteria, while propidium iodide can penetrate only bacteria with damaged membranes [11].
2.6. Functionalization of Cork with MoSe
The M. oleifera seeds were provided by the Polytechnic University of Catalonia, and collected in the Canary Islands, Spain. Initially, the seeds were crushed, dried at 60 °C for 24 h and sieved to obtain a particle size of 0.9 mm. The stock solution was made of dried M. oleifera seeds in distilled water and NaCl (0.85%), achieving two concentrations of 5% and 10%, which were consecutively filtered using 40 µm filter paper [34]. Additionally, the granulated cork was cleaned with hydrochloric acid (HCl, pH 2), sodium hydroxide (NaOH, pH 10) and ethanol (96%) followed by three rinses with distilled water to remove the sediment. We worked with two particle sizes (3 mm and 5 mm), smaller and larger, respectively. The material was dried at 50 °C for 12 h and consecutively sterilized (15 min at 121 °C). Two hundred millilitres of the seed extract stock solution were added and the mixture stirred with a magnet, ensuring that the solution covered the filter material for 2 h. Subsequently, the material was washed with distilled water until part of the solution was removed and dried at 50 °C for 12 h [23].
2.7. Biofiltration System with Filter Cartridges
The filter cartridge system was constructed with PVC pipes (diameter 0.032 m; length 0.20 m), with a polyethylene grid at the upper end (0.0808 m2) and a piece of geotextile with two holes at the lower end. The filter cartridge was additionally enclosed by two plugs (diameter 0.032 m) attached to a section of polyethylene pipe (diameter 0.06 m). The upper tube was connected to a peristaltic pump (ET-PBV4336574, Etatron) which was connected to the inlet tank under constant stirring at 250 rpm; the lower part of the tube allowed collection of the treated water in the effluent container and the entry of aeration into the system. Each filter cartridge had a volumetric capacity of 160 mL, of which 110 mL corresponded to the volume displaced by the cork. Therefore, it had a useful volume of 50 mL. The filter cartridges were organized in a system of four filters (Figure 1), replicates of cork functionalized with MoSe and granulated cork not functionalized with MoSe as control. To determine the efficiency of E. coli cell death, the influent flow was equally divided between the cartridges at a constant flow of 3 mL/min [35].
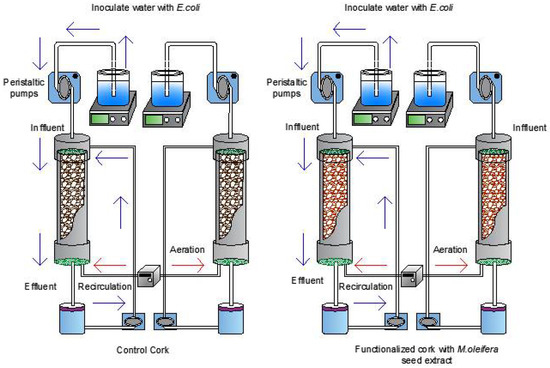
Figure 1.
Biofilter system functionalizated with MoSe.
The operation of the biofiltration system was carried out with the conditions established in each treatment from the 27−4 fractional factorial experimental design (see Section 2.7.1). Initially, the filtering cartridges were filled with f-cork functionalized with MoSe and not-functionalized cork (control) and were incorporated in the filtering system (Figure 1). To test the viability of the bacteria, the inffluent flow was injected with aliquots of 50 mL of water inoculated with E. coli (~1 × 108 CFU/mL). Then, water contaminated was incubated in the filtering cartridges as hydraulic retention time at room temperature for 6 or 12 h. Additionally, bacterial load of the inffluent water circulated constantly through filtering systems at a flow rate between 3 and 7 (mL/min). Once the test had been completed the number of viable bacteria from effluent were taken at 0, 6 and 12 h, seeding took place in Chromocult coliform agar with serial dilutions, and then plate cells were counted using the micro-drop technique and incubated at 37 °C for 12 h.
2.7.1. Fractional 27−4 Experimental Design Using Cork as Filter Material
A 27−4 fractional factorial design was used to study the effect of cork functionalized with MoSe on InhEc for a total of eight treatments [36] as shown in Table 1. The fixed factors were recirculation (Rec), aeration (Air), seed extract concentration (Conc), electric conductivity (EC), hydraulic retention time (HRT), cork particle size (PS) and lysis time (LT). The matrix of treatments and independent factors is presented in Table 2.

Table 1.
Independent factors and their low and high values of a 27−4 fractional factorial design for the reduction of E. coli in trickling cork biofilters functionalized with MoSe.

Table 2.
Experimental matrix of a 27−4 fractional factorial design with the levels of the independent factors in eight treatments.
2.8. Antimicrobial Activity of Cork Functionalized with MoSe
E. coli was grown at 37 °C for 12 h in TSB broth culture. Subsequently, the amount of microorganism was sufficient to reach a concentration of 1.5–3.0 × 108 CFU/mL. Thus, the bacterial suspension simulated those conditions of the waters to be treated in terms of conductivity and pH to evaluate the effect of cork functionalized with MoSe. Samples were taken at 0, 6 and 12 h and were grown on Chromocult coliform agar from serial dilutions incubated at 37 °C for 12 h. The E. coli inhibition results were expressed as a percentage reduction of microorganisms, using the following formula: the difference between the CFU/mL at time zero and the CFU/mL at the end of 6 and 12 h [37].
2.9. Statistical Analysis
Analysis of variance (ANOVA) was used and determined for 95% reliability (p < 0.05). Additionally, generalized minimum regression analysis and the LSD (least significant difference) test of comparison of means were used after applying Student’s t-test for the comparison of two means with weighted variance when the ANOVA presented significant differences. The analysis of LSD means allowed the determination of treatments that presented significant differences, classifying them into different groups depending on the similarity between the means. All statistical analyses were conducted using the software SAS (version 9).
3. Results
3.1. Growth Kinetic of E. coli and Amplification of LacZ Gen
The microorganism grew exponentially for 8 h and reached the lag phase at t = 12 h (Figure S1). The cell concentration obtained at 8 h was 4.5 × 108 CFU/mL, although at 6 h it was 1.5 × 108 CFU/mL, high enough to extract a significant quantity of DNA; a cell concentration above 1 x 105 CFU/mL is required to achieve better suppression of DNA amplification by PMA treatment in the PMA-qPCR assay to avoid overestimating the total amount of viable E. coli.
The electrophoresis gel obtained from the PCR product of LacZ gene amplification can be observed in Figure S2. A positive result was obtained for the amplification of a 142 bp product. Likewise, sequencing analysis of the LacZ/pJET1.2 construct of two candidate clones showed 100% homology with the LacZ gene from E. coli NBRC 102203. Figure S3 shows the electrophoretic analysis of the PCR products for identification of the candidate clones containing the LacZ/pJET1.2 construct. Positive clones, those that amplified with the primers of pJET1.2/blunt, obtaining a molecular size of 262 bp (120 bp + 142 bp = 262 bp). The 120 bp added to the LacZ gene during amplification corresponds to a fragment from the pJET1.2/blunt vector, due to the primers pJET-F and pJET-R.
A standard curve for the LacZ gene was obtained to quantify the cell concentration present in the dilutions and the viability of E. coli. The concentration of microorganisms in the sample was inversely proportional to the amount of target DNA and hence the limits of detection were from 101 to 108 cells/mL, which gave mean CT values of 33.95 and 19.11, respectively. Linear regression determined R2 = 0.996. The LacZ gene induces β-d-galactosidase activity and it has been proved a good marker and a rapid method for detecting E. coli in environmental waters.
3.2. Functionalization of Cork with MoSe and Effect on InhEc
The biofilter of granulated cork was functionalized with MoSe (f-cork) and was used applying a 27−4 fractional factorial design to optimize the reduction of the logarithmic units of E. coli. (Figure 2).
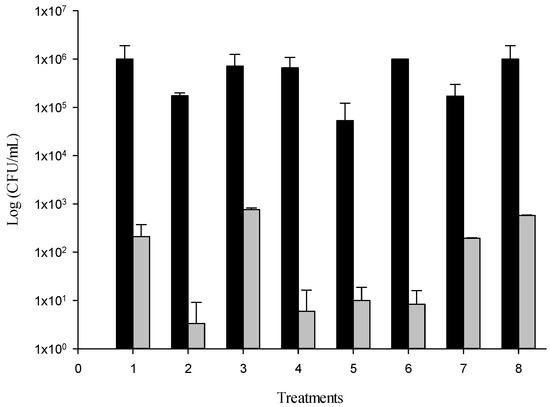
Figure 2.
Bacterial load of the effluent water circulated through non-modified cork (control, black bars) and through the f-cork (grey bars) in the eight treatments. The initial E. coli count for the contaminated water was ~1 × 108 CFU/mL.
The effect of f-cork indicated a notable bacterial inhibition. In contrast, the reduction bacterial count cartridges filled with cork granulated using the biofiltration system that did not show any bacteria inhibition. The bacterial contamination of water decreased by 6 and 7 log CFU/mL between treatments with f-cork. The greatest bacterial reduction was evidenced in treatment 2, 4 and 6.
The biofilter of granulated cork was functionalized with MoSe (f-cork) and was used applying a 27−4 fractional factorial design to optimize the reduction of the logarithmic units of E. coli. When performing the statistical analysis, a significance level of p < 0.0001 was obtained for ANOVA of the generalized linear procedure (GLM), presenting a moderately high positive correlation; the independent factors that presented a significant effect were Air, Conc, PS and LT. The effect of functionalized cork on InhEc was observed in eight treatments, as shown in Table 3. Additionally, the linear regression model values were p < 0.0001 and R2 = 0.8; the equation model was: Inh (%) = 98.42 + 0.20 Air − 0.12 TP − 0.35 TL, with Air (high level), PS (low level) and LT (low level) having a significant effect.

Table 3.
Effect of cork functionalized with MoSe on the inhibition of E. coli.
When comparing means between treatments using the LSD test, significant differences were observed, forming two groups: treatments 6, 4, 2, 1 and 5 on the one side and 7, 3 and 8 on the other. Treatments 6, 4 and 2 presented the highest percentage removal of E. coli (>99.99%), unlike the other treatments, and the independent factor for which a significant effect was observed a low level of Air (without aeration). Likewise, the independent factor of Recirculation a high level (7 mL/min) was common among these treatments. Treatment 6 was made up of the following factors: 7 mL/min Rec, without Air, 10% MoSe, 4000 µs/cm EC, 12 h HRT, 3 mm PS and 2 h LT, while treatment 4 was characterized by 7 mL/min Rec, 4 L/min Air, 10% MoSe, 4000 µs/cm EC, 6 h HRT, 3 mm PS and 2 h LT. Finally, treatment 2 was selected for the vqPCR assay, with the following factors: 7 mL/min Rec, without Air (0 L/min), 5% MoSe, 1700 µs/cm EC, 6 h HRT, 5 mm PS and 3 h LT.
3.3. LacZ vqPCR Assay
The effect of the cork biofilter functionalized with MoSe on the viability of E. coli at two times (0 and 6 h) evaluated with a qPCR assay is shown in Figure 3; the conditions in the biofiltration system included a concentration in the bacterial suspension of 108 cells/mL in the solution with treatment 2, selected from a 27−4 fractional factorial design with the following factors: 7 mL/s Rec, without Air (0 L/min), 5% MoSe, 1700 µs/cm EC, 6 h HRT, 5 mm PS and 3 h LT. The qPCR assay was performed after application of PMA and DNA extraction.
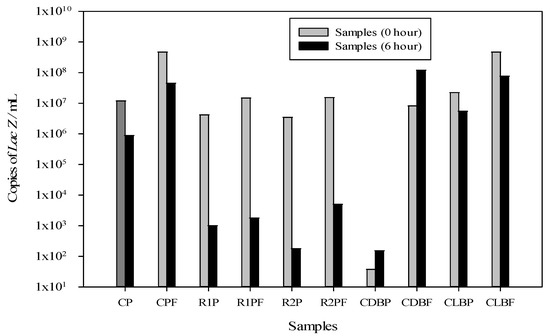
Figure 3.
Inhibitory effect of f-cork on E. coli using PMA staining. Gene copy numbers of LacZ gene/µL present in the biolfiltration system at 0 and 6 h. Biofilter control without f-cork with PMA (CP), Biofilter control without f-cork and without PMA (CPF), Replicate of biofilter with f-cork and PMA (R1P), Replicate of biofilter with f-cork and without PMA (R1PF), Control of dead bacteria with PMA (CDBP), Control of dead bacteria without PMA (CDBF), Control of live bacteria with PMA (CLBP) and Control of live bacteria without PMA (CLBF).
According to the CT displacement with PMA treatment and the qPCR assay, the results presented wide variation between the samples obtained for Control of dead bacteria with PMA (CDBP) and Control of dead bacteria without PMA (CDBF) at two times (0 and 6 h) (see Supplementary Material, shown in Tables S3 and S4). The CT value indicates the number of cycles in which the fluorescence generated in a reaction crosses the fluorescence threshold and, therefore, there is amplification of the LacZ gene.
When performing statistical analysis of the genomic copy number, ANOVA yielded values of p > 0.0001 and R2 = 0.96, evidencing that there was no significant difference at the initial time (0 h) between the control with PMA treatment and the replicates, in contrast to those obtained for the final time (6 h; p < 0.0001 and R2 = 0.79). The CT values for Control of live bacteria with PMA (CLBP) were slightly higher than those for Control of live bacteria without PMA (CLBF). This is due to the portion of dead cells that occurs in a microbial culture. This CT change generally varied between 1 and 3 CT values. On the other hand, CDBP exhibited, as expected, a prominent CT shift compared to CDBF. This CT shift made very clear the distinction between dead and live cells.
In addition, there were not large differences presented between the number of genomic copies at t = 0 h, remaining in the range of 106–108 genomic copies/mL as shown in Figure 3. However, for CDBP, a large difference in gene concentration was indeed obtained, with a change from 108 to 101 genomic copies/mL, evidencing the effect of PMA; it should be noted that in this killing control, heat was used to eliminate bacteria. The values obtained at t = 6 h presented a significant change between the number of replicates for treatment with cork biofunctionalized with MoSe: 1–8 × 102 and 1–2.5 × 102 for R1P and R2P, respectively, different to the 1–4 × 105 genomic copies/mL using the cork biofilter control without MoSe functionalization with PMA treatment (CP). Thus, in this study a decrease in the number of E. coli genomic copies/mL was obtained using treatment 2 with factors that affected the bacterial growth, among them oxygen limitation (0 L/min) and larger granulated cork particles (5 mm).
Subsequently, to assess the differences in the number of genomic copies between treatments, analysis of means was performed with the Fisher LDS test, and five groups were obtained in which there was no difference among them (R1PF and R2PF) (p > 0.0001). Concerning the CT values, three groups were obtained, and they did not present a significant difference (R2SP, CPF and CDBF). Additionally, a large decrease in the number of gene copies was observed in the replicates R1CP and R2CP with respect to CP and CDBP.
Quantitative PCR of samples treated in the biofiltration system with PMA (vqPCR) and without PMA was carried out to evaluate the efficiency of the f-cork, and the results showed notable differences between treatments (Table 4). The number of copies/mL was constant over time in untreated samples (approximately 108 copies/mL), unlike in the samples treated with PMA. Thus, at 0 h, we measured similar values in the treated and untreated samples, while at 6 h, a large decrease in the copy number/mL was always observed in the samples treated with PMA, resulting in 1.03 × 103 and 1.80 × 102 copies/mL for f-cork and PMA (R1P and R2P), respectively. The PCR amplification efficiency (% vqPCR) was then strongly reduced in the replicates with f-cork and cells exposed to PMA, with values of 0.024% and 0.005% of viable microorganisms, respectively.

Table 4.
Quantitative evaluation of LacZ gene at different times in treatments with and without PMA (vqPCR). qPCR results are expressed as copies/mL. Data represent means of two independent experiments performed in duplicate.
3.4. Live/Dead Bacterial Cell Viability Test Using Cork as Filter Material
Live/dead staining of the bacterial strain was conducted to determine the effect of cellular inhibition by f-cork. Figure 4 shows the difference in InhEc between the control treatment without functionalization of cork and that with f-cork, with a greater than 99.99% reduction of bacteria evident after 6 h. Treatment 2 was selected (see Section 3.2.) to carry out the live/dead test.
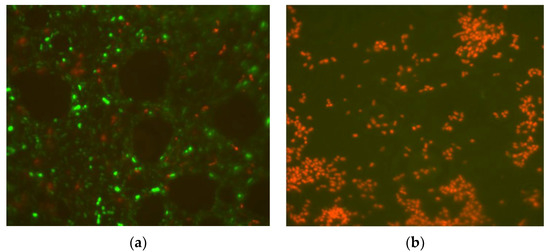
Figure 4.
LIVE/DEAD Baclight test carried out with fluorescence microscopy from samples with cork functionalized with MoSe. Biofilter without f-cork (a), Biofilter functionalized with f-cork (b). Dead cells are stained in red, while green-stained cells are alive.
An improvement of InhEc was observed for the treatment with f-cork and the limitation of aeration had a strong effect with respect to the bacterial inhibition. The removal of pathogens elicited a 6–7 log CFU/mL reduction of bacteria; the final microbial count using treatment 2 was about 1 × 101 CFU/mL, (See Section 3.2), confirmed by epifluorescence microscope.
4. Discussion
4.1. Growth Kinetic of E. coli and Amplification of LacZ Gene
In this study, a large enough amount of E. coli cells was obtained to extract a significant quantity of DNA according to the studies by Yuan et al. [29], who recommended a cell concentration above 1 × 105 CFU/mL to achieve better suppression of DNA amplification by PMA treatment in the PMA-qPCR assay to avoid overestimating the total amount of viable E. coli. The study of Fiksdal and Tryland [38] indicated that the LacZ gene induces β-D-galactosidase activity, and it has been proved a good marker and a rapid method for detecting E. coli in environmental waters.
4.2. Functionalization of Cork with MoSe and InhEc
The main factors that had a highly significant (p < 0.001) effect were aeration, MoSe concentration, cork particle size and lysis time. These factors, combined with surface modification of the cork filter medium with MoSe possessing inhibitory properties, presented a significant (p < 0.001) effect on InhEc, similar to the results obtained by Mohanty et al. [39], who indicated that surface modification with a disinfectant agent of the polycationic coatings utilized for coating the collector surface confers antimicrobial properties and facilitates microbial attachment to filter media, contributing to a net negative charge on the cell membrane. Although increasing the concentrations between 5 and 10% of MoSe used in treatments did not present difference on bacterial inhibition, the conditions that presented the greatest inhibitory effect were obtained in treatment 2 with a concentration of MoSe (5%). Possibly, this optime dose was enough to achieve the critical micellar concentration (CMC) a main characteristics of surfactant macromolecules, such as proteins of the M. oleifera seeds, with coagulant activity as well as hydrophobic interactions [40]. This behaviour of the seed extract might be due to the increase in the entropy that occurs when the hydrophobic region of the surfactant is isolated from the water, after which there is a loss of the structure of the water molecules. Therefore, as the CMC increases, the surface tense decreases until it remains constant, when the surfactant reaches micelles spontaneously, and it is at this point that the CMC is reached [41]. Additionality, the increase the concentration of protein with hydrophobic sections affect the antibacterial effect, explained by several factors, such as the properties of the tensoactive agent (adsorption, surface forces, physicochemical properties, protein emulsifier concentration and internal osmotic stresses) and hydrophobic interactions between the bacterial cell surface charge, microbial surface-active compounds, bacterial aggregates, outer membrane protein and bacteria attachment [42,43,44,45].
In addition, the efficiency of InhEc using the system functionalized with MoSe was maintained during the test, probably because MoSe was physically adsorbed on the cork and not removed by the washings using the recirculation factor and stock solution with cell aggregates. These results are supported by the study of Garcia et al. [35], in which they evaluated the antimicrobial effect of chitosan and silver nanoparticles (CS/AgNPs) embedded on cork and found a 4 and 5 log CFU/mL decrease of bacterial contamination in water; the amount of nanoparticles explained the relation between their retention on cork and the durability of antibacterial efficiency, nanoparticles achieving better and more stable adhesion when the coating was more uniform. However, dissolution of the physically adsorbed CS/AgNPs from the cork surface could have affected the antibacterial performance of the material. In contrast, the work by De Oliveira et al. [46] evaluated the functionalization with M. oleifera of magnetic nanoparticles (Fe3O4) for S. aureus removal and showed that the antibacterial effect was more efficient when using a combination of M. oleifera and magnetite (Fe-Fe2O4) with constant removal rates compared to Fe-Fe2O4 or M. oleifera acting alone, since the functionalization of Fe-Fe2O4 with M. oleifera produced a coagulant within the surface area containing functional groups and minerals. That antibacterial effect was consistent with our study, where the combination of MoSe and cork improved the adsorption and InhEc efficiency. Additionally, some of the reasons why the seed extract increased the filter’s antibacterial potential may be justified by the study carried out by Francesco et al. [23], who assembled a biopolymer with laccase and silver nanoparticles on cork for the removal of pathogens in wastewater and noticed that the composition of cork could cause an enhanced antibacterial effect. These authors evidenced that its composition depends on its origin and can contain 40–60% suberin, 10–50% lignin, 5–15% polysaccharides and 10–25% removable components, including waxes and tannins. Lignin, suberin and tannin molecules have many phenolic residues, which can be enzymatically oxidized into reactive quinones that can react with the amino groups of biopolymers; thus, the covalent immobilization of biopolymers in cork can supply a durable antibacterial effect and synergistically enhance their activity.
4.3. LacZ vqPCR Assay
In this study, a decrease in the number of E. coli genomic copies/mL was obtained using treatment 2 (see Section 3.2.); these results could be supported by Domingues et al. [47], who determined some physical and chemical surface properties of cork granulates (dimensions 1–2 and 3–4 mm), indicating the presence of macropores. Likewise, an increase in particle size reduces the accessibility to internal pores and diminishes the surface area available for the uptake of pollutants in a possible biosorbent. In addition, with respect to oxygen limitation, Losen et al. [48] evaluated the oxygen parameters for growth of E. coli, especially in culture with the lowest oxygen transfer capacity or respiratory activity; they indicated that oxygen is a very sensitive parameter and can yield highly valuable information about the carbon metabolism of the cultured cells. The primary metabolism of E. coli responds quickly to oxygen limitation, changes in pH and acetate concentration.
Additionally, a large decrease in the number of gene copies was observed in the replicates R1CP and R2CP with respect to the biofilter control (CP) and the killed control (CDBP) for the treatment using PMA. Once it has penetrated into the cells, it inserts into the DNA and when illuminated with visible light, it binds to the genetic material. This is due to a PMA photolysis process in which the azide group becomes a highly reactive radical that can form a covalent bond with DNA. The PMA that remains free in the medium is simultaneously inactivated by reacting with the water present in molecules. Binding of PMA to DNA results in strong inhibition of amplification during PCR, such that only genetic material from unmodified cells (living cells with uncommitted membrane) can be amplified [49,50].
The decrease in the number of gene copies at the end of the treatment (6 h) in contrast to that at the beginning (0 h) can be supported by the study carried out by Kubota et al. [51], in which it was suggested that the lipopolysaccharide layer of Gram-negative bacteria could have an affinity with the surface of cork. However, it should be noted that both the cork and MoSe contain phytochemical compounds that could interfere in the affinity of the attachment process. Bacterial inhibition potential was exposed by Rashid et al. [52] and Nazzaro et al. [53], who reported that the fatty acid profile derived from M. oleifera oil showed a high content of oleic acid (>70%), generating biochemical and antibacterial activity, respectively, against E. coli.
These results reveal the disruption of the cell wall in E. coli. Likewise, the use of PMA was indispensable since it allowed us to determine this effect. In this study, we evaluated the use of cork as an organic matrix with biosorbent function and MoSe with active antimicrobial compounds, generating the main physiological responses occurring in immobilized cells which may contribute to improving the efficiency of immobilization techniques. The qPCR assay indicated a reduction in the number of copies of the LacZ gene in E. coli and a loss of viability with the PMA treatment, confirming that the efficiency of the immobilization (adsorption) of microbial cells depends on the properties of the support, microbial cell surface and environmental conditions [54]. Thus, the metabolic response of immobilized cells was resistance to inhibitors that make alterations to the composition and organization of the cell wall and cell membrane. This potential is associated with the compound benzyl-isothiocyanate which inhibits bacterial growth by disrupting the mechanisms of membrane and enzyme synthesis [55].
4.4. Live/Dead Bacterial Cell Viability Test Using Cork as Filter Material
Cork might contain a resin that potentiates antimicrobial activity, together with some components that possibly generate a synergy and a greater effect of chemisorption of bacteria, similarly to the results reported by Francesco et al. [23]. Recently, Xiong et al. [25] obtained a biofilter functionalized with MoSe and sand that decreased the bacterial load. The mechanism was also explained by Jerri et al. [11], who stated that the cationic protein MOCP present in the seeds contains a glutamine-rich portion that can be adsorbed in the sand through electrostatic attraction, while maintaining coagulant-flocculation capacity, improving antimicrobial activity and the removal of pathogens and avoiding the release of organic matter. They reported a 3–4 log reduction of bacteria with the functionalized sand. Likewise, the f-sand (sand functionalized with MoSe) samples presented the capacity to attract bacteria through electrostatic forces and destroy them by means of a protruding hydrophobic loop on the protein that may penetrate the bacterial cell wall [56].
5. Conclusions
Biological filter systems have the capacity to significantly reduce pathogen microorganisms in effluents with nature-based solutions, treating wastewater and supplying microbiologically safe water. This study showed an attractive potential antibacterial system using cartridges of granulated cork functionalized with MoSe that effectively reduced the growth of E. coli by > 99.99%. Likewise, the vqPCR technique used at t = 0 and 6 h indicated the usefulness of this fast method and its sensitivity to show the functionalization effect of MoSe concerning bacterial viability (% vqPCR), which was reduced to 0.024% and 0.005% for the replicates with f-cork and cells exposed to PMA, respectively. Additionally, the live/dead test using fluorescence microscopy confirmed this antibacterial effect. Thus, we encourage the use of natural products to reduce microbiological parameters in water that are harmful to the environment and human health.
The concept of using functionalized biomaterials from natural extracts with antimicrobial properties justifies future research with a biotechnological application and to improve water quality in human settlements. Even though the present study results are preliminary, using M. oleifera seeds as a disinfectant agent for improving E. coli removal by cork-based biofilters appears promising.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/w13192731/s1, Table S1: Primer sequences of LacZ gene, Table S2: Viability test of biofilter functionalized with MoSe, Table S3: Effect of the biofiltration system on E. coli at the initial time zero h using qPCR with and without PMA, Table S4: Effect of the biofiltration system on E. coli in the final time 6 h using qPCR with and without PMA, Figure S1: Growth kinetic of E. coli strain NBRC 102203 as Log CFU/mL, Figure S2: Electrophoresis of the amplification from LacZ gene, Figure S3: Electrophoretic analysis of the PCR products obtained by “Colony PCR” to detect the candidate clones carrying out the construct LacZ/pJET1.2.
Author Contributions
Conceptualization, N.I., R.R. and Y.B.; methodology, N.I., Y.B., O.S., I.S., L.B. and J.M.; software, I.S. and L.B.; validation, J.M.; formal analysis, N.I., Y.B., O.S. and I.S.; investigation, N.I., Y.B., O.S., I.S., L.B. and J.M.; resources, R.R., O.S. and J.M.; data curation, N.I.; writing—original draft preparation, N.I., R.R., Y.B. and O.S.; Writing—review & editing, N.I., R.R., Y.B., O.S., I.S., L.B. and J.M.; Visualization, N.I., R.R. and J.M.; supervision, N.I., R.R. and J.M.; project administration, R.R. and J.M.; Funding acquisition, R.R. and J.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Mexico-CONACYT, grant number 3006060 and grant by the UNESCO Chair on Sustainability of the Polytechnic University of Catalonia (Spain).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank the technical assistance and collaboration of the Environmental Pollution Control Laboratory (INTEXTER) at the Polytechnic University of Catalonia, Universitat Autònoma de Barcelona and Institute of Marine Sciences (ICM-CSIC) Barcelona.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Guidelines for Drinking-Water Quality. 2017. Available online: https://www.who.int/publications/i/item/9789241549950 (accessed on 3 March 2021).
- Rusiñol, M.; Hundesa, A.; Cárdenas-Youngs, Y.; Fernández-Bravo, A.; Pérez-Cataluña, A.; Moreno-Mesonero, L.; Moreno, Y.; Calvo, M.; Alonso, J.; Figueras, M.J.; et al. Microbiological contamination of conventional and reclaimed irrigation water: Evaluation and management measures. Sci. Total Environ. 2020, 710, 136298. [Google Scholar] [CrossRef]
- Motlagh, A.M.; Yang, Z. Detection and occurrence of indicator organisms and pathogens. Water Environ. Res. 2019, 91, 1402–1408. [Google Scholar] [CrossRef] [Green Version]
- Wen, X.; Chen, F.; Lin, Y.; Zhu, H.; Yuan, F.; Kuang, D.; Jia, Z.; Yuan, Z. Microbial Indicators and Their Use for Monitoring Drinking Water Quality—A Review. Sustainability 2020, 12, 2249. [Google Scholar] [CrossRef] [Green Version]
- Brennan, F.P.; O’Flaherty, V.; Kramers, G.; Grant, J.; Richards, K.G. Long-Term Persistence and Leaching of Escherichia coli in Temperate Maritime Soils. Appl. Environ. Microbiol. 2010, 76, 1449–1455. [Google Scholar] [CrossRef] [Green Version]
- Alegbeleye, O.O.; Sant’Ana, A.S. Manure-borne pathogens as an important source of water contamination: An update on the dynamics of pathogen survival/transport as well as practical risk mitigation strategies. Int. J. Hyg. Environ. Health. 2020, 227, 113524. [Google Scholar] [CrossRef]
- Oral, H.V.; Carvalho, P.; Gajewska, M.; Ursino, N.; Masi, F.; van Hullebusch, E.D.; Kazak, J.; Exposito, A.; Cipolletta, G.; Andersen, T.R.; et al. A review of nature-based solutions for urban water management in European circular cities: A critical assessment based on case studies and literature. Blue-Green Syst. 2020, 2, 112–136. [Google Scholar] [CrossRef] [Green Version]
- Choy, S.Y.; Prasad, N.; Wu, T.Y.; Raghunandan, M.E.; Ramanan, R.N. Utilization of plant-based natural coagulants as future alternatives towards sustainable water clarification. J. Environ. Sci. 2014, 26, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Okuda, T.; Baes, A.U.; Nishijima, W.; Okada, M. Coagulation Mechanism of Salt Solution-Extracted Active Component in Moringa oleifera Seeds. Water Res. 2001, 35, 830–834. [Google Scholar] [CrossRef] [Green Version]
- Camacho, F.P.; Sousa, V.S.; Bergamasco, R.; Teixeira, M.R. The use of Moringa oleifera as a natural coagulant in surface water treatment. Chem. Eng. J. 2017, 313, 226–237. [Google Scholar] [CrossRef]
- Jerri, H.A.; Adolfsen, K.J.; McCullough, L.R.; Velegol, D.; Velegol, S.B. Antimicrobial Sand via Adsorption of Cationic Moringa oleifera Protein. Langmuir 2011, 28, 2262–2268. [Google Scholar] [CrossRef]
- Rajan, S.; Thirunalasundari, T.; Jeeva, S. Anti—enteric bacterial activity and phytochemical analysis of the seed kernel extract of Mangifera indica Linnaeus against Shigella dysenteriae (Shiga, corrig.) Castellani and Chalmers. Asian Pac. J. Trop. Med. 2011, 4, 294–300. [Google Scholar] [CrossRef] [Green Version]
- Gomaa, E.Z. In vitro antioxidant, antimicrobial, and antitumor activities of bitter almond and sweet apricot (Prunus armeniaca L.) kernels. Food Sci. Biotechnol. 2013, 22, 455–463. [Google Scholar] [CrossRef]
- Chaudhary, D.S.; Vigneswaran, S.; Ngo, H.H.; Shim, W.G.; Moon, H. Biofilter in water and wastewater treatment. Korean J. Chem. Eng. 2003, 20, 1054–1065. [Google Scholar] [CrossRef]
- Maurya, A.; Singh, M.K.; Kumar, S. Biofiltration technique for removal of waterborne pathogens. Waterborne Pathog. 2020, 123–141. [Google Scholar] [CrossRef]
- Fernandes, E.M.; Correlo, V.M.; Chagas, J.A.; Mano, J.F.; Reis, R.L. Cork based composites using polyolefin’s as matrix: Morphology and mechanical performance. Compos. Sci. Technol. 2010, 70, 2310–2318. [Google Scholar] [CrossRef]
- Rodriguez, O.; Peralta-Hernandez, J.M.; Goonetilleke, A.; Bandala, E.R. Treatment technologies for emerging contaminants in water: A review. Chem. Eng. J. 2017, 323, 361–380. [Google Scholar] [CrossRef] [Green Version]
- Patel, M.; Kumar, R.; Kishor, K.; Mlsna, T.; Pittman, C.U., Jr.; Mohan, D. Pharmaceuticals of Emerging Concern in Aquatic Systems: Chemistry, Occurrence, Effects, and Removal Methods. Chem. Rev. 2019, 119, 3510–3673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castellar, J.; Formosa, J.; Fernandez, A.I.; Jové, P.; Bosch, M.G.; Morató, J.; Brix, H.; Arias, C.A. Cork as a sustainable carbon source for nature-based solutions treating hydroponic wastewaters—Preliminary batch studies. Sci. Total Environ. 2019, 650, 267–276. [Google Scholar] [CrossRef]
- Pintor, A.M.A.; Ferreira, C.I.A.; Pereira, J.C.; Correia, P.; Silva, S.P.; Vilar, V.J.P.; Botelho, C.M.S.; Boaventura, R.A.R. Use of cork powder and granules for the adsorption of pollutants: A review. Water Res. 2012, 46, 3152–3166. [Google Scholar] [CrossRef]
- Chubar, N.; Carvalho, J.M.R.; Correia, M.N. Cork biomass as biosorbent for Cu(II), Zn(II) and Ni(II). Colloids Surf. A Physicochem. Eng. Asp. 2003, 230, 57–65. [Google Scholar] [CrossRef]
- Olivella, M.; Jové, P.; Bianchi, A.; Bazzicalupi, C.; Cano, L. An integrated approach to understanding the sorption mechanism of phenanthrene by cork. Chemosphere 2013, 90, 1939–1944. [Google Scholar] [CrossRef]
- Francesko, A.; Blandón, L.; Vázquez, M.; Petkova, P.S.P.; Morató, J.; Pfeifer, A.; Heinze, T.; Mendoza, E.; Tzanov, T. Enzymatic Functionalization of Cork Surface with Antimicrobial Hybrid Biopolymer/Silver Nanoparticles. ACS Appl. Mater. Interfaces 2015, 7, 9792–9799. [Google Scholar] [CrossRef] [Green Version]
- Nabinejad, A. Antibacterial effects of Saponaria officinalis extracts against avian pathogenic Escherichia coli (APEC). Afr. J. Agric. Res. 2013, 8, 2068–2071. [Google Scholar] [CrossRef]
- Xiong, B.; Piechowicz, B.; Wang, Z.; Marinaro, R.; Clement, E.; Carlin, T.; Uliana, A.; Kumar, M.; Velegol, S.B. Moringa oleifera f-sand Filters for Sustainable Water Purification. Environ. Sci. Technol. Lett. 2017, 5, 38–42. [Google Scholar] [CrossRef]
- Al_husnan, L.A.; Alkahtani, M.D.F. Impact of Moringa aqueous extract on pathogenic bacteria and fungi in vitro. Ann. Agric. Sci. 2016, 61, 247–250. [Google Scholar] [CrossRef]
- Taskin, B.; Gozen, A.G.; Duran, M. Selective Quantification of Viable Escherichia coli Bacteria in Biosolids by Quantitative PCR with Propidium Monoazide Modification. Appl. Environ. Microbiol. 2011, 77, 4329–4335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nocker, A.; Sossa-Fernandez, P.; Burr, M.D.; Camper, A.K. Use of Propidium Monoazide for Live/Dead Distinction in Microbial Ecology. Appl. Environ. Microbiol. 2007, 73, 5111–5117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.; Zheng, G.; Lin, M.; Mustapha, A. Detection of viable Escherichia coli in environmental water using combined propidium monoazide staining and quantitative PCR. Water Res. 2018, 145, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Foulds, I.; Granacki, A.; Xiao, C.; Krull, U.; Castle, A.; Horgen, P. Quantification of microcystin-producing cyanobacteria and E. coli in water by 5’-nuclease PCR. J. Appl. Microbiol. 2002, 93, 825–834. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, L.E.; Dalposso, K.; Hackbarth, B.B.; Gonçalves, A.R.; Westphal, G.A.; De França, P.H.C.; Pinho, M.D.S.L. Molecular panel for detection of sepsis-related microorganisms. Rev. Bras. Ter. Intensiv. 2011, 23, 36–40. [Google Scholar] [CrossRef]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. Current protocols in molecular biology. In Molecular Reproduction & Development; John Wiley & Sons: New York, NY, USA, 1988; Volume 1, p. 146. [Google Scholar]
- Rocha, J.; Fernandes, T.; Riquelme, M.V.; Zhu, N.; Pruden, A.; Manaia, C.M. Comparison of Culture- and Quantitative PCR-Based Indicators of Antibiotic Resistance in Wastewater, Recycled Water, and Tap Water. Int. J. Environ. Res. Public Health 2019, 16, 4217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madrona, G.S.; Serpelloni, G.B.; Vieira, A.M.S.; Nishi, L.; Cardoso, K.C.; Bergamasco, R. Study of the Effect of Saline Solution on the Extraction of the Moringa oleifera Seed’s Active Component for Water Treatment. Water Air Soil Pollut. 2010, 211, 409–415. [Google Scholar] [CrossRef]
- Peña, L.V.G.; Petkova, P.; Margalef-Marti, R.; Vives, M.; Aguilar, L.; Gallegos, A.; Francesko, A.; Perelshtein, I.; Gedanken, A.; Mendoza, E.; et al. Hybrid Chitosan–Silver Nanoparticles Enzymatically Embedded on Cork Filter Material for Water Disinfection. Ind. Eng. Chem. Res. 2017, 56, 3599–3606. [Google Scholar] [CrossRef]
- Montgomery, D.C. Design and Analysis of Experiments, 8th ed.; John Wiley & Sons: Sedona, AZ, USA, 2012. [Google Scholar]
- ASTM E2149-01: Standard Test Method for Determining the Antimicrobial Activity of Immobilized Antimicrobial Agents under Dynamic Contact Conditions (Withdrawn 2010); ASTM International: West Conshohochen, PA, USA, 2001. [CrossRef]
- Fiksdal, L.; Tryland, I. Application of rapid enzyme assay techniques for monitoring of microbial water quality. Curr. Opin. Biotechnol. 2008, 19, 289–294. [Google Scholar] [CrossRef]
- Mohanty, S.; Torkelson, A.A.; Dodd, H.; Nelson, K.L.; Boehm, A.B. Engineering Solutions to Improve the Removal of Fecal Indicator Bacteria by Bioinfiltration Systems during Intermittent Flow of Stormwater. Environ. Sci. Technol. 2013, 47, 10791–10798. [Google Scholar] [CrossRef] [Green Version]
- Kwaambwa, H.M.; Hellsing, M.S.; Rennie, A.R.; Barker, R. Interaction of Moringa oleifera seed protein with a mineral surface and the influence of surfactants. J. Colloid Interface Sci. 2015, 448, 339–346. [Google Scholar] [CrossRef]
- Sheng, J.J. Modern Chemical Enhanced Oil Recovery, 1st ed.; Gulf Professional Publishing: Houston, TX, USA, 2011. [Google Scholar] [CrossRef]
- Dickson, J.S.; Koohmaraie, M. Cell surface charge characteristics and their relationship to bacterial attachment to meat surfaces. Appl. Environ. Microbiol. 1989, 55, 832–836. [Google Scholar] [CrossRef] [Green Version]
- Goulter, R.; Gentle, I.; Dykes, G. Issues in determining factors influencing bacterial attachment: A review using the attachment of Escherichia coli to abiotic surfaces as an example. Lett. Appl. Microbiol. 2009, 49, 1–7. [Google Scholar] [CrossRef]
- Neu, T.R. Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol. Rev. 1996, 60, 151–166. [Google Scholar] [CrossRef]
- Ukuku, D.O.; Fett, W.F. Relationship of Cell Surface Charge and Hydrophobicity to Strength of Attachment of Bacteria to Cantaloupe Rind†. J. Food Prot. 2002, 65, 1093–1099. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, A.M.; Mateus, G.A.P.; Dos Santos, T.R.T.; Filho, B.A.D.A.; Gomes, R.G.; Bergamasco, R. Functionalized magnetite nanoparticles with Moringa oleifera with potent antibacterial action in wastewater. Environ. Technol. 2020, 1–22. [Google Scholar] [CrossRef]
- Domingues, V.; Alves, A.; Cabral, M.; Delerue-Matos, C. Sorption behaviour of bifenthrin on cork. J. Chromatogr. A 2005, 1069, 127–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Losen, M.; Frolich, B.; Pohl, M.; Büchs, J. Effect of oxygen limitation and medium composition on Escherichia coli fermentation in shake-flask cultures. Biotechnol. Prog. 2004, 20, 1062–1068. [Google Scholar] [CrossRef] [PubMed]
- Nogva, H.K.; Drømtorp, S.M.; Nissen, H.; Rudi, K. Ethidium Monoazide for DNA-Based Differentiation of Viable and Dead Bacteria by 5′-Nuclease PCR. BioTechniques 2003, 34, 804–813. [Google Scholar] [CrossRef] [PubMed]
- Rudi, K.; Moen, B.; Drømtorp, S.M.; Holck, A.L. Use of Ethidium Monoazide and PCR in Combination for Quantification of Viable and Dead Cells in Complex Samples. Appl. Environ. Microbiol. 2005, 71, 1018–1024. [Google Scholar] [CrossRef] [Green Version]
- Kubota, M.; Nakabayashi, T.; Matsumoto, Y.; Shiomi, T.; Yamada, Y.; Ino, K.; Yamanokuchi, H.; Matsui, M.; Tsunoda, T.; Mizukami, F.; et al. Selective adsorption of bacterial cells onto zeolites. Colloids Surf. B Biointerfaces 2008, 64, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Rashid, U.; Anwar, F.; Moser, B.; Knothe, G. Moringa oleifera oil: A possible source of biodiesel. Bioresour. Technol. 2008, 99, 8175–8179. [Google Scholar] [CrossRef]
- Nazzaro, F.; Fratianni, F.; Cozzolino, R.; Martignetti, A.; Malorni, L.; De Feo, V.; Cruz, A.G.; D’Acierno, A. Antibacterial Activity of Three Extra Virgin Olive Oils of the Campania Region, Southern Italy, Related to Their Polyphenol Content and Composition. Microorganisms 2019, 7, 321. [Google Scholar] [CrossRef] [Green Version]
- Klein, M.P.; Scheeren, C.W.; Lorenzoni, A.S.G.; Dupont, J.; Frazzon, J.; Hertz, P.F. Ionic liquid-cellulose film for enzyme immobilization. Process. Biochem. 2011, 46, 1375–1379. [Google Scholar] [CrossRef]
- Żur, J.; Wojcieszyńska, D.; Guzik, U. Metabolic Responses of Bacterial Cells to Immobilization. Molecules 2016, 21, 958. [Google Scholar] [CrossRef] [Green Version]
- Suarez, M.; Haenni, M.; Canarelli, S.; Fisch, F.; Chodanowski, P.; Servis, C.; Michielin, O.; Freitag, R.; Moreillon, P.; Mermod, N. Structure-Function Characterization and Optimization of a Plant-Derived Antibacterial Peptide. Antimicrob. Agents Chemother. 2005, 49, 3847–3857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).