Using Optimal Environmental DNA Method to Improve the Fish Diversity Survey—From Laboratory to Aquatic Life Reserve
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
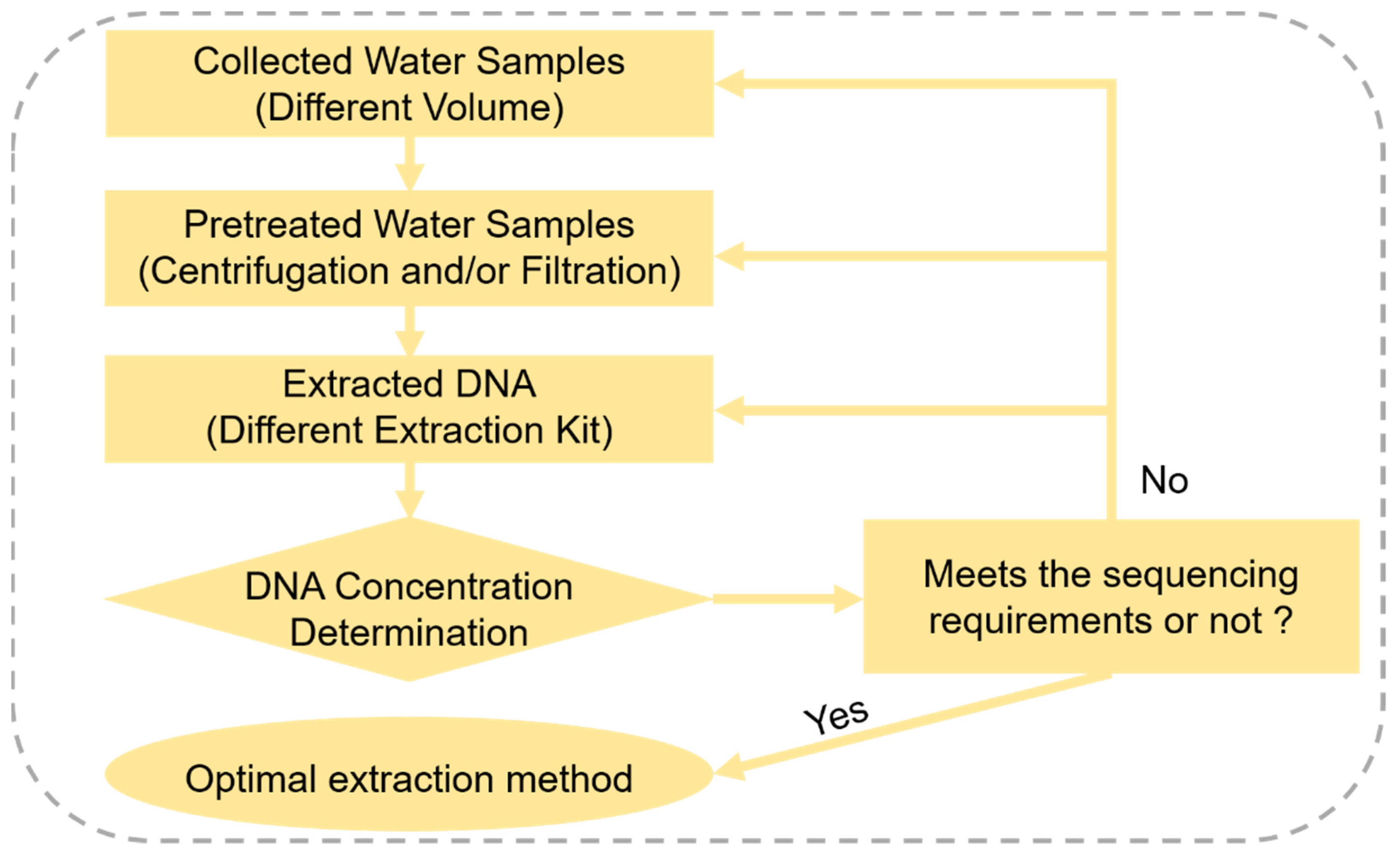
2.2. Optimization of Environmental DNA Method in the Laboratory
2.2.1. Water Sample Collection and Processing
2.2.2. eDNA Extraction
2.2.3. Primer Comparison
2.3. Application of Optimized Environmental DNA Method in The Aquatic Reserve
2.3.1. Sampling and DNA Extraction Methods
2.3.2. DNA Amplification and Sequencing
2.3.3. Quantitative Analysis of Target Fish eDNA
2.3.4. Calculation of Fish Diversity
3. Results and Discussion
3.1. Optimization of Environmental DNA in the Laboratory
3.1.1. DNA Concentration of Different Sampling Methods in Laboratory
3.1.2. Sequencing Results of Different DNA Primers
3.2. Application of Optimized Environmental DNA Method in the Aquatic Reserve
3.2.1. Fish Diversity from Trawling and eDNA Survey
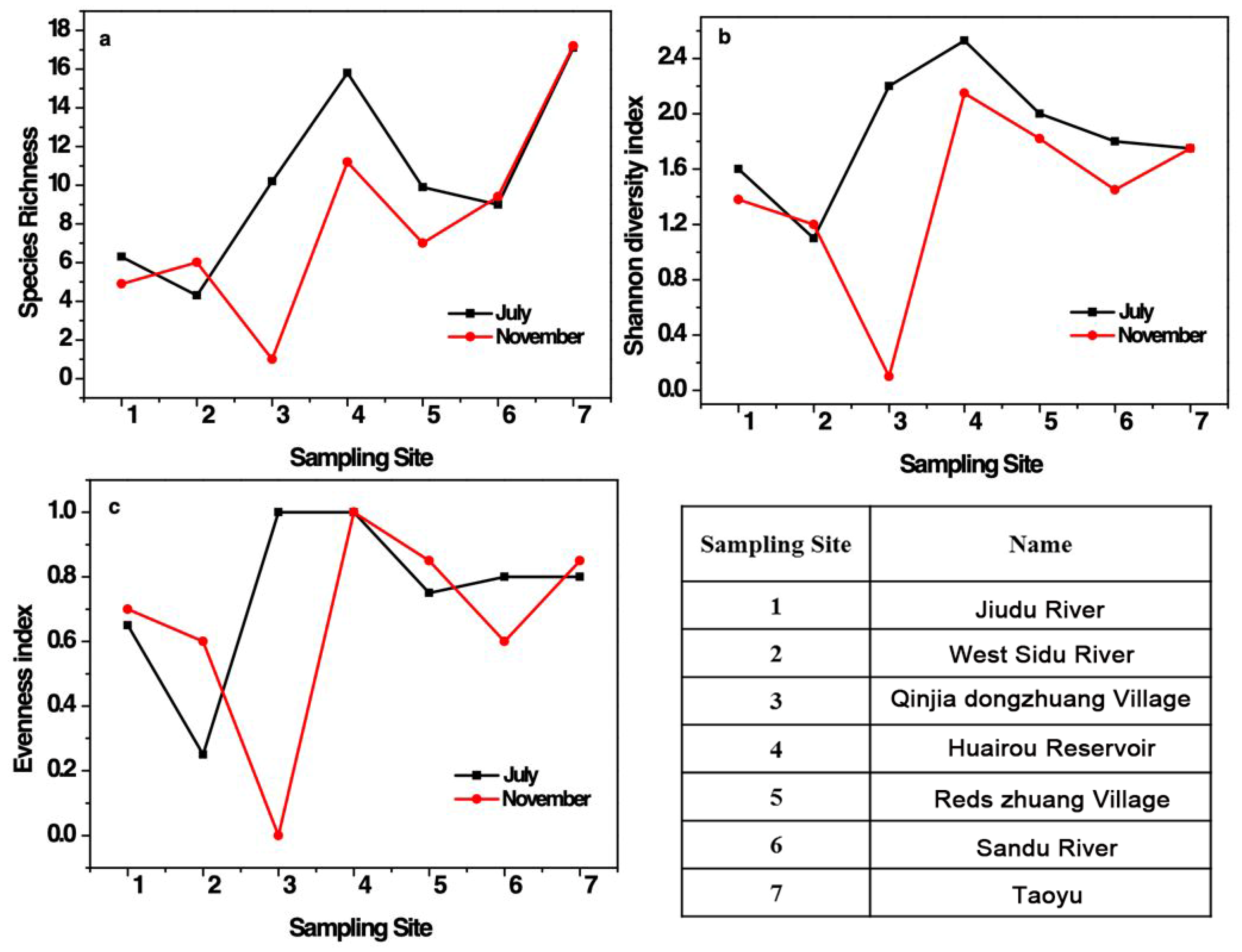
3.2.2. Fish Diversity
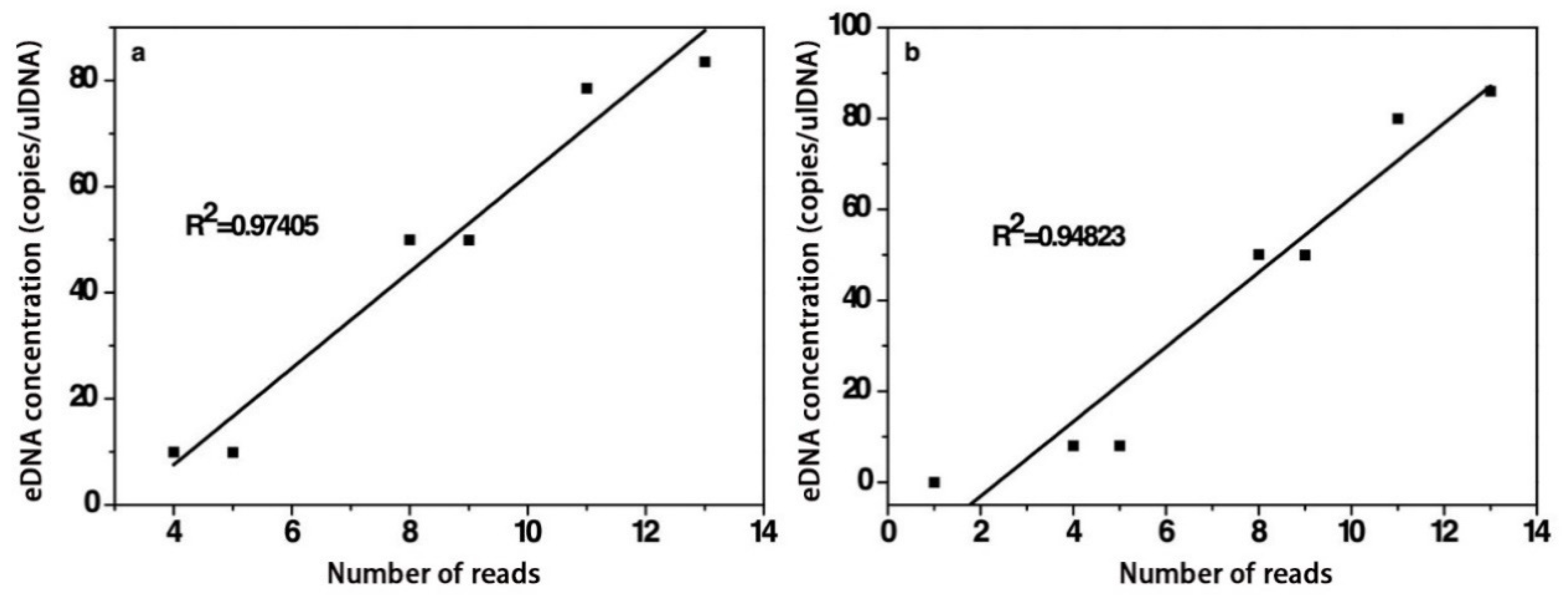
3.2.3. Quantitative Analysis of Target Fish eDNA
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Primer | Sequence | Fragment Length | Reference |
|---|---|---|---|
| 12S_30F | 5′-CACTGAAGMTGYTAAGAYG-3′ | 700 bp | [29] |
| 12S_1380R | 5′-CTKGCTAAATCATGATGC-3′ | ||
| Fish2degCBF | 5′-ACAACTTCACCCCTGCRAAY-3′ | 540 bp | [30] |
| Fish2CBR | 5′-GATGGCGTAGGCAAATAGGA-3′ | ||
| 16sF | 5′-CGCCTGTTTATCAAAAACAT-3′ | 600 bp | [31] |
| 16sR | 5′-CCGGTCTGAACTCAGATCACGT-3′ | ||
| COIF | 5′-TTCTCCACCAACCACAARGAYATYGG-3′ | 608 bp | [32] |
| COIR | 5′-CACCTCAGGGTGTCCGAARAAYCARAA-3′ |
| Primer | Primer Sequences (5′ to 3′) |
|---|---|
| AB741878.1 forward primer | AACATCGCCTCCTGCAAC |
| AB741878.1 reverse primer | GTTTAGCCATTCATACAGGTCTC |
| KF305680.1 forward primer | ATTGATCTACCCGTGCAGAAG |
| KF305680.1 reverse primer | AGGGTAACTCGGTCCGTTG |
References
- Pimm, S.L.; Jenkins, C.N.; Abell, R.; Brooks, T.; Gittleman, J.L.; Joppa, L.N.; Raven, P.H.; Roberts, C.M.; Sexton, J.O. The biodiversity of species and their rates of extinction, distribution, and protection. Science 2014, 344, 1246752. [Google Scholar] [CrossRef]
- Pimm, S.L.; Russell, G.J.; Gittleman, J.L.; Brooks, T. The Future of Biodiversity. Science 1995, 269, 347–350. [Google Scholar] [CrossRef]
- Barnosky, A.D.; Matzke, N.; Tomiya, S.; Wogan, G.O.U.; Swartz, B.; Quental, T.B.; Marshall, C.R.; McGuire, J.L.; Lindsey, E.L.; Maguire, K.C.; et al. Has the Earth’s sixth mass extinction already arrived? Nat. Cell Biol. 2011, 471, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Yasuhara, M.; Wei, C.-L.; Kucera, M.; Costello, M.J.; Tittensor, D.P.; Kiessling, W.; Bonebrake, T.C.; Tabor, C.R.; Feng, R.; Baselga, A.; et al. Past and future decline of tropical pelagic biodiversity. Proc. Natl. Acad. Sci. USA 2020, 117, 12891–12896. [Google Scholar] [CrossRef] [PubMed]
- Trathan, P.N.; García-Borboroglu, P.; Boersma, D.; Bost, C.; Crawford, R.J.M.; Crossin, G.T.; Cuthbert, R.J.; Dann, P.; Davis, L.S.; De La Puente, S.; et al. Pollution, habitat loss, fishing, and climate change as critical threats to penguins. Conserv. Biol. 2015, 29, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Stafford, R.; Jones, P.J. Viewpoint—Ocean plastic pollution: A convenient but distracting truth? Mar. Policy 2019, 103, 187–191. [Google Scholar] [CrossRef]
- Jackson, J.B.C.; Kirby, M.X.; Berger, W.H.; Bjorndal, K.A.; Botsford, L.W.; Bourque, B.J.; Bradbury, R.H.; Cooke, R.; Erlandson, J.; Estes, J.A.; et al. Historical Overfishing and the Recent Collapse of Coastal Ecosystems. Science 2001, 293, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Kaimuddin, A.H.; Laë, R.; De Morais, L.T. Fish Species in a Changing World: The Route and Timing of Species Migration between Tropical and Temperate Ecosystems in Eastern Atlantic. Front. Mar. Sci. 2016, 3, 162. [Google Scholar] [CrossRef]
- Lapointe, N.W.R.; Corkum, L.D.; Mandrak, N.E. A Comparison of Methods for Sampling Fish Diversity in Shallow Offshore Waters of Large Rivers. N. Am. J. Fish. Manag. 2006, 26, 503–513. [Google Scholar] [CrossRef]
- Desiderà, E.; Guidetti, P.; Panzalis, P.; Navone, A.; Valentini-Poirrier, C.; Boissery, P.; Gervaise, C.; Di Iorio, L. Acoustic fish communities: Sound diversity of rocky habitats reflects fish species diversity. Mar. Ecol. Prog. Ser. 2019, 608, 183–197. [Google Scholar] [CrossRef]
- Lodge, D.M.; Williams, S.; MacIsaac, H.J.; Hayes, K.R.; Leung, B.; Reichard, S.; Mack, R.N.; Moyle, P.B.; Smith, M.; Andow, D.A.; et al. Biological invasions: Recommendations for U.S. policy and management. Ecol. Appl. 2006, 16, 2035–2054. [Google Scholar] [CrossRef]
- Jerde, C.L.; Mahon, A.R.; Chadderton, W.L.; Lodge, D.M. “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv. Lett. 2011, 4, 150–157. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Miaud, C.; Pompanon, F.; Taberlet, P. Species detection using environmental DNA from water samples. Biol. Lett. 2008, 4, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Hansen, B.K.; Jacobsen, M.W.; Middelboe, A.L.; Preston, C.M.; Marin, R.; Bekkevold, D.; Knudsen, S.W.; Møller, P.R.; Nielsen, E.E. Remote, autonomous real-time monitoring of environmental DNA from commercial fish. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rees, H.C.; Maddison, B.C.; Middleditch, D.J.; Patmore, J.R.; Gough, K. REVIEW: The detection of aquatic animal species using environmental DNA—A review of eDNA as a survey tool in ecology. J. Appl. Ecol. 2014, 51, 1450–1459. [Google Scholar] [CrossRef]
- Laramie, M.B.; Pilliod, D.S.; Goldberg, C.S. Characterizing the distribution of an endangered salmonid using environmental DNA analysis. Biol. Conserv. 2015, 183, 29–37. [Google Scholar] [CrossRef]
- Xu, N.; Zhu, B.; Shi, F.; Shao, K.; Que, Y.; Li, W.; Li, W.; Jiao, W.; Tian, H.; Xu, D.; et al. Monitoring seasonal distribution of an endangered anadromous sturgeon in a large river using environmental DNA. Naturwissenschaften 2018, 105, 62. [Google Scholar] [CrossRef] [PubMed]
- Takahara, T.; Minamoto, T.; Doi, H. Using Environmental DNA to Estimate the Distribution of an Invasive Fish Species in Ponds. PLoS ONE 2013, 8, e56584. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, P.F.; Willerslev, E. Environmental DNA—An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Qu, C.; Stewart, K.A.; Clemente-Carvalho, R.; Zheng, J.; Wang, Y.; Gong, C.; Ma, L.; Zhao, J.; Lougheed, S.C. Comparing fish prey diversity for a critically endangered aquatic mammal in a reserve and the wild using eDNA metabarcoding. Sci. Rep. 2020, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.L.; Clarke, L.J.; Wedderburn, S.D.; Barnes, T.C.; Weyrich, L.S.; Cooper, A. Comparison of environmental DNA metabarcoding and conventional fish survey methods in a river system. Biol. Conserv. 2016, 197, 131–138. [Google Scholar] [CrossRef]
- Wang, S.; Yan, Z.; Hänfling, B.; Zheng, X.; Wang, P.; Fan, J.; Li, J. Methodology of fish eDNA and its applications in ecology and environment. Sci. Total Environ. 2021, 755, 142622. [Google Scholar] [CrossRef]
- Muha, T.P.; Rodriguez-Barreto, D.; O’Rorke, R.; de Leaniz, C.G.; Consuegra, S. Using eDNA Metabarcoding to Monitor Changes in Fish Community Composition After Barrier Removal. Front. Ecol. Evol. 2021, 9, 28. [Google Scholar] [CrossRef]
- Valdivia-Carrillo, T.; Rocha-Olivares, A.; Reyes-Bonilla, H.; Domínguez-Contreras, J.F.; Munguia-Vega, A. Integrating eDNA metabarcoding and simultaneous underwater visual surveys to describe complex fish communities in a marine biodiversity hotspot. Mol. Ecol. Resour. 2021. [Google Scholar] [CrossRef] [PubMed]
- Sales, N.G.; Wangensteen, O.S.; Carvalho, D.C.; Deiner, K.; Præbel, K.; Coscia, I.; McDevitt, A.D.; Mariani, S. Space-time dynamics in monitoring neotropical fish communities using eDNA metabarcoding. Sci. Total Environ. 2021, 754, 142096. [Google Scholar] [CrossRef] [PubMed]
- Lahoz-Monfort, J.J.; Guillera-Arroita, G.; Tingley, R. Statistical approaches to account for false-positive errors in environmental DNA samples. Mol. Ecol. Resour. 2016, 16, 673–685. [Google Scholar] [CrossRef]
- Scott, K.M. Using BLAST to Teach “E-value-tionary” Concepts. PLoS Biol. 2011, 9, e1001014. [Google Scholar] [CrossRef]
- Xing, Y.; Zhao, Y.; Li, G.; Yang, Y.; Hu, Y. Fish species diversity and resource conservation in Huaisha-Huaijiuhe municipal aquatic wildlife reserve, Beijing. Chin. J. Zool. 2007, 42, 29–37. [Google Scholar] [CrossRef]
- Ficht, T.A.; Adams, G.L. Probes and Method for Identifying Species and Biovars of Brucella. U.S. Patent 5,348,857, 20 September 1994. [Google Scholar]
- Kyle, C.; Wilson, C. Mitochondrial DNA identification of game and harvested freshwater fish species. Forensic Sci. Int. 2007, 166, 68–76. [Google Scholar] [CrossRef]
- Palumbi, S.R. Nucleic Acids II: The Polymerase Chain Reaction. In Molecular Systematics, 2nd ed.; Hillis, D.M., Moritz, C., Mable, B.K., Eds.; Sinauer Associates: Sunderland, MA, USA, 1996; pp. 205–247. [Google Scholar]
- Ivanova, N.V.; Zemlak, T.S.; Hanner, R.H.; Hebert, P.D.N. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes 2007, 7, 544–548. [Google Scholar] [CrossRef]
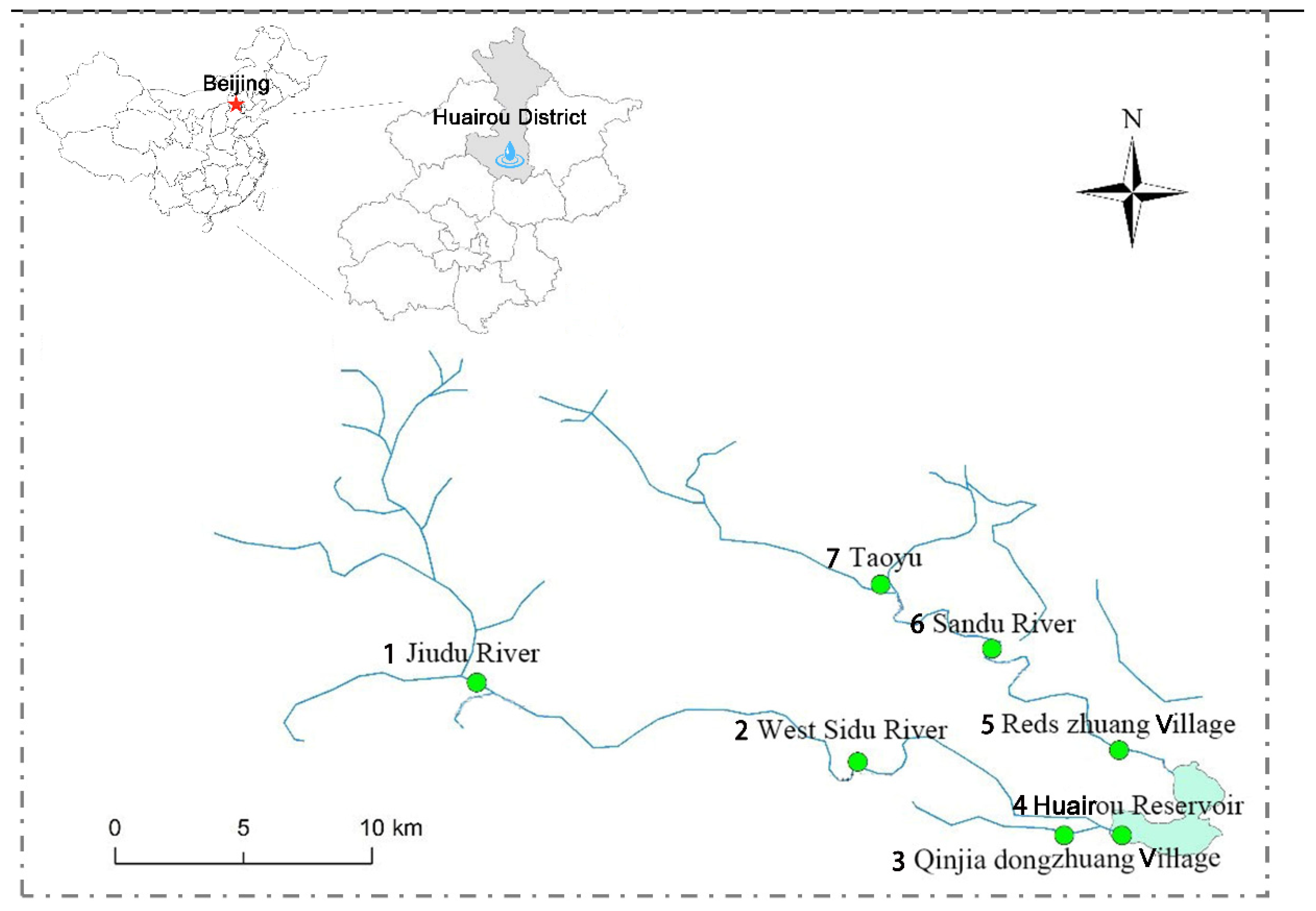
| Number | Site | Location |
|---|---|---|
| 1 | Yidu River | 40°20′1″ N, 116°29′7″ E |
| 2 | Sidu River | 40°20′3″ N, 116°28′4″ E |
| 3 | Jiudu River | 40°20′5″ N; 116°29′5″ E |
| Number | Pretreated Method | Average Concentration of Nucleic Acid (ng/µL) | Average OD260/OD280 | Main Ingredients |
|---|---|---|---|---|
| 1 | 15 mL + Centrifugation + DTK | 21.1 | 1.52 < 1.60 | Protein |
| 2 | 15 mL + Centrifugation + QDK | 36.4 | 1.50 < 1.60 | Protein |
| 3 | 1 L + Filtration + DTK | 60.7 | 1.60 (1.60–1.90) | DNA |
| 4 | 1 L + Filtration + QDK | 79.3 | 1.96 ≥ 1.90 | RNA |
| 5 | 1 L + Filtration + MPK | 102.8 | 1.90 ≥ 1.90 | RNA |
| 6 | 1 L + Filtration + QSK | 43.4 | 1.61 (1.60–1.90) | DNA |
| 7 | 2 L + Filtration + DTK | 62.1 | 1.61 (1.60–1.90) | DNA |
| 8 | 2 L + Filtration + QDK | 100.2 | 1.93 ≥ 1.90 | RNA |
| 9 | 2 L + Filtration + MPK | 105.7 | 1.93 ≥ 1.90 | RNA |
| 10 | 2 L + Filtration + QSK | 66.4 | 1.60 (1.60–1.90) | DNA |
| Primers | Species | Number of Reads |
|---|---|---|
| COI | Ramlibacter tataouinensis TTB310 | 4 |
| Polaromonas sp. JS666 | 4 | |
| Acidovorax avenae subsp. ATCC 19860 | 11 | |
| 12s | Epinephelus fuscoguttatus | 2 |
| 16s | Rhodeus ocellatus | 2 |
| Carassius gibelio | 4 | |
| Corbicula colorata | 4 | |
| Tachysurus fulvidraco | 1 | |
| Pseudorasbora parva | 13 | |
| Sarcocheilichthys nigripinnis | 1 | |
| Micropterus salmoides | 1 | |
| Zacco platypus | 5 |
| Detected Species | eDNA (2017) | Trawling (2005) | Beijing Fish Record (1984) |
|---|---|---|---|
| Abbottina obtusirostris | 2 | P | N |
| Abbottina rivularis | 1 | P | P |
| Acheilognathus chankaensis | 2 | P | N |
| Acipenser gueldenstaedtii | 24 | N | N |
| Acipenser mikadoi | 2 | N | N |
| Acipenser fulvescens | 21 | N | N |
| Acipenser schrenckii | 11 | N | N |
| Carassius gibelio | 30 | N | P |
| Channa argus | 2 | N | P |
| Ctenopharyngodon idella | 1 | N | P |
| Culter erythropterus | 2 | N | P |
| Cyprinus carpio | 1 | N | P |
| Hypomesus olidus | 2 | P (identified in genus level) | N |
| Hypophthalmichthys sp. | 48 | N | P |
| Micropercops swinhonis | 8 | N | N |
| Misgurnus sp. | 5 | N | P |
| Misgurnus anguillicaudatus | 1 | P | P |
| Odontobutis potamophila | 4 | N | N |
| Oncorhynchus mykiss | 7 | P | P |
| Oreochromis aureus | 1 | N | P |
| Pelteobafrus fulvidraco | 1 | N | P |
| Pseudobagrus ussuriensis | 1 | N | N |
| Pseudorasbora parva | 15 | P | P |
| Rhinogobius giurinus | 4 | P | N |
| Rhodeus sp. | 11 | P | N |
| Sarcocheilichthys nigripinnis | 2 | P | P |
| Silurus sp. | 3 | N | N |
| Squalidus gracilis | 1 | N | N |
| Zacco platypus | 58 | P | N |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.-P.; Liu, Z.-F.; Guo, T.; Chen, H.; Xie, X. Using Optimal Environmental DNA Method to Improve the Fish Diversity Survey—From Laboratory to Aquatic Life Reserve. Water 2021, 13, 1468. https://doi.org/10.3390/w13111468
Li W-P, Liu Z-F, Guo T, Chen H, Xie X. Using Optimal Environmental DNA Method to Improve the Fish Diversity Survey—From Laboratory to Aquatic Life Reserve. Water. 2021; 13(11):1468. https://doi.org/10.3390/w13111468
Chicago/Turabian StyleLi, Wen-Pan, Zi-Fang Liu, Tong Guo, He Chen, and Xin Xie. 2021. "Using Optimal Environmental DNA Method to Improve the Fish Diversity Survey—From Laboratory to Aquatic Life Reserve" Water 13, no. 11: 1468. https://doi.org/10.3390/w13111468
APA StyleLi, W.-P., Liu, Z.-F., Guo, T., Chen, H., & Xie, X. (2021). Using Optimal Environmental DNA Method to Improve the Fish Diversity Survey—From Laboratory to Aquatic Life Reserve. Water, 13(11), 1468. https://doi.org/10.3390/w13111468







