Mobile Genetic Elements Drive the Antibiotic Resistome Alteration in Freshwater Shrimp Aquaculture
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. High-Throughput Sequencing
2.3. Bioinformatics Analysis
2.4. Statistical Analysis
3. Results
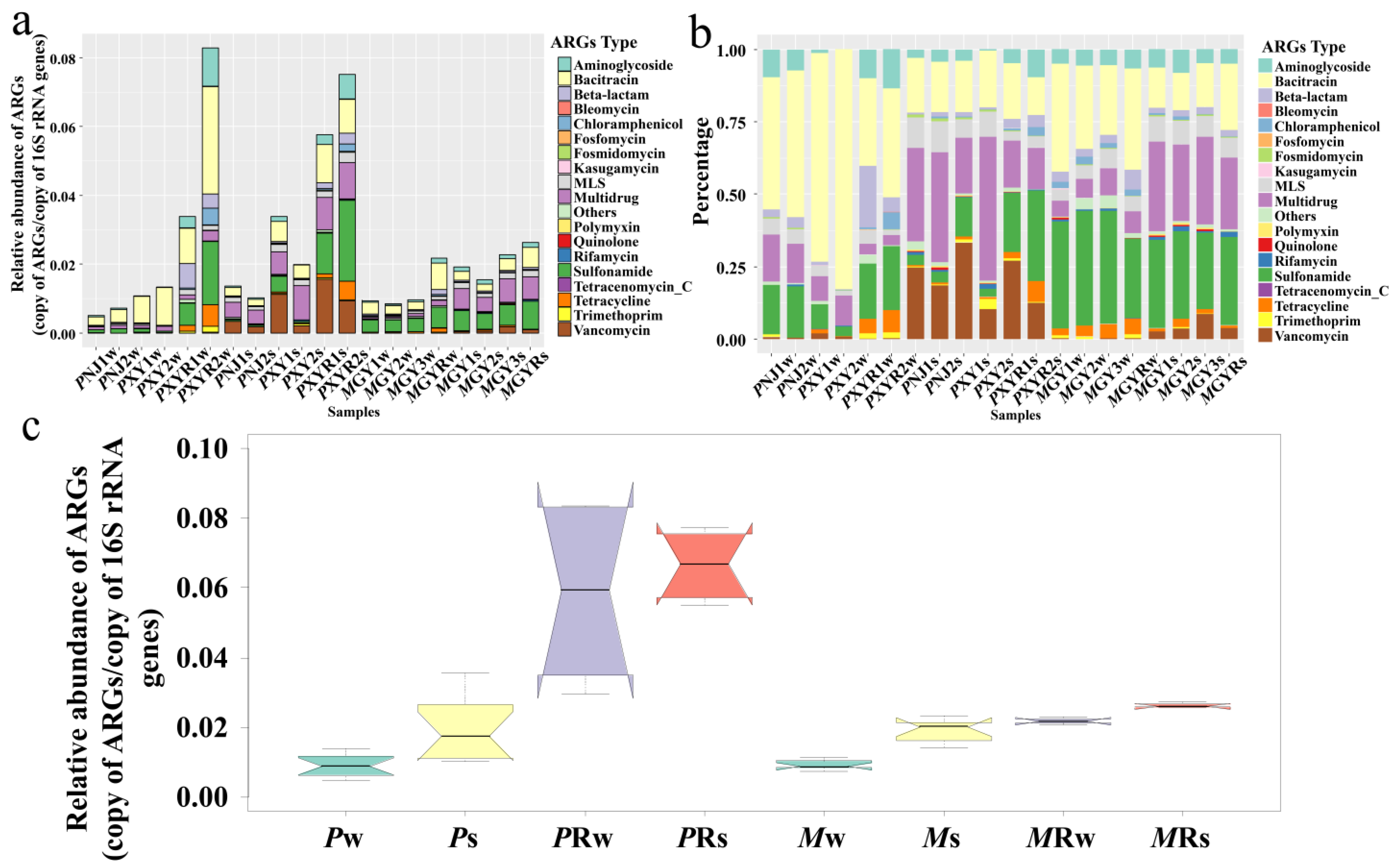
3.1. Abundance and Diversity of ARGs in the Shrimp Aquaculture Environment
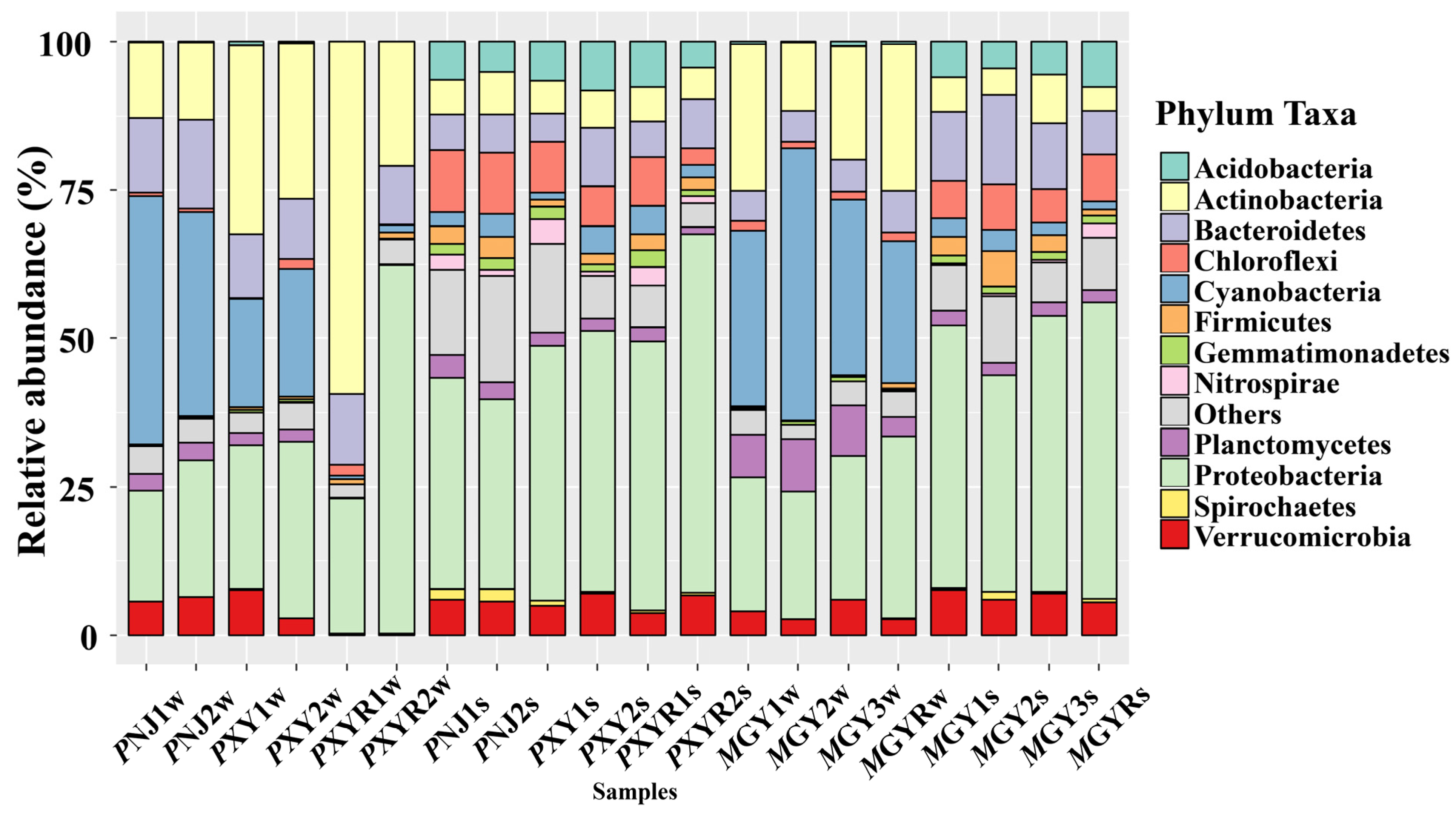
3.2. Diversity and Structure of Bacterial Communities in the Shrimp Aquaculture Environment
3.3. Linkages between Bacterial Community (Including Pathogens) and ARGs
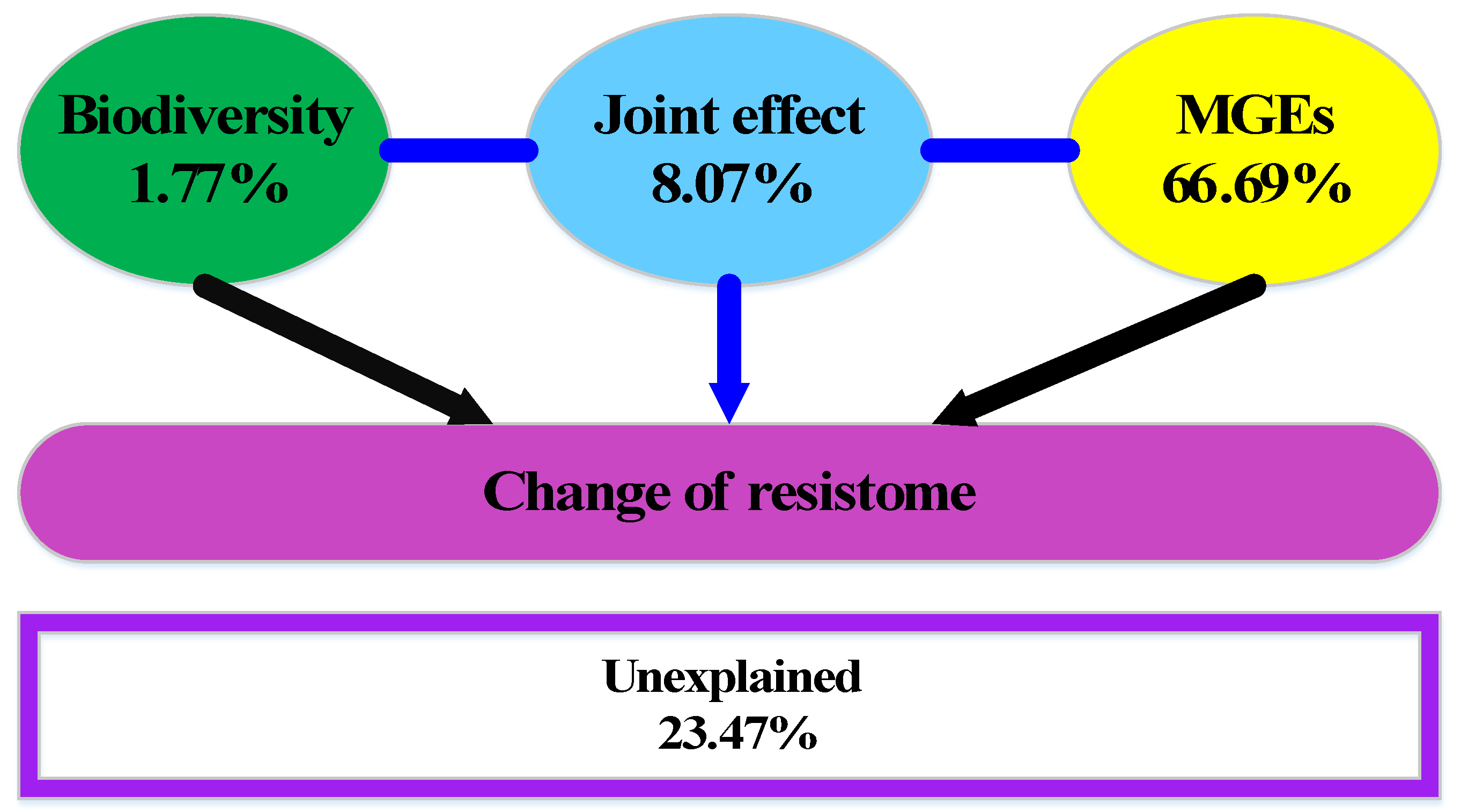
3.4. Correlations among Biodiversity, MGEs, and ARGs
4. Discussion
4.1. ARGs Distribution in Freshwater Shrimp Aquaculture Environment
4.2. Associations between ARGs and Bacteria
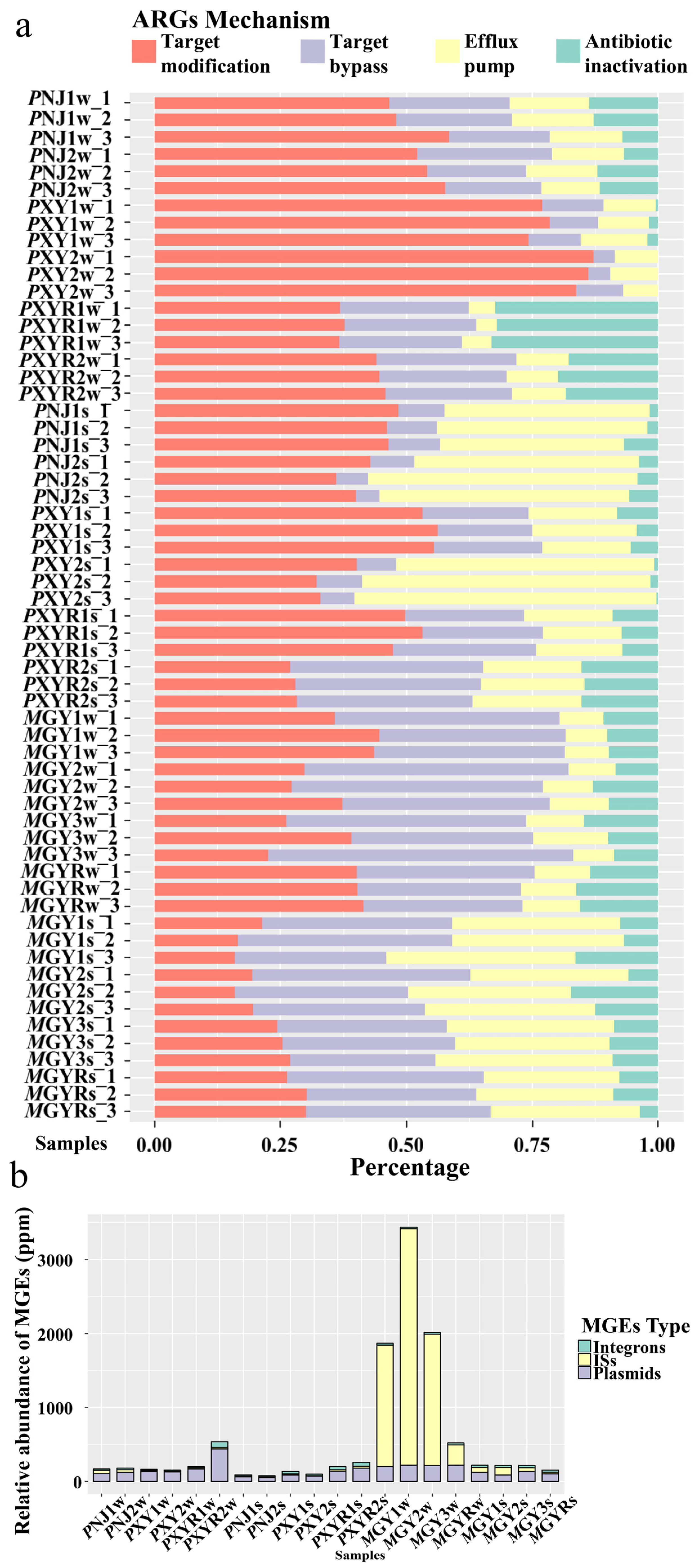
4.3. Associations among ARGs, Biodiversity and MGEs
4.4. Practical Implications of This Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, Q.Q.; Yang, G.F.; Sun, K.K.; Tian, G.M.; Jin, R.C. Insights into the effects of bio-augmentation on the granule-based anammox process under continuous oxytetracycline stress: Performance and microflora structure. Chem. Eng. J. 2018, 348, 503–513. [Google Scholar] [CrossRef]
- Santos, L.; Ramos, F. Antimicrobial resistance in aquaculture: Current knowledge and alternatives to tackle the problem. Int. J. Antimicrob. Agents 2018, 52, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Hu, Y.; Cheng, J.; Chen, Y. Research progress on distribution, migration, transformation of antibiotics and antibiotic resistance genes (ARGs) in aquatic environment. Crit. Rev. Biotechnol. 2018, 38, 1195–1208. [Google Scholar] [CrossRef] [PubMed]
- He, L.Y.; Liu, Y.S.; Su, H.C.; Zhao, J.L.; Liu, S.S.; Chen, J.; Liu, W.R.; Ying, G.G. Dissemination of antibiotic resistance genes in representative broiler feedlots environments: Identification of indicator ARGs and correlations with environmental variables. Environ. Sci. Technol. 2014, 48, 13120–13129. [Google Scholar] [CrossRef]
- Jia, S.; Shi, P.; Hu, Q.; Li, B.; Zhang, T.; Zhang, X.X. Bacterial community shift drives antibiotic resistance promotion during drinking water chlorination. Environ. Sci. Technol. 2015, 49, 12271–12279. [Google Scholar] [CrossRef]
- Zhang, H.; He, H.; Chen, S.; Huang, T.; Lu, K.; Zhang, Z.; Wang, R.; Zhang, X.; Li, H. Abundance of antibiotic resistance genes and their association with bacterial communities in activated sludge of wastewater treatment plants: Geographical distribution and network analysis. J. Environ. Sci. 2019, 82, 24–38. [Google Scholar] [CrossRef]
- Zhang, T.; Li, X.; Wang, M.; Chen, H.; Yang, Y.; Chen, Q.; Yao, M. Time-resolved spread of antibiotic resistance genes in highly polluted air. Environ. Int. 2019, 127, 333–339. [Google Scholar] [CrossRef]
- Dong, P.; Cui, Q.; Fang, T.; Huang, Y.; Wang, H. Occurrence of antibiotic resistance genes and bacterial pathogens in water and sediment in urban recreational water. J. Environ. Sci. 2019, 77, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.G.; Zhao, Y.; Li, B.; Huang, C.L.; Zhang, S.Y.; Yu, S.; Chen, Y.-S.; Zhang, T.; Gillings, M.R.; Su, J.-Q. Continental-scale pollution of estuaries with antibiotic resistance genes. Nat. Microbiol. 2017, 2, 16270. [Google Scholar] [CrossRef]
- Tacon, A.G.J. Trends in global aquaculture and aquafeed production: 2000–2017. Rev. Fish. Sci. 2020, 28, 43–56. [Google Scholar] [CrossRef]
- Watts, J.E.M.; Schreier, H.J.; Lanska, L.; Hale, M.S. The rising tide of antimicrobial resistance in aquaculture: Sources, sinks and solutions. Mar. Drugs 2017, 15, 158. [Google Scholar] [CrossRef] [PubMed]
- Chuah, L.O.; Effarizah, M.E.; Goni, A.M.; Rusul, G. Antibiotic application and emergence of multiple antibiotic resistance (MAR) in global catfish aquaculture. Curr. Environ. Health Rep. 2016, 3, 118–127. [Google Scholar] [CrossRef]
- Zhou, R.; Zeng, S.; Hou, D.; Liu, J.; Weng, S.; He, J.; Huang, Z. Occurrence of human pathogenic bacteria carrying antibiotic resistance genes revealed by metagenomic approach: A case study from an aquatic environment. J. Environ. Sci. 2019, 80, 248–256. [Google Scholar] [CrossRef]
- Liu, X.; Steele, J.C.; Meng, X.Z. Usage, residue, and human health risk of antibiotics in Chinese aquaculture: A review. Environ. Pollut. 2017, 223, 161–169. [Google Scholar] [CrossRef]
- Zhang, Y.B.; Li, Y.; Sun, X.L. Antibiotic resistance of bacteria isolated from shrimp hatcheries and cultural ponds on Donghai Island, China. Mar. Pollut. Bull. 2011, 62, 2299–2307. [Google Scholar] [CrossRef]
- Tiedje, J.M.; Wang, F.; Manaia, C.M.; Virta, M.; Sheng, H.; Ma, L.; Zhang, T.; Topp, E. Antibiotic resistance genes in the human-impacted environment: A One Health perspective. Pedosphere 2019, 29, 273–282. [Google Scholar] [CrossRef]
- Xiong, W.; Sun, Y.; Zhang, T.; Ding, X.; Li, Y.; Wang, M.; Zeng, Z. Antibiotics, antibiotic resistance genes, and bacterial community composition in fresh water aquaculture environment in China. Microb. Ecol. 2015, 70, 425–432. [Google Scholar] [CrossRef]
- Zhou, R.; Zeng, S.; Hou, D.; Liu, J.; Weng, S.; He, J.; Huang, Z. Temporal variation of antibiotic resistance genes carried by culturable bacteria in the shrimp hepatopancreas and shrimp culture pond water. Ecotoxicol. Environ. Saf. 2020, 199, 110738. [Google Scholar] [CrossRef]
- Zhang, R.Q.; Ying, G.G.; Su, H.C.; Zhou, L.J.; Liu, Y.S. Antibiotic resistance and genetic diversity of Escherichia coli isolates from traditional and integrated aquaculture in South China. J. Environ. Sci. Health Part B 2013, 48, 999–1013. [Google Scholar] [CrossRef]
- He, Y.; Jin, L.; Sun, F.; Hu, Q.; Chen, L. Antibiotic and heavy-metal resistance of Vibrio parahaemolyticus isolated from fresh shrimps in Shanghai fish markets, China. Environ. Sci. Pollut. Res. 2016, 23, 15033–15040. [Google Scholar] [CrossRef]
- Zhou, Z.C.; Lin, Z.J.; Shuai, X.Y.; Zheng, J.; Meng, L.X.; Zhu, L.; Sun, Y.-J.; Shang, W.-C.; Chen, H. Temporal variation and sharing of antibiotic resistance genes between water and wild fish gut in a peri-urban river. J. Environ. Sci. 2021, 103, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Lu, J.; Wu, J.; Zhang, Y.; Zhang, C. Proliferation of antibiotic resistance genes in coastal recirculating mariculture system. Environ. Pollut. 2019, 248, 462–470. [Google Scholar] [CrossRef]
- Zeng, S.; Hou, D.; Liu, J.; Ji, P.; Weng, S.; He, J.; Huang, Z. Antibiotic supplement in feed can perturb the intestinal microbial composition and function in Pacific white shrimp. Appl. Microbiol. Biotechnol. 2019, 103, 3111–3122. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, X.; Zhao, Z.; Duan, C.; Chen, H.; Wang, M.; Ren, H.; Yin, Y.; Ye, L. Metagenomic analysis revealed the prevalence of antibiotic resistance genes in the gut and living environment of freshwater shrimp. J. Hazard. Mater. 2018, 350, 10–18. [Google Scholar] [CrossRef]
- Liu, K.; Han, J.; Li, S.; Liu, L.; Lin, W.; Luo, J. Insight into the diversity of antibiotic resistance genes in the intestinal bacteria of shrimp Penaeus vannamei by culture-dependent and independent approaches. Ecotoxicol. Environ. Saf. 2019, 172, 451–459. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Yin, X.; Jiang, X.T.; Chai, B.; Li, L.; Yang, Y.; Cole, J.R.; Tjedje, J.M.; Zhang, T. ARGs-OAP v2.0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes. Bioinformatics 2018, 34, 2263–2270. [Google Scholar] [CrossRef]
- Fang, H.; Huang, K.; Yu, J.; Ding, C.; Wang, Z.; Zhao, C.; Yuan, H.; Wang, Z.; Wang, S.; Hu, J.; et al. Metagenomic analysis of bacterial communities and antibiotic resistance genes in the Eriocheir sinensis freshwater aquaculture environment. Chemosphere 2019, 224, 202–211. [Google Scholar] [CrossRef]
- Truong, D.T.; Franzosa, E.A.; Tickle, T.L.; Scholz, M.; Weingart, G.; Pasolli, E.; Tett, A.; Huttenhower, C.; Segata, N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 2015, 12, 902–903. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.; Gowtage-Sequeria, S.; Evans, B. T16: Quantitative Analysis of the Characteristics of Emerging and Re-Emerging Human Pathogens. Centre for Infectious Diseases, University of Edinburgh. 2015. Available online: http://www.bis.gov.uk/assets/foresight/docs/infectious-diseases/t16.pdf (accessed on 25 July 2015).
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261. [Google Scholar] [CrossRef] [PubMed]
- Barberán, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef]
- Feng, J.; Li, B.; Jiang, X.; Yang, Y.; Wells, G.F.; Zhang, T.; Li, X. Antibiotic resistome in a large-scale healthy human gut microbiota deciphered by metagenomic and network analyses. Environ. Microbiol. 2018, 20, 355–368. [Google Scholar] [CrossRef]
- Ju, F.; Xia, Y.; Guo, F.; Wang, Z.; Zhang, T. Taxonomic relatedness shapes bacterial assembly in activated sludge of globally distributed wastewater treatment plants. Environ. Microbiol. 2014, 16, 2421–2432. [Google Scholar] [CrossRef]
- Devarajan, N.; Laffite, A.; Graham, N.D.; Meijer, M.; Prabakar, K.; Mubedi, J.I.; Elongo, V.; Mpiana, P.T.; Ibelings, B.W.; Wildi, W.; et al. Accumulation of clinically relevant antibiotic-resistance genes, bacterial load, and metals in freshwater lake sediments in central Europe. Environ. Sci. Technol. 2015, 49, 6528–6537. [Google Scholar] [CrossRef]
- Pruden, A.; Arabi, M.; Storteboom, H.N. Correlation between upstream human activities and riverine antibiotic resistance genes. Environ. Sci. Technol. 2012, 46, 11541–11549. [Google Scholar] [CrossRef]
- Hu, Y.; Yang, X.; Qin, J.; Lu, N.; Cheng, G.; Wu, N.; Pan, Y.; Li, J.; Zhu, L.; Wang, X.; et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 2013, 4, 2151. [Google Scholar] [CrossRef] [PubMed]
- Mascher, T.; Margulis, N.G.; Wang, T.; Ye, R.W.; Helmann, J.D. Cell wall stress responses in Bacillus subtilis: The regulatory network of the bacitracin stimulon. Mol. Microbiol. 2003, 50, 1591–1604. [Google Scholar] [CrossRef]
- Siewert, G.; Strominger, J.L. Bacitracin: An inhibitor of the dephosphorylation of lipid pyrophosphate, an intermediate in the biosynthesis of the peptidoglycan of bacterial cell walls. Proc. Natl. Acad. Sci. USA 1967, 57, 767–773. [Google Scholar] [CrossRef]
- Gao, Q.; Li, Y.; Qi, Z.; Yue, Y.; Min, M.; Peng, S.; Shi, Z.; Gao, Y. Diverse and abundant antibiotic resistance genes from mariculture sites of China’s coastline. Sci. Total Environ. 2018, 630, 117–125. [Google Scholar] [CrossRef]
- Yang, Y.; Li, Z.; Song, W.; Du, L.; Ye, C.; Zhao, B.; Liu, W.; Deng, D.; Pan, Y.; Lin, H.; et al. Metagenomic insights into the abundance and composition of resistance genes in aquatic environments: Influence of stratification and geography. Environ. Int. 2019, 127, 371–380. [Google Scholar] [CrossRef]
- Sherpa, M.T.; Najar, I.N.; Das, S.; Thakur, N. Distribution of antibiotic and metal resistance genes in two glaciers of North Sikkim, India. Ecotoxicol. Environ. Saf. 2020, 203, 111037. [Google Scholar] [CrossRef]
- Li, Y.; Cao, W.; Liang, S.; Yamasaki, S.; Chen, X.; Shi, L.; Ye, L. Metagenomic characterization of bacterial community and antibiotic resistance genes in representative ready-to-eat food in southern China. Sci. Rep. 2020, 10, 15175. [Google Scholar] [CrossRef]
- Phuong Hoa, P.T.; Nonaka, L.; Hung Viet, P.; Suzuki, S. Detection of the sul1, sul2, and sul3 genes in sulfonamide-resistant bacteria from wastewater and shrimp ponds of north Vietnam. Sci. Total Environ. 2008, 405, 377–384. [Google Scholar] [CrossRef]
- Wang, L.; Su, H.; Hu, X.; Xu, Y.; Xu, W.; Huang, X.; Li, Z.; Cao, Y.; Wen, G. Abundance and removal of antibiotic resistance genes (ARGs) in the rearing environments of intensive shrimp aquaculture in South China. J. Environ. Sci. Health Part B 2019, 54, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Ying, C.; Chang, M.J.; Hu, C.H.; Chang, Y.T.; Chao, W.L.; Yeh, S.L.; Chang, S.J.; Hsu, J.T. The effects of marine farm-scale sequentially integrated multi-trophic aquaculture systems on microbial community composition, prevalence of sulfonamide-resistant bacteria and sulfonamide resistance gene sul1. Sci. Total Environ. 2018, 643, 681–691. [Google Scholar] [CrossRef]
- Echeverria-Palencia, C.M.; Thulsiraj, V.; Tran, N.; Ericksen, C.A.; Melendez, I.; Sanchez, M.G.; Walpert, D.; Yuan, T.; Ficara, E.; Senthilkumar, N.; et al. Disparate antibiotic resistance gene quantities revealed across 4 major cities in California: A survey in drinking water, air, and soil at 24 public parks. ACS Omega 2017, 2, 2255–2263. [Google Scholar] [CrossRef]
- Yuan, J.; Ni, M.; Liu, M.; Zheng, Y.; Gu, Z. Occurrence of antibiotics and antibiotic resistance genes in a typical estuary aquaculture region of Hangzhou Bay, China. Mar. Pollut. Bull. 2019, 138, 376–384. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Su, H.; Liu, S.; Hu, X.; Xu, X.; Xu, W.; Xu, Y.; Li, Z.; Wen, G.; Liu, Y.; Cao, Y. Occurrence and temporal variation of antibiotic resistance genes (ARGs) in shrimp aquaculture: ARGs dissemination from farming source to reared organisms. Sci. Total Environ. 2017, 607–608, 357–366. [Google Scholar] [CrossRef]
- Wang, J.H.; Lu, J.; Zhang, Y.X.; Wu, J.; Luo, Y.; Liu, H. Metagenomic analysis of antibiotic resistance genes in coastal industrial mariculture systems. Bioresour. Technol. 2018, 253, 235–243. [Google Scholar] [CrossRef]
- Becerra-Castro, C.; Macedo, G.; Silva, A.M.T.; Manaia, C.M.; Nunes, O.C. Proteobacteria become predominant during regrowth after water disinfection. Sci. Total Environ. 2016, 573, 313–323. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, Y.; Wu, J. Continental-scale spatio-temporal distribution of antibiotic resistance genes in coastal waters along coastline of China. Chemosphere 2020, 247, 125908. [Google Scholar] [CrossRef]
- Peterson, E.; Kaur, P. Antibiotic resistance mechanisms in bacteria: Relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef]
- Han, Y.; Wang, J.; Zhao, Z.; Chen, J.; Lu, H.; Liu, G. Fishmeal application induces antibiotic resistance gene propagation in mariculture sediment. Environ. Sci. Technol. 2017, 51, 10850–10860. [Google Scholar] [CrossRef]
- Willems, A.; Gillis, M. Hydrogenophaga. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2015. [Google Scholar]
- Yi, Y.; Zhang, Z.; Zhao, F.; Liu, H.; Yu, L.; Zha, J.; Wang, G. Probiotic potential of Bacillus velezensis JW: Antimicrobial activity against fish pathogenic bacteria and immune enhancement effects on Carassius auratus. Fish Shellfish Immunol. 2018, 78, 322–330. [Google Scholar] [CrossRef]
- Kuebutornye, F.K.A.; Abarike, E.D.; Lu, Y.; Hlordzi, V.; Sakyi, M.E.; Afriyie, G.; Wang, Z.; Li, Y.; Xie, C.X. Mechanisms and the role of probiotic Bacillus in mitigating fish pathogens in aquaculture. Fish Physiol. Biochem. 2020, 46, 819–841. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wu, Y.; Jiang, L.; Tan, A.; Zhang, R.; Luo, L. Multi-drug resistance mediated by Class 1 integrons in Aeromonas isolated from farmed freshwater animals. Front. Microbiol. 2016, 7, 935. [Google Scholar] [CrossRef] [PubMed]
- Igbinosa, I.H.; Igumbor, E.U.; Aghdasi, F.; Tom, M.; Okoh, A.I. Emerging Aeromonas species infections and their significance in public health. Sci. World J. 2012, 2012, 625023. [Google Scholar] [CrossRef] [PubMed]
- Adderson, E.E.; Boudreaux, J.W.; Hayden, R.T. Infections caused by coryneform bacteria in pediatric oncology patients. Pediatr. Infect. Dis. J. 2008, 27, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Evtushenko, L.I. Microbacteriaceae. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2015. [Google Scholar]
- Su, H.; Hu, X.; Wang, L.; Xu, W.; Xu, Y.; Wen, G.; Li, Z.; Cao, Y. Contamination of antibiotic resistance genes (ARGs) in a typical marine aquaculture farm: Source tracking of ARGs in reared aquatic organisms. J. Environ. Sci. Health Part B 2020, 55, 220–229. [Google Scholar] [CrossRef]
- Jia, J.; Guan, Y.; Cheng, M.; Chen, H.; He, J.; Wang, S.; Wang, Z. Occurrence and distribution of antibiotics and antibiotic resistance genes in Ba River, China. Sci. Total Environ. 2018, 642, 1136–1144. [Google Scholar] [CrossRef] [PubMed]
- Harrison, E.; Brockhurst, M.A. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 2012, 20, 262–267. [Google Scholar] [CrossRef]
- Pham, T.T.H.; Rossi, P.; Dinh, H.D.K.; Pham, N.T.A.; Tran, P.A.; Ho, T.T.K.M.; Dinh, Q.T.; De Alencastro, L.F. Analysis of antibiotic multi-resistant bacteria and resistance genes in the effluent of an intensive shrimp farm (Long An, Vietnam). J. Environ. Manag. 2018, 214, 149–156. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, Y.; Wu, J.; Luo, Y. Effects of microplastics on distribution of antibiotic resistance genes in recirculating aquaculture system. Ecotoxicol. Environ. Saf. 2019, 184, 109631. [Google Scholar] [CrossRef]
- Yuan, K.; Wang, X.; Chen, X.; Zhao, Z.; Fang, L.; Chen, B.; Jiang, J.; Luan, T.; Chen, B. Occurrence of antibiotic resistance genes in extracellular and intracellular DNA from sediments collected from two types of aquaculture farms. Chemosphere 2019, 234, 520–527. [Google Scholar] [CrossRef]
- Hu, Q.; Zhang, X.X.; Jia, S.; Huang, K.; Tang, J.; Shi, P.; Ye, L.; Ren, H. Metagenomic insights into ultraviolet disinfection effects on antibiotic resistome in biologically treated wastewater. Water Res. 2016, 101, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Su, J.Q.; Wei, B.; OuYang, W.Y.; Huang, F.Y.; Zhao, Y.; Xu, H.J.; Zhu, Y.G. Antibiotic resistome and its association with bacterial communities during sewage sludge composting. Environ. Sci. Technol. 2015, 49, 7356–7363. [Google Scholar] [CrossRef] [PubMed]

| Sampling Site Sitesites | City | GPS | Size | Sampling Date |
|---|---|---|---|---|
| P. clarkii | ||||
| NJ1 | Nanjing | 32°10′56″ N 118°50′59″ E | 1.47 ha | November, 2018 |
| NJ2 | Nanjing | 32°11′7″ N 118°51′34″ E | 1.33 ha | November, 2018 |
| XY1 | Xuyi | 32°57′35″ N 118°31′2″ E | 4.00 ha | May, 2019 |
| XY2 | Xuyi | 32°45′14″ N 118°33′38″ E | 1.33 ha | May, 2019 |
| M. rosenbergii | ||||
| GY1 | Gaoyou | 32°54′32″ N 119°33′35″ E | 1.13 ha | July, 2019 |
| GY2 | Gaoyou | 32°54′30″ N 119°33′32″ E | 1.67 ha | July, 2019 |
| GY3 | Gaoyou | 32°54′30″ N 119°33′32″ E | 1.67 ha | July, 2019 |
| Samples | Reads | 3% Cutoff | |||
|---|---|---|---|---|---|
| Effective | Normalized | OTUs | Chao1 | Shannon | |
| P. clarkii | |||||
| PNJ1w | 74,943 | 42,536 | 5748 | 24,684.75 | 8.04 |
| PNJ2w | 75,447 | 42,536 | 5504 | 24,172.09 | 8.21 |
| PXY1w | 64,800 | 42,536 | 5375 | 22,560.42 | 8.02 |
| PXY2w | 69,032 | 42,536 | 7041 | 32,291.63 | 8.88 |
| PXYR1w | 58,791 | 42,536 | 3172 | 12,715.01 | 6.96 |
| PXYR2w | 75,569 | 42,536 | 6045 | 24,304.46 | 7.35 |
| PNJ1s | 68,900 | 42,536 | 11,747 | 32,961.82 | 11.87 |
| PNJ2s | 67,133 | 42,536 | 13,008 | 39,020.42 | 12.06 |
| PXY1s | 59,419 | 42,536 | 17,639 | 107,074.68 | 12.51 |
| PXY2s | 74,825 | 42,536 | 9752 | 33,679.55 | 10.96 |
| PXYR1s | 74,483 | 42,536 | 10,655 | 27,083.08 | 11.51 |
| PXYR2s | 70,192 | 42,536 | 7799 | 22,096.71 | 9.26 |
| M. rosenbergii | |||||
| MGY1w | 104,391 | 42,536 | 4532 | 17,284.90 | 7.48 |
| MGY2w | 102,415 | 42,536 | 4016 | 16,575.88 | 6.34 |
| MGY3w | 94,063 | 42,536 | 4796 | 20,204.21 | 7.45 |
| MGYRw | 88,096 | 42,536 | 8496 | 36,354.26 | 9.73 |
| MGY1s | 98,335 | 42,536 | 8878 | 31,662.12 | 10.45 |
| MGY2s | 93,417 | 42,536 | 9771 | 31,961.08 | 10.95 |
| MGY3s | 101,897 | 42,536 | 9618 | 35,152.96 | 10.41 |
| MGYRs | 89,043 | 42,536 | 9744 | 25,425.28 | 11.13 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, H.; Ye, N.; Huang, K.; Yu, J.; Zhang, S. Mobile Genetic Elements Drive the Antibiotic Resistome Alteration in Freshwater Shrimp Aquaculture. Water 2021, 13, 1461. https://doi.org/10.3390/w13111461
Fang H, Ye N, Huang K, Yu J, Zhang S. Mobile Genetic Elements Drive the Antibiotic Resistome Alteration in Freshwater Shrimp Aquaculture. Water. 2021; 13(11):1461. https://doi.org/10.3390/w13111461
Chicago/Turabian StyleFang, Hao, Nan Ye, Kailong Huang, Junnan Yu, and Shuai Zhang. 2021. "Mobile Genetic Elements Drive the Antibiotic Resistome Alteration in Freshwater Shrimp Aquaculture" Water 13, no. 11: 1461. https://doi.org/10.3390/w13111461
APA StyleFang, H., Ye, N., Huang, K., Yu, J., & Zhang, S. (2021). Mobile Genetic Elements Drive the Antibiotic Resistome Alteration in Freshwater Shrimp Aquaculture. Water, 13(11), 1461. https://doi.org/10.3390/w13111461






