Comparison of Prokaryotic Communities Associated with Different TOC Concentrations in Dianchi Lake
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sites Description and Sampling Procedure
2.2. DNA Extraction and PCR Amplification
2.3. Illumina MiSeq Sequencing
2.4. Sequencing Data Processing
2.5. Detection and Visualization of Microbiological Interactions
2.6. Statistical Analysis
3. Results
3.1. Variability of Cardinal Environmental Factors of the Sediments
3.2. Prokaryotic Community Richness and Diversity
3.3. Prokaryotic Community Overall Variation in Sediments
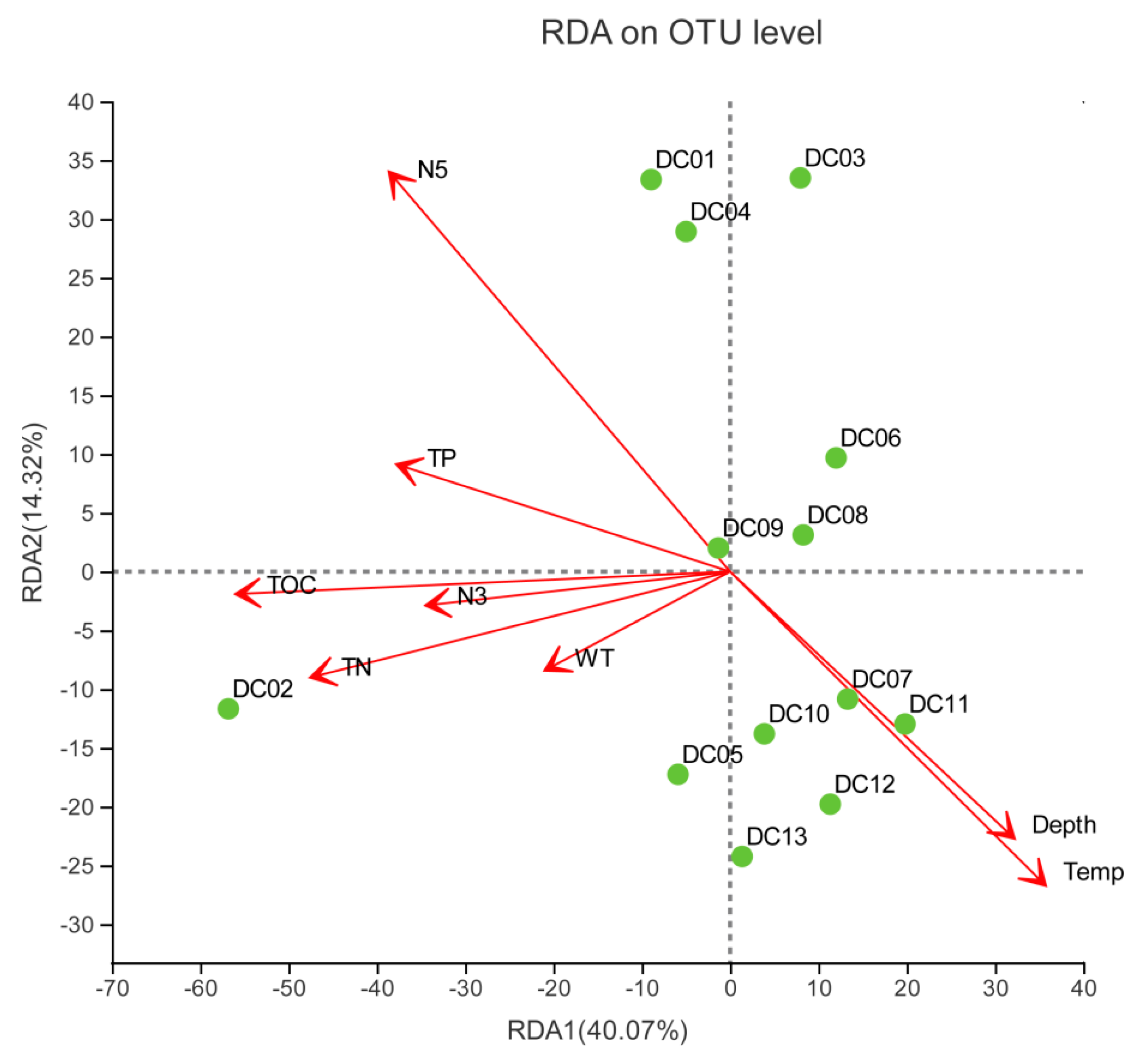
3.4. Linking Sediment Prokaryotic Communities to Environmental Factors
3.5. Prokaryotic β-Diversity among Samples
3.6. Diversity of the Prokaryotic Communities among Groups
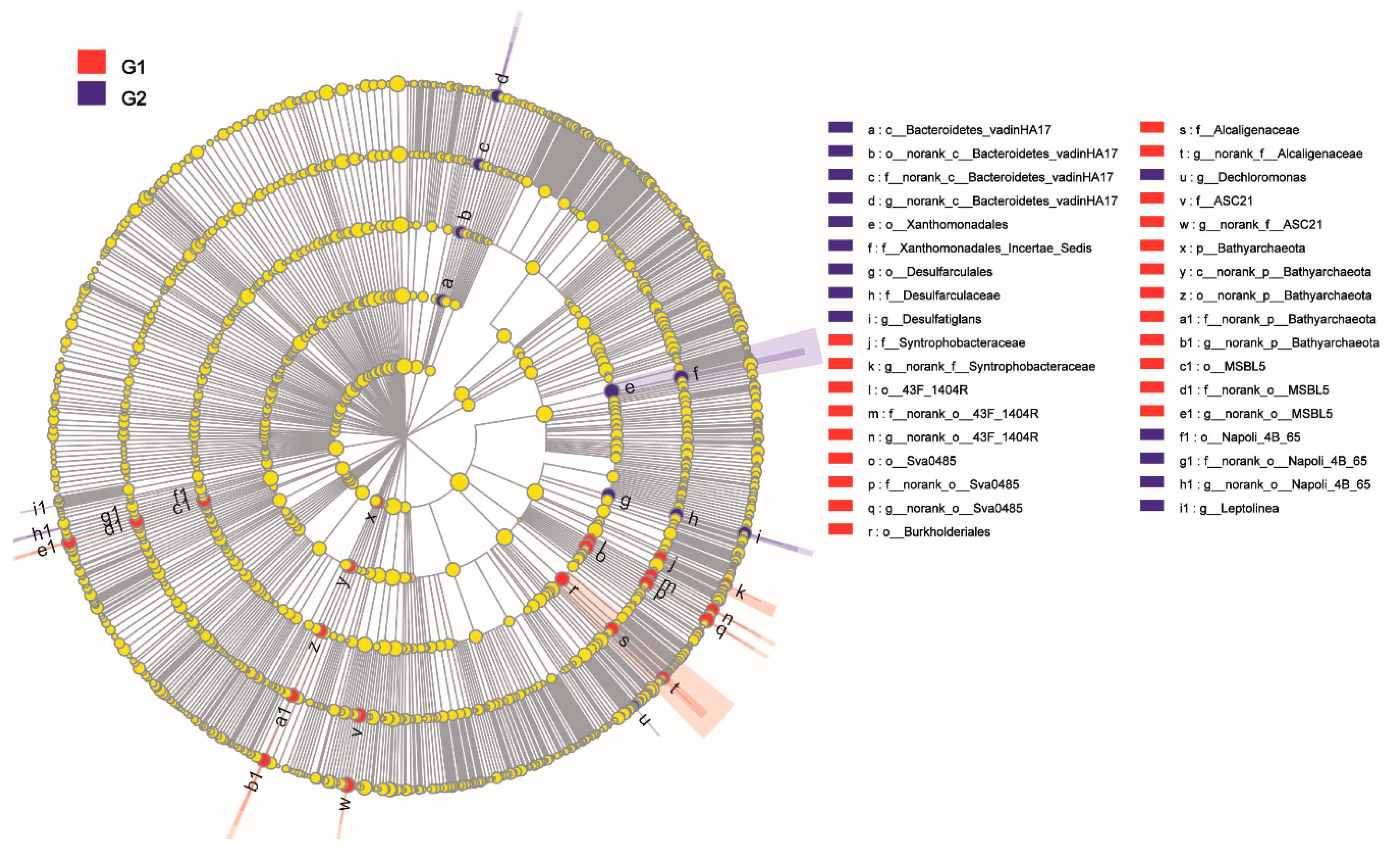
3.7. Prokaryotic Interaction Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Liu, F.H.; Lin, G.H.; Gao, G.; Qin, B.Q.; Zhang, J.S.; Zhao, G.P.; Zhou, Z.H.; Shen, J.H. Bacterial and archaeal assemblages in sediments of a large shallow freshwater lake, Lake Taihu, as revealed by denaturing gradient gel electrophoresis. J. Appl. Microbiol. 2009, 106, 1022–1032. [Google Scholar] [CrossRef] [PubMed]
- Jiao, N.; Herndl, G.J.; Hansell, D.A.; Benner, R.; Kattner, G.; Wilhelm, S.W.; Kirchman, D.L.; Weinbauer, M.G.; Luo, T.; Chen, F. Microbial production of recalcitrant dissolved organic matter: Long-term carbon storage in the global ocean. Nat. Rev. Microbiol. 2010, 8, 593. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Song, C.; Cao, X.; Zhou, Y. Shifts between ammonia-oxidizing bacteria and archaea in relation to nitrification potential across trophic gradients in two large Chinese lakes (Lake Taihu and Lake Chaohu). Water Res. 2013, 47, 2285–2296. [Google Scholar] [CrossRef] [PubMed]
- Kolmonen, E.; Haukka, K.; Rantala-Ylinen, A.; Rajaniemi-Wacklin, P.; Lepistö, L.; Sivonen, K. Bacterioplankton community composition in 67 Finnish lakes differs according to trophic status. Aquat. Microb. Ecol. 2011, 62, 241–250. [Google Scholar] [CrossRef]
- Bai, Y.; Shi, Q.; Wen, D.; Li, Z.; Jefferson, W.A.; Feng, C.; Tang, X. Bacterial communities in the sediments of Dianchi Lake, a partitioned eutrophic waterbody in China. PLoS ONE. 2012, 7, e37796. [Google Scholar] [CrossRef]
- Li, W.; Zhang, Y.; Wu, N.; Zhao, Z.; Xu, W.; Ma, Y.; Niu, Z. Colonization Characteristics of Bacterial Communities on Plastic Debris Influenced by Environmental Factors and Polymer Types in the Haihe Estuary of Bohai Bay, China. Environ. Sci. Technol. 2019, 53, 10763–10773. [Google Scholar] [CrossRef]
- Zhang, L.; Cheng, Y.; Gao, G.; Jiang, J. Spatial-Temporal Variation of Bacterial Communities in Sediments in Lake Chaohu, a Large, Shallow Eutrophic Lake in China. Int. J. Environ. Res. Public Health 2019, 16. [Google Scholar] [CrossRef]
- Zhang, J.X.; Yang, Y.Y.; Zhao, L.; Li, Y.Z.; Xie, S.G.; Liu, Y. Distribution of sediment bacterial and archaeal communities in plateau freshwater lakes. Appl. Microbiol. Biotechnol. 2015, 99, 3291–3302. [Google Scholar] [CrossRef]
- Swan, B.; Ehrhardt, C.; Reifel, K.; Moreno, L.; Valentine, D. Archaeal and Bacterial Communities Respond Differently to Environmental Gradients in Anoxic Sediments of a California Hypersaline Lake, the Salton Sea. Appl. Environ. Microbiol. 2010, 76, 757–768. [Google Scholar] [CrossRef]
- Sutcliffe, B.; Hose, G.C.; Harford, A.J.; Midgley, D.J.; Greenfield, P.; Paulsen, I.T.; Chariton, A.A. Microbial communities are sensitive indicators for freshwater sediment copper contamination. Environ. Pollut. 2019, 247, 1028–1038. [Google Scholar] [CrossRef]
- Yu, D.; Yang, Y.; Zhen, W.; Feng, Q.; Xie, S.; Yong, L. Spatiotemporal variation of planktonic and sediment bacterial assemblages in two plateau freshwater lakes at different trophic status. Appl. Microbiol. Biotechnol. 2016, 100, 4161–4175. [Google Scholar]
- Mora-Ruiz, M.D.R.; Cifuentes, A.; Font-Verdera, F.; Perez-Fernandez, C.; Farias, M.E.; Gonzalez, B.; Orfila, A.; Rossello-Mora, R. Biogeographical patterns of bacterial and archaeal communities from distant hypersaline environments. Syst. Appl. Microbiol. 2018, 41, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Liu, Y.; Lin, X.; Zhang, H.; Zeng, J.; Hou, J.; Yang, Y.; Yao, T.; Knight, R.; Chu, H. Geographic distance and pH drive bacterial distribution in alkaline lake sediments across Tibetan Plateau. Environ. Microbiol. 2012, 14, 2457–2466. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.J.; Pan, J.; Duan, C.H.; Wang, Y.M.; Liu, Y.; Sun, J.; Zhou, H.C.; Song, X.; Li, M. Prokaryotic Diversity in Mangrove Sediments across Southeastern China Fundamentally Differs from That in Other Biomes. mSystems 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Yang, J.S.; Qu, J.H.; Li, H.F.; Liu, W.J.; Li, B.Z.; Wang, E.T.; Yuan, H.L. Sediment prokaryote communities in different sites of eutrophic Lake Taihu and their interactions with environmental factors. World J. Microbiol. Biotechnol. 2015, 31, 883–896. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Liu, H.; Zhang, C.; Ding, K.; Chen, R.; Liu, S. Joint response of chemistry and functional microbial community to oxygenation of the reductive confined aquifer. Sci. Total Environ. 2020, 720, 137587. [Google Scholar] [CrossRef]
- Yang, C.; Wang, Q.; Simon, P.N.; Liu, J.; Liu, L.; Dai, X.; Zhang, X.; Kuang, J.; Igarashi, Y.; Pan, X.; et al. Distinct Network Interactions in Particle-Associated and Free-Living Bacterial Communities during a Microcystis aeruginosa Bloom in a Plateau Lake. Front. Microbiol. 2017, 8, 1202. [Google Scholar] [CrossRef]
- Zhang, M.; Lu, T.; Paerl, H.W.; Chen, Y.; Zhang, Z.; Zhou, Z.; Qian, H. Feedback Regulation between Aquatic Microorganisms and the Bloom-Forming Cyanobacterium Microcystis aeruginosa. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef]
- Li, Q.; Lin, F.; Yang, C.; Wang, J.; Lin, Y.; Shen, M.; Park, M.S.; Li, T.; Zhao, J. A Large-Scale Comparative Metagenomic Study Reveals the Functional Interactions in Six Bloom-Forming Microcystis-Epibiont Communities. Front. Microbiol. 2018, 9, 746. [Google Scholar] [CrossRef]
- Jeong, S.Y.; Choi, J.Y.; Kim, T.G. Coordinated Metacommunity Assembly and Spatial Distribution of Multiple Microbial Kingdoms within a Lake. Microb. Ecol. 2019. [Google Scholar] [CrossRef]
- Xue, M.; Guo, Z.; Gu, X.; Gao, H.; Weng, S.; Zhou, J.; Gu, D.; Lu, H.; Zhou, X. Rare rather than abundant microbial communities drive the effects of long-term greenhouse cultivation on ecosystem functions in subtropical agricultural soils. Sci. Total Environ. 2019, 706, 136004. [Google Scholar] [CrossRef] [PubMed]
- Duchinski, K.; Moyer, C.L.; Hager, K.; Fullerton, H. Fine-Scale Biogeography and the Inference of Ecological Interactions Among Neutrophilic Iron-Oxidizing Zetaproteobacteria as Determined by a Rule-Based Microbial Network. Front. Microbiol. 2019, 10, 2389. [Google Scholar] [CrossRef] [PubMed]
- Filippini, G.; Bugnot, A.B.; Johnston, E.L.; Ruszczyk, J.; Potts, J.; Scanes, P.; Ferguson, A.; Ostrowski, M.; Varkey, D.; Dafforn, K.A. Sediment bacterial communities associated with environmental factors in Intermittently Closed and Open Lakes and Lagoons (ICOLLs). Sci. Total Environ. 2019, 693, 133462. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.-Y.; Han, X.-X.; Huang, X.-H.; Wu, Y.-L.; Yang, H.; Huang, T.; Yu, Y.-H.; Huang, C.-C. Distribution Characteristics of n-alkanes in Sediment Core and Implication of Environment in Different Lakes of Dianchi. Huan Jing Ke Xue 2016, 37, 4605–4614. [Google Scholar] [PubMed]
- Yu, Y.; Lee, C.; Kim, J.; Hwang, S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 2005, 89, 670–679. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Yoav, B.; Krieger, A.M.; Daniel, Y. Adaptive linear step-up procedures that control the false discovery rate. Biometrika 2006, 3, 491–507. [Google Scholar]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the Third International AAAI Conference on Weblogs and Social Media, San Jose, CA, USA, 17–20 May 2009. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Zeng, J.; Yang, L.Y.; Liang, Y.; Li, J.Y.; Xiao, L.; Jiang, L.J.; Zhao, D.Y. Spatial distribution of bacterial communities in sediment of a eutrophic lake revealed by denaturing gradient gel electrophoresis and multivariate analysis. Can. J. Microbiol. 2008, 54, 1053. [Google Scholar] [CrossRef]
- Huang, W.; Chen, X.; Jiang, X.; Zheng, B. Characterization of sediment bacterial communities in plain lakes with different trophic statuses. Microbiologyopen 2017, 6, e00503. [Google Scholar] [CrossRef]
- Shao, K.; Gao, G.; Qin, B.; Tang, X.; Wang, Y.; Chi, K.; Dai, J. Comparing sediment bacterial communities in the macrophyte-dominated and algae-dominated areas of eutrophic Lake Taihu, China. Can. J. Microbiol. 2011, 57, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Zhang, J.; Wu, L.; Liu, Y.; Ge, G. Effects of Heavy Metal and Nutrients on Benthic Microbial Communities in Freshwater Sediment of Poyang Lake (China). J. Residuals Sci. Technol. 2015, 12, 105–111. [Google Scholar] [CrossRef]
- Xiong, W.; Xie, P.; Wang, S.; Niu, Y.; Yang, X.; Chen, W. Sources of organic matter affect depth-related microbial community composition in sediments of Lake Erhai, Southwest China. J. Limnol. 2015, 74, 310–323. [Google Scholar] [CrossRef]
- Laurence, H.; Mauro, T.; Jakob, Z.; Raffaele, P.; Walter, W.; John, P. Composition of bacterial and archaeal communities in freshwater sediments with different contamination levels (Lake Geneva, Switzerland). Water Res. 2011, 45, 1213–1228. [Google Scholar]
- Song, H.; Li, Z.; Du, B.; Wang, G.; Ding, Y. Bacterial communities in sediments of the shallow Lake Dongping in China. J. Appl. Microbiol. 2015, 112, 79–89. [Google Scholar] [CrossRef]
- Lenihan, H.S.; Peterson, C.H.; Kim, S.L.; Conlan, K.E.; Fairey, R.; McDonald, C.; Grabowski, J.H.; Oliver, J.S. Variation in marine benthic community composition allows discrimination of multiple stressors. Mar. Ecol. Prog. Ser. 2003, 261, 63–73. [Google Scholar] [CrossRef]
- Giovannoni, S.J.; Cameron Thrash, J.; Temperton, B. Implications of streamlining theory for microbial ecology. ISME J. 2014, 8, 1553–1565. [Google Scholar] [CrossRef]
- Monier, A.; Comte, J.; Babin, M.; Forest, A.; Matsuoka, A.; Lovejoy, C. Oceanographic structure drives the assembly processes of microbial eukaryotic communities. ISME J. 2015, 9, 990–1002. [Google Scholar] [CrossRef]
- Mas, A.; Jamshidi, S.; Lagadeuc, Y.; Eveillard, D.; Vandenkoornhuyse, P. Beyond the Black Queen Hypothesis. ISME J. 2016, 10, 2085–2091. [Google Scholar] [CrossRef]




| Samples | DC01 | DC02 | DC03 | DC04 | DC05 | DC06 | DC07 | DC08 | DC09 | DC10 | DC11 | DC12 | DC13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Longitude (E) | 102°39.934′ | 102°38.807′ | 102°38.529′ | 102°40.742′ | 102°39.852′ | 102°41.893′ | 102°40.419′ | 102°45.063′ | 102°42.58′ | 102°44.603′ | 102°36.932′ | 102°36.785′ | 102°39.823′ |
| Latitude (N) | 25°1.167′ | 25°0.304′ | 24°58.871′ | 24°57.154′ | 24°55.997′ | 24°53.333′ | 24°51.799′ | 24°49.445′ | 24°49.137′ | 24°48.057′ | 24°46.164′ | 24°41.733′ | 24°41.324′ |
| Depth (m) | 3 | 3 | 3.2 | 4.2 | 5 | 5.5 | 5.5 | 6 | 5.5 | 6 | 4.5 | 4 | 4 |
| Temp (°C) | 11 | 11 | 11 | 16.5 | 16 | 17 | 17 | 15.2 | 15 | 15.2 | 15 | 15.1 | 15.1 |
| WT (%) | 83.81 | 85.97 | 83.66 | 67.11 | 82.07 | 80.45 | 78.71 | 73.67 | 84.07 | 83.78 | 81.37 | 75.04 | 78.27 |
| TOC (g/kg) | 87.3 | 131.2 | 49.1 | 32.5 | 56.9 | 45 | 41.1 | 36.1 | 53.6 | 49 | 54.4 | 50.4 | 40.8 |
| TN (N, %) | 0.678 | 0.92 | 0.472 | 0.24 | 0.519 | 0.424 | 0.39 | 0.343 | 0.504 | 0.448 | 0.537 | 0.517 | 0.391 |
| TP (P, %) | 0.477 | 0.437 | 0.18 | 0.221 | 0.229 | 0.22 | 0.145 | 0.211 | 0.084 | 0.188 | 0.263 | 0.305 | 0.214 |
| NO3−-N (mg/kg) | 12.67 | 13.59 | 3.33 | 15.74 | 2.43 | 9.49 | 4.36 | 6.51 | 6.51 | 2.6 | 0.87 | 6.49 | 5.79 |
| NO2−-N (mg/kg) | 0.032 | 0.032 | 0.023 | 0.023 | 0.034 | 0.009 | 0.009 | 0.018 | 0.012 | 0.014 | 0.011 | 0.014 | 0.046 |
| Sample | DC01 | DC02 | DC03 | DC04 | DC05 | DC06 | DC07 | DC08 | DC09 | DC10 | DC11 | DC12 | DC13 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequences | 26,005 | 25,853 | 21,952 | 27,606 | 26,523 | 31,895 | 30,264 | 24,990 | 33,898 | 36,736 | 29,570 | 24,665 | 35,213 | 375,170 |
| No. of OTUs | 1557 | 1465 | 1424 | 1655 | 1443 | 1479 | 1366 | 1440 | 1679 | 1458 | 1280 | 1433 | 1136 | 3312 |
| Simpson | 0.009 | 0.036 | 0.012 | 0.008 | 0.013 | 0.01 | 0.013 | 0.011 | 0.008 | 0.014 | 0.02 | 0.017 | 0.016 | _ |
| Chao | 2042.274 | 1917.7 | 1849.58 | 2100.918 | 2041.724 | 2064.937 | 2048.141 | 1989.004 | 2319.198 | 2074.249 | 1856.914 | 1994.204 | 1704.119 | _ |
| ACE | 2084.332 | 2002.065 | 1919.384 | 2183.52 | 2108.839 | 2121.869 | 2074.313 | 2084.492 | 2377.044 | 2141.484 | 1947.488 | 2087.683 | 2105.406 | _ |
| Shannon | 5.889 | 5.421 | 5.608 | 5.991 | 5.508 | 5.659 | 5.532 | 5.6 | 6.01 | 5.532 | 5.253 | 5.512 | 5.122 | _ |
| Coverage | 0.966 | 0.967 | 0.969 | 0.964 | 0.964 | 0.963 | 0.963 | 0.964 | 0.96 | 0.962 | 0.966 | 0.964 | 0.969 | _ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.-P.; Li, Y.-P.; Huo, Q.-Q.; Xiao, W.; Duan, C.-Q.; Wang, Y.-X.; Cui, X.-L. Comparison of Prokaryotic Communities Associated with Different TOC Concentrations in Dianchi Lake. Water 2020, 12, 2557. https://doi.org/10.3390/w12092557
Li C-P, Li Y-P, Huo Q-Q, Xiao W, Duan C-Q, Wang Y-X, Cui X-L. Comparison of Prokaryotic Communities Associated with Different TOC Concentrations in Dianchi Lake. Water. 2020; 12(9):2557. https://doi.org/10.3390/w12092557
Chicago/Turabian StyleLi, Cheng-Peng, Ya-Ping Li, Qing-Qing Huo, Wei Xiao, Chang-Qun Duan, Yong-Xia Wang, and Xiao-Long Cui. 2020. "Comparison of Prokaryotic Communities Associated with Different TOC Concentrations in Dianchi Lake" Water 12, no. 9: 2557. https://doi.org/10.3390/w12092557
APA StyleLi, C.-P., Li, Y.-P., Huo, Q.-Q., Xiao, W., Duan, C.-Q., Wang, Y.-X., & Cui, X.-L. (2020). Comparison of Prokaryotic Communities Associated with Different TOC Concentrations in Dianchi Lake. Water, 12(9), 2557. https://doi.org/10.3390/w12092557






