Monitoring Waterborne Pathogens in Surface and Drinking Waters. Are Water Treatment Plants (WTPs) Simultaneously Efficient in the Elimination of Enteric Viruses and Fecal Indicator Bacteria (FIB)?
Abstract
1. Introduction
2. Materials and Methods
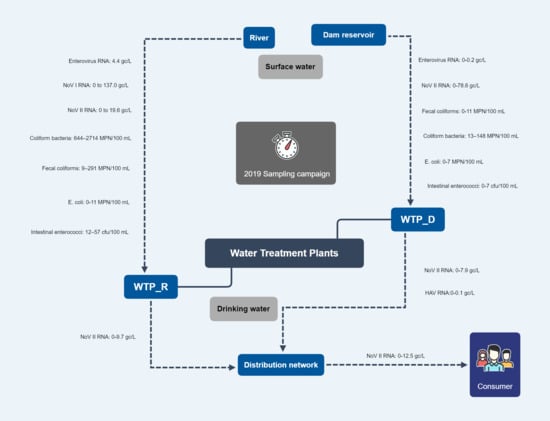
2.1. Sampling Sites of Surface Water and Drinking Water
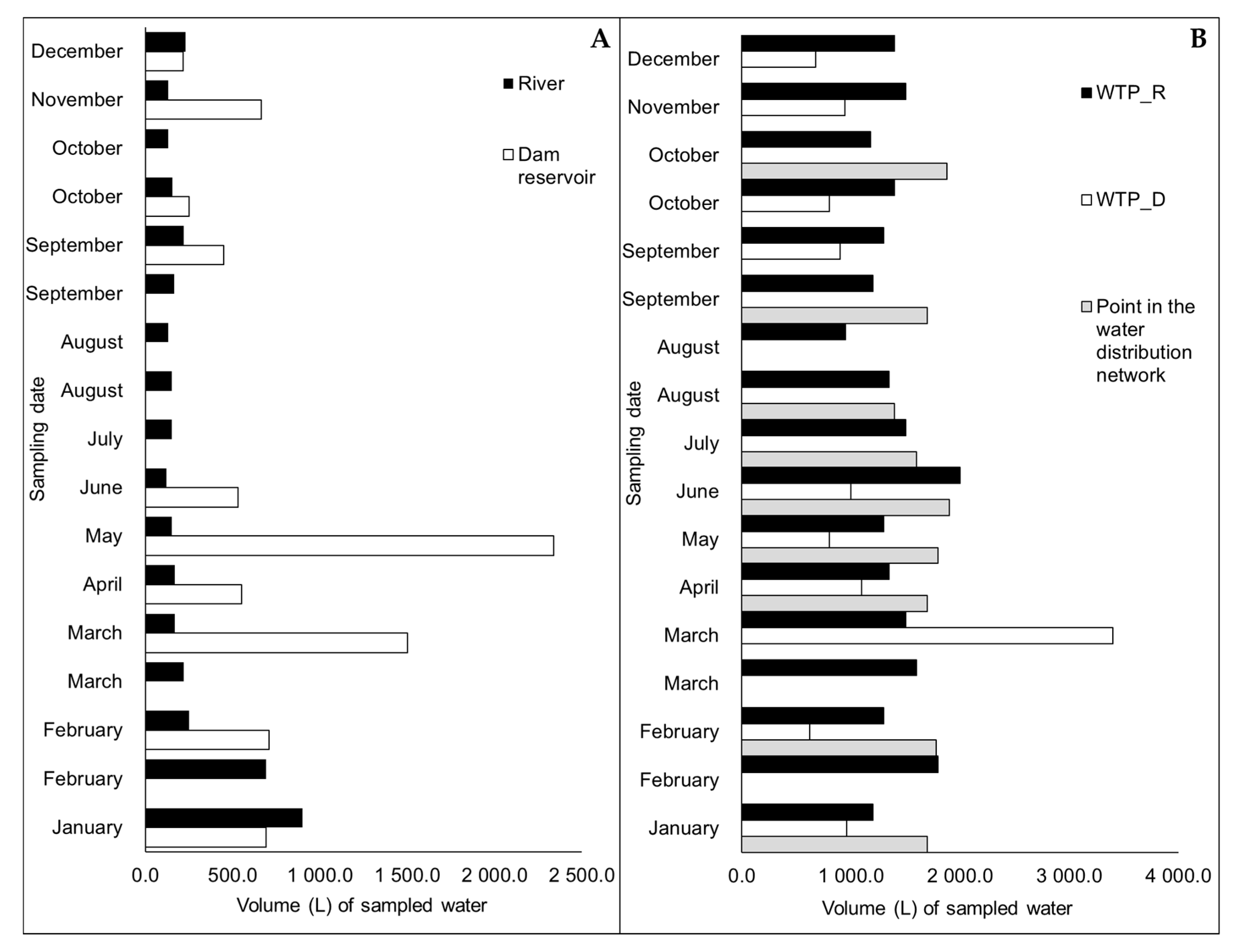
2.2. Sample Collection
2.3. Detection and Quantification of Enteric Viruses’ Genomic RNAs
2.4. Quantification of Fecal Indicator Bacteria (FIB)
2.4.1. Coliform Bacteria and E. Coli
2.4.2. Fecal Coliforms
2.4.3. Intestinal Enterococci
2.5. Determination of Physical-Chemical Parameters
2.6. Statistical Analyses
3. Results
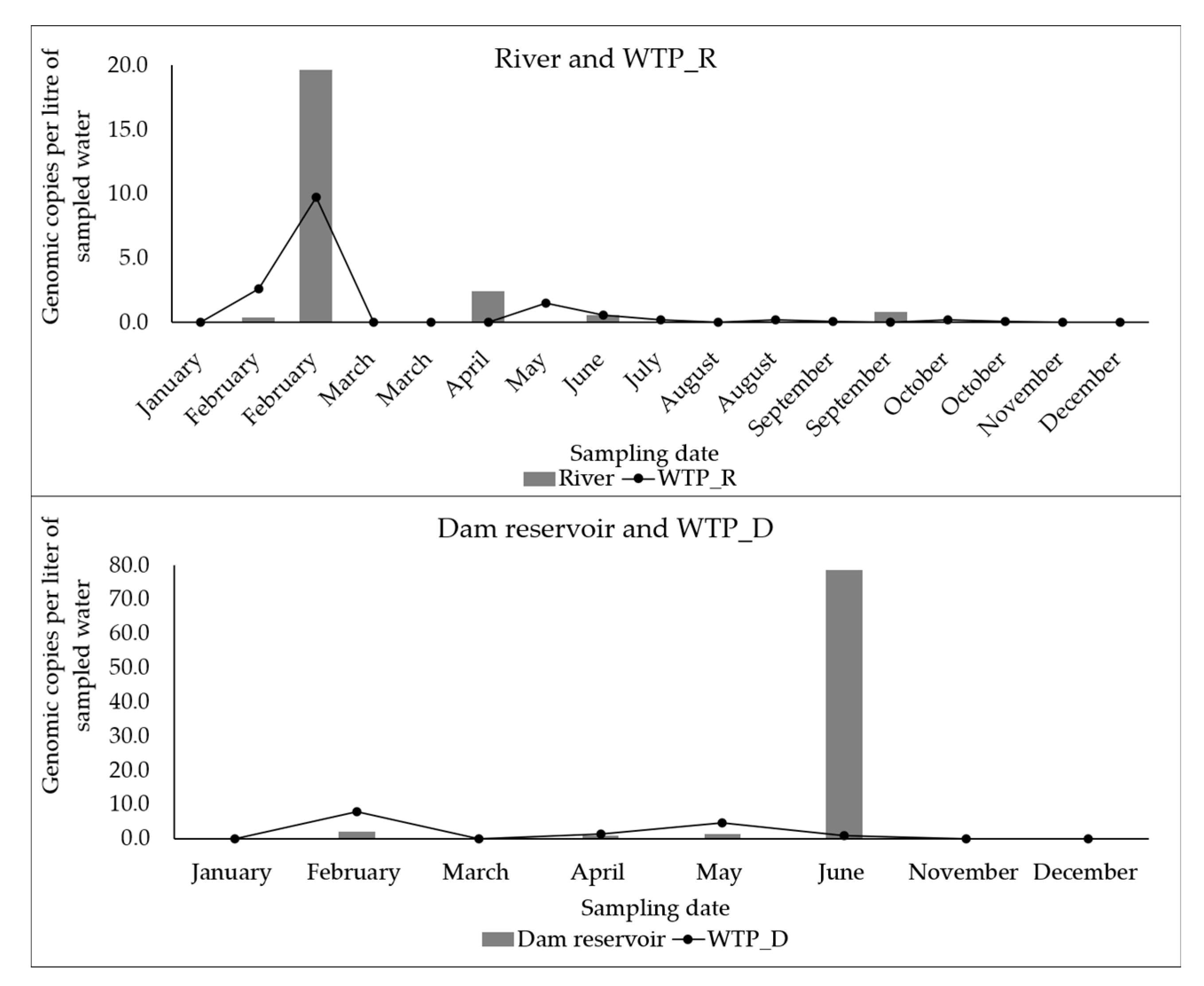
3.1. Detection and Quantification of Enteric Viruses
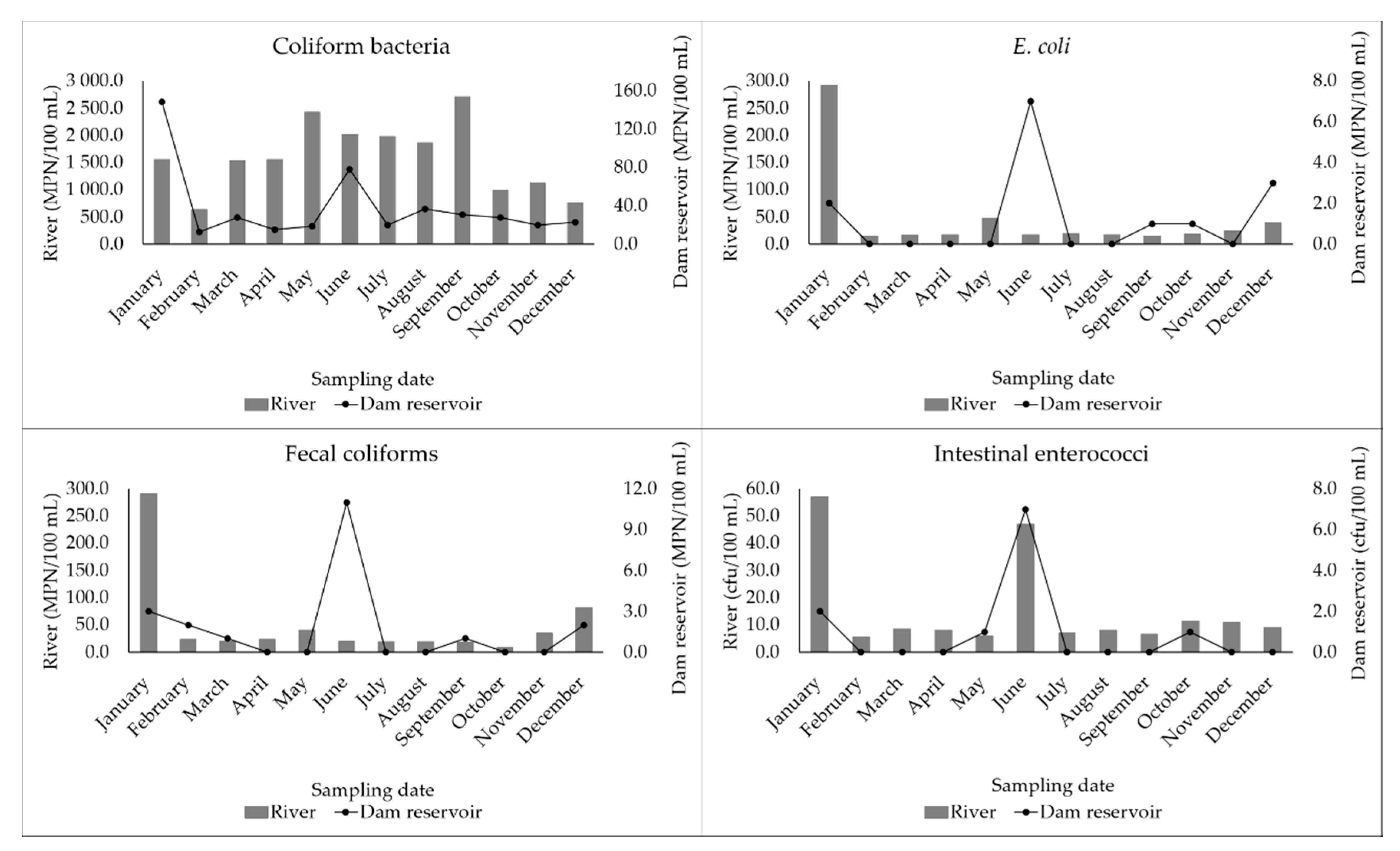
3.2. Fecal Indicator Bacteria (FIB) Quantified in Water Samples
3.3. Evaluation of the Effectiveness of the WTPs in the Reduction of Enteric Viruses and FIB
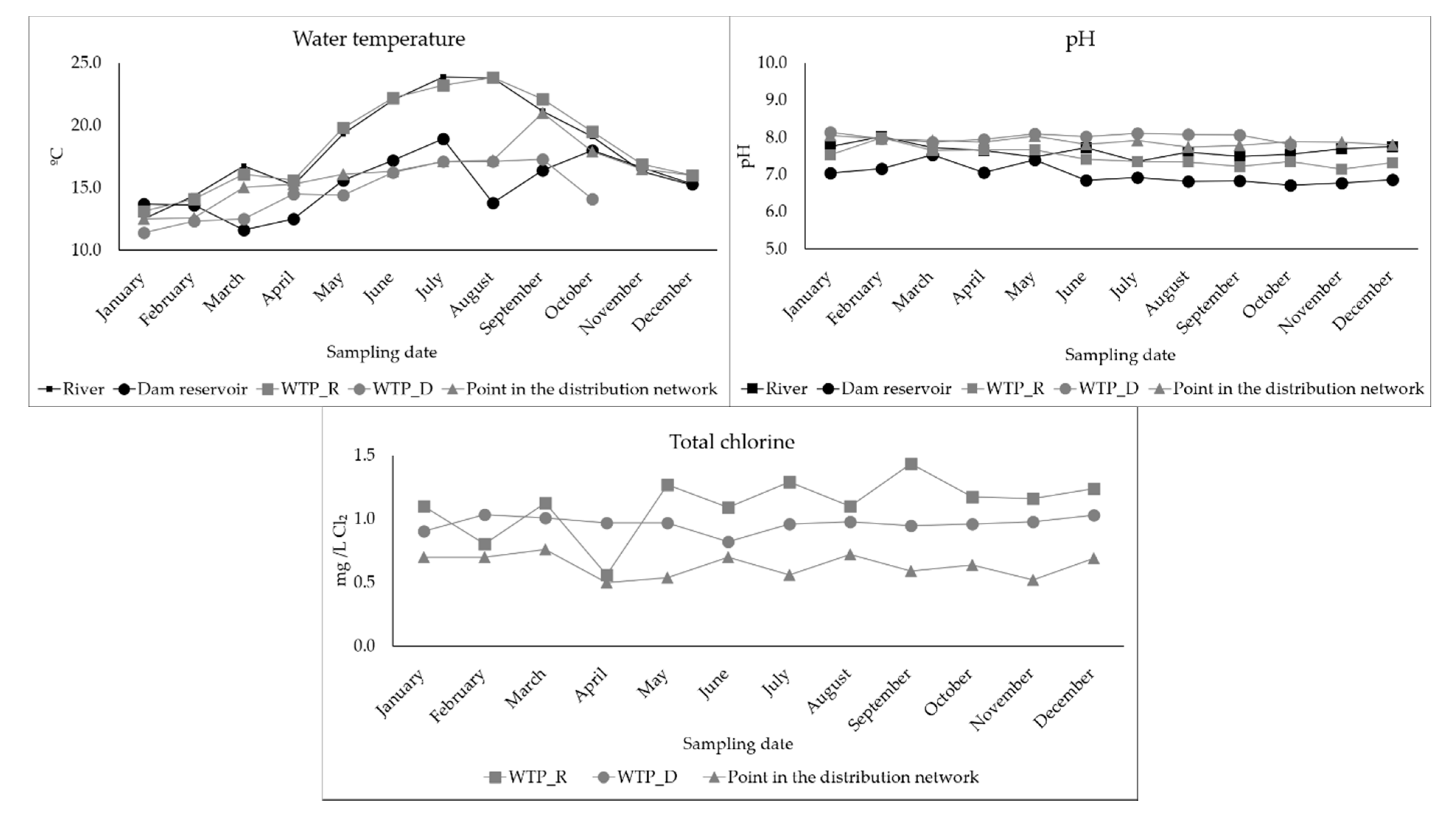
3.4. Determination of Physical-Chemical Parameters in the Waters Surveyed
3.5. Evaluation of Correlations between the Various Parameters under Study
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Drinking-Water. Available online: https://www.who.int/news-room/fact-sheets/detail/drinking-water (accessed on 23 May 2020).
- Macy, J.T.; Quick, R.E. Transmission and prevention of water-related diseases. In Water and Health; Grabow, W., Ed.; EOLSS Publications: Oxford, UK, 2009; Volume I, pp. 104–107. [Google Scholar]
- Bouseettine, R.; Hassou, N.; Bessi, H.; Ennaji, M. Waterborne Transmission of Enteric Viruses and Their Impact on Public Health. In Emerging and Reemerging Viral Pathogens; Academic Press: Cambridge, MA, USA, 2020; Volume 21, pp. 907–932. ISBN 9789896540821. [Google Scholar]
- Ashbolt, N.J. Microbial Contamination of Drinking Water and Human Health from Community Water Systems. Curr. Environ. Health Rep. 2015, 2, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Sibanda, T.; Okoh, A.I. Real-time PCR quantitative assessment of hepatitis A virus, rotaviruses and enteroviruses in the Tyume River located in the Eastern Cape Province, South Africa. Water SA 2013, 39, 295–304. [Google Scholar] [CrossRef]
- Eifan, S.A. Enteric Viruses And Aquatic Environment. Internet J. Microbiol. 2013, 12, 1–7. [Google Scholar]
- La Rosa, G.; Fratini, M.; della Libera, S.; Iaconelli, M.; Muscillo, M. Emerging and potentially emerging viruses in water environments. Ann. Ist. Super Sanità 2012, 48, 397–406. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Guidelines for Drinking-Water Quality, 4th ed.; WHO: Geneva, Switzerland, 2017; pp. 1–541. [Google Scholar]
- USEPA Final CCL 4 Microbial Contaminants. Available online: https://www.epa.gov/ccl/contaminant-candidate-list-4-ccl-4-0 (accessed on 15 July 2020).
- Teixeira, P.; Salvador, D.; Brandão, J.; Ahmed, W.; Sadowsky, M.J.; Valério, E. Environmental and adaptive changes necessitate a paradigm shift for indicators of fecal contamination. Microbiol. Spectr. 2020, 8. [Google Scholar] [CrossRef]
- ICTV Picornaviridae. Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/positive-sense-rna-viruses/picornavirales/w/picornaviridae (accessed on 25 May 2020).
- Holm-Hansen, C.C.; Midgley, S.E.; Fischer, T.K. Global emergence of enterovirus D68: A systematic review. Lancet Infect. Dis. 2016, 16, 64–75. [Google Scholar] [CrossRef]
- Palminha, P.; Ribeiro, C.; Roque, C.; Vinagre, E. Vigilância laboratorial da infeção a Enterovirus entre 2010 e 2013. Bol. Epidemiológico Obs. 2015, 5, 19–21. [Google Scholar]
- Sinclair, R.G.; Jones, E.L.; Gerba, C.P. Viruses in recreational water-borne disease outbreaks: A review. J. Appl. Microbiol. 2009, 107, 1769–1780. [Google Scholar] [CrossRef]
- Lee, C.S.; Lee, C.; Marion, J.; Wang, Q.; Saif, L.; Lee, J. Occurrence of human enteric viruses at freshwater beaches during swimming season and its link to water inflow. Sci. Total Environ. 2014, 472, 757–766. [Google Scholar] [CrossRef]
- Haramoto, E.; Katayama, H.; Oguma, K.; Ohgaki, S. Application of cation-coated filter method to detection of noroviruses, enteroviruses, adenoviruses, and torque teno viruses in the Tamagawa River in Japan. Appl. Environ. Microbiol. 2005, 71, 2403–2411. [Google Scholar] [CrossRef]
- Schets, F.M.; Van Wijnen, J.H.; Schijven, J.F.; Schoon, H.; De Roda Husman, A.M. Monitoring of waterborne pathogens in surface waters in Amsterdam, the Netherlands, and the potential health risk associated with exposure to Cryptosporidium and Giardia in these waters. Appl. Environ. Microbiol. 2008, 74, 2069–2078. [Google Scholar] [CrossRef] [PubMed]
- Updyke, E.A.; Wang, Z.; Sun, S.; Connell, C.; Kirs, M.; Wong, M.; Lu, Y. Human enteric viruses–potential indicators for enhanced monitoring of recreational water quality. Virol. Sin. 2015, 30, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Singh, A. Detection of human enteric viruses in Umgeni River, Durban, South Africa. J. Water Health 2015, 13, 1098–1112. [Google Scholar] [CrossRef] [PubMed]
- Prevost, B.; Goulet, M.; Lucas, F.S.; Joyeux, M.; Moulin, L.; Wurtzer, S. Viral persistence in surface and drinking water: Suitability of PCR pre-treatment with intercalating dyes. Water Res. 2016, 91, 68–76. [Google Scholar] [CrossRef]
- Peláez-Carvajal, D.; Guzmán, B.L.; Rodríguez, J.; Acero, F.; Nava, G. Presence of enteric viruses in water samples for consumption in Colombia: Challenges for supply systems. Biomedica 2016, 36, 169–172. [Google Scholar]
- ICTV Genus: Hepatovirus. Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/positive-sense-rna-viruses/picornavirales/w/picornaviridae/709/genus-hepatovirus (accessed on 1 June 2020).
- World Health Organization. Global Hepatitis Report, 2017; WHO: Geneva, Switzerland, 2017; pp. 1–68. [Google Scholar]
- DGS. Boletim Epidemiológico. In Hepatite A em Portugal; Direção-Geral da Saúde: Lisboa, Portugal, 2017. [Google Scholar]
- Jiang, S.C.; Chu, W. PCR detection of pathogenic viruses in southern California urban rivers. J. Appl. Microbiol. 2004, 97, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Elmahdy, E.M.; Fongaro, G.; Schissi, C.D.; Petrucio, M.M.; Barardi, C.R.M. Enteric viruses in surface water and sediment samples from the catchment area of Peri Lagoon, Santa Catarina State, Brazil. J. Water Health 2016, 14, 142–154. [Google Scholar] [CrossRef] [PubMed]
- López-Gálvez, F.; Truchado, P.; Sánchez, G.; Aznar, R.; Gil, M.I.; Allende, A. Occurrence of enteric viruses in reclaimed and surface irrigation water: Relationship with microbiological and physicochemical indicators. J. Appl. Microbiol. 2016, 121, 1180–1188. [Google Scholar] [CrossRef]
- O’Brien, E.; Nakyazze, J.; Wu, H.; Kiwanuka, N.; Cunningham, W.; Kaneene, J.B.; Xagoraraki, I. Viral diversity and abundance in polluted waters in Kampala, Uganda. Water Res. 2017, 127, 41–49. [Google Scholar] [CrossRef]
- ICTV Genus: Norovirus. Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/positive-sense-rna-viruses/w/caliciviridae/1167/genus-norovirus (accessed on 27 May 2020).
- Lopman, B.; Atmar, R.; Baric, R.; Estes, M.; Green, K.; Glass, R.; Hall, A.; Iturriza-Gomara, M.; Kang, C.; Lee, B.; et al. Global burden of norovirus and prospects for vaccine development. Cent. Dis. Control Prev. 2015, 1–46. [Google Scholar]
- Costa, I.; Mesquita, J.R.; Veiga, E.; Oleastro, M.; Nascimento, M.J.S. Surveillance of norovirus in Portugal and the emergence of the Sydney variant, 2011–2013. J. Clin. Virol. 2015, 70, 26–28. [Google Scholar] [CrossRef]
- Maunula, L.; Söderberg, K.; Vahtera, H.; Vuorilehto, V.P.; Von Bonsdorff, C.H.; Valtari, M.; Laakso, T.; Lahti, K. Presence of human noro- and adenoviruses in river and treated wastewater, a longitudinal study and method comparison. J. Water Health 2012, 10, 87–99. [Google Scholar] [CrossRef]
- Grøndahl-rosado, R.; Yarovitsyna, E.; Trettenes, E.; Myrmel, M.; Robertson, L.J. A One Year Study on the Concentrations of Norovirus and Enteric Adenoviruses in Wastewater and A Surface Drinking Water Source in Norway. Food Environ. Virol. 2014, 6, 232–245. [Google Scholar] [CrossRef]
- Lodder, W.J.; de Roda Husman, A.M. Presence of Noroviruses and Other Enteric Viruses in Sewage and Surface Waters in The Netherlands. Appl. Environ. Microbiol. 2005, 71, 1453–1461. [Google Scholar] [CrossRef]
- Kukkula, M.; Maunula, L.; Silvennoinen, E. Outbreak of Viral Gastroenteritis Due to Drinking Water Contaminated by Norwalk-like Viruses. J. Infect. Dis. 1999, 180, 1771–1776. [Google Scholar] [CrossRef]
- Blanco, A.; Guix, S.; Fuster, N.; Fuentes, C.; Bartolomé, R.; Cornejo, T.; Pintó, R.M.; Bosch, A. Norovirus in Bottled Water Associated with Gastroenteritis Outbreak, Spain, 2016. Emerg. Infect. Dis. 2017, 23, 1531–1534. [Google Scholar] [CrossRef]
- Gall, A.M.; Mariñas, B.J.; Lu, Y.; Shisler, J.L. Waterborne Viruses: A Barrier to Safe Drinking Water. PLoS Pathog. 2015, 11, 1–7. [Google Scholar] [CrossRef]
- Goyal, S.M.; Cannon, J.L. The Molecular Virology of Enteric Viruses. In Viruses in Foods; Springer: Berlin/Heidelberg, Germany, 2006; pp. 471–503. ISBN 978-3-319-30721-3. [Google Scholar]
- Rzezutka, A.; Cook, N. Survival of human enteric viruses in the environment and food. FEMS Microbiol. Rev. 2004, 28, 441–453. [Google Scholar] [CrossRef]
- Guerrero-Latorre, L.; Gonzales-Gustavson, E.; Hundesa, A.; Sommer, R.; Rosina, G. UV disinfection and flocculation-chlorination sachets to reduce hepatitis E virus in drinking water. Int. J. Hyg. Environ. Health 2016, 219, 405–411. [Google Scholar] [CrossRef]
- Yeh, H.Y.; Hwang, Y.C.; Yates, M.V.; Mulchandani, A.; Chen, W. Detection of hepatitis A virus by using a combined cell culture-molecular beacon assay. Appl. Environ. Microbiol. 2008, 74, 2239–2243. [Google Scholar] [CrossRef][Green Version]
- República Portuguesa. Decreto-Lei n. 152/2017; Diário da República: Lisboa, Portugal, 2017.
- Espinosa, A.C.; Arias, C.F.; Snchez-Colán, S.; Mazari-Hiriart, M. Comparative study of enteric viruses, coliphages and indicator bacteria for evaluating water quality in a tropical high-altitude system. Environ. Health A Glob. Access Sci. Source 2009, 8, 1–10. [Google Scholar] [CrossRef]
- Teixeira, P.; Costa, S.; Brown, B.; Silva, S.; Rodrigues, R.; Valério, E. Quantitative PCR detection of enteric viruses in wastewater and environmental water sources by the Lisbon municipality: A case study. Water 2020, 12, 544. [Google Scholar] [CrossRef]
- Zhu, H.; Yuan, F.; Yuan, Z.; Liu, R.; Xie, F.; Huang, L.; Liu, X.; Jiang, X.; Wang, J.; Xu, Q.; et al. Monitoring of Poyang lake water for sewage contamination using human enteric viruses as an indicator. Virol. J. 2018, 15, 1–9. [Google Scholar] [CrossRef]
- SNIRH Recursos Hídricos. Available online: https://snirh.apambiente.pt/ (accessed on 20 May 2020).
- APA Planos de Ordenamento de Albufeiras. Available online: https://apambiente.pt/index.php?ref=16&subref=7 (accessed on 20 July 2020).
- Salvador, D.; Neto, C.; Benoliel, M.J.; Filomena Caeiro, M. Assessment of the presence of hepatitis e virus in surface water and drinking water in Portugal. Microorganisms 2020, 8, 761. [Google Scholar] [CrossRef]
- Ikner, L.A.; Gerba, C.P.; Bright, K.R. Concentration and Recovery of Viruses from Water: A Comprehensive Review. Food Environ. Virol. 2012, 4, 41–67. [Google Scholar] [CrossRef]
- Fout, G.S.; Brinkman, N.E.; Cashdollar, J.L.; Griffin, M.; McMinn, B.R.; Rhodes, E.R.; Varughese, E.A.; Karim, M.R.; Grimm, A.C.; Spencer, S.K.; et al. Method 1615—Measurement of Enterovirus and Norovirus Occurrence in Water by Culture and RT-Qpcr; U.S. Environmental Protection Agency: Cincinnati, OH, USA, 2014; pp. 1–105.
- ISO. ISO 9308-2:2012. Available online: https://www.iso.org/standard/52246.html (accessed on 15 May 2020).
- ISO. ISO 9308-1:2014. Available online: https://www.iso.org/standard/55832.html (accessed on 18 May 2020).
- ISO. ISO 7899-2:2000. Available online: https://www.iso.org/standard/14854.html (accessed on 18 May 2020).
- Standard Methods for the Examination of Water and Wastewater. Chlorine (Residual). Available online: https://www.standardmethods.org/doi/abs/10.2105/SMWW.2882.078 (accessed on 1 June 2020).
- Standard Methods for the Examination of Water and Wastewater. pH Value. Available online: https://www.standardmethods.org/doi/abs/10.2105/SMWW.2882.082 (accessed on 1 June 2020).
- Prevost, B.; Lucas, F.S.; Goncalves, A.; Richard, F.; Moulin, L.; Wurtzer, S. Large scale survey of enteric viruses in river and waste water underlines the health status of the local population. Environ. Int. 2015, 79, 42–50. [Google Scholar] [CrossRef]
- Pang, X.; Qiu, Y.; Gao, T.; Zurawell, R.; Neumann, N.F.; Craik, S.; Lee, B.E. Prevalence, levels and seasonal variations of human enteric viruses in six major rivers in Alberta, Canada. Water Res. 2019, 153, 349–356. [Google Scholar] [CrossRef]
- Elmahdy, M.E.I.; Fongaro, G.; Magri, M.E.; Petruccio, M.M.; Barardi, C.R.M. Spatial distribution of enteric viruses and somatic coliphages in a Lagoon used as drinking water source and recreation in Southern Brazil. Int. J. Hyg. Environ. Health 2016, 219, 617–625. [Google Scholar] [CrossRef]
- Pordata Valor da Produção Agrícola: Total e Por Tipo. Available online: https://www.pordata.pt/Municipios/Valor+da+produção+agrícola+total+e+por+tipo-956 (accessed on 26 March 2020).
- Tandukar, S.; Sherchand, J.B.; Id, D.B.; Sherchan, S.P.; Malla, B.; Shrestha, R.G.; Haramoto, E. Presence of Human Enteric Viruses, Protozoa, and Indicators of Pathogens in the Bagmati River, Nepal. Pathogens 2018, 7, 38. [Google Scholar] [CrossRef]
- Megchún-García, J.V.; Landeros-Sánchez, C.; Soto-Estrada, A.; del Refugio Castañeda-Chávez, M.; Martínez-Dávila, J.P.; Nikolskii-Gavrilov, I.; Galaviz-Villa, I.; Lango-Reynoso, F. Total Coliforms and Escherichia coli in Surface and Subsurface Water from a Sugarcane Agroecosystem in Veracruz, Mexico. J. Agric. Sci. 2015, 7, 110–119. [Google Scholar] [CrossRef]
- Onyango, A.E.; Okoth, M.W.; Kunyanga, C.N.; Aliwa, B.O. Microbiological Quality and Contamination Level of Water Sources in Isiolo County in Kenya. J. Environ. Public Health 2018, 2018, 1–10. [Google Scholar] [CrossRef]
- Olasoji, S.O.; Oyewole, N.O.; Abiola, B.; Edokpayi, J.N. Water quality assessment of surface and groundwater sources using a water quality index method: A case study of a peri-urban town in southwest, Nigeria. Environments 2019, 6, 23. [Google Scholar] [CrossRef]
- Yuan, T. A consecutive centrifugal method for concentration of human enteric viruses in water samples. Arch. Virol. 2016, 161, 3323–3330. [Google Scholar] [CrossRef]
- Haramoto, E.; Kitajima, M.; Hata, A.; Torrey, J.R.; Masago, Y.; Sano, D.; Katayama, H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018, 135, 168–186. [Google Scholar] [CrossRef]
- Da Silva, A.K.; Le Saux, J.C.; Parnaudeau, S.; Pommepuy, M.; Elimelech, M.; Le Guyader, F.S. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: Different behaviors of genogroups I and II. Appl. Environ. Microbiol. 2007, 73, 7891–7897. [Google Scholar] [CrossRef]
- Wurtzer, S.; Prevost, B.; Lucas, F.S.; Moulin, L. Detection of enterovirus in environmental waters: A new optimized method compared to commercial real-time RT-qPCR kits. J. Virol. Methods 2014, 209, 47–54. [Google Scholar] [CrossRef]
- Gregory, J.B.; Litaker, R.W.; Noble, R.T. Rapid one-step quantitative reverse transcriptase PCR assay with competitive internal positive control for detection of enteroviruses in environmental samples. Appl. Environ. Microbiol. 2006, 72, 3960–3967. [Google Scholar] [CrossRef]
- Fout, G.S.; Cashdollar, J.L.; Varughese, E.A.; Parshionikar, S.U.; Grimm, A.C. EPA Method 1615. Measurement of Enterovirus and Norovirus Occurrence in Water by Culture and RT-qPCR. I. Collection of Virus Samples. J. Vis. Exp. 2016, 107, 1–13. [Google Scholar] [CrossRef]
- Seo, M.; Lee, H.; Kim, Y. Relationship between Coliform bacteria and water quality factors at weir stations in the Nakdong River, South Korea. Water 2019, 11, 1171. [Google Scholar] [CrossRef]
- Addo, M.G.; Woods, C.A.; Andoh, L.A.; Obiri-Danso, K. Effect of varying storage temperatures on the microbiological quality of street sold water in the Kumasi metropolis, Ghana. Res. J. Microbiol. 2016, 11, 56–63. [Google Scholar] [CrossRef]
| Step | Time | Temperature | Number of Cycles | |
|---|---|---|---|---|
| Enterovirus | Reverse Transcription | 10 min | 55 °C | 1 |
| Enzyme activation | 2 min | 95 °C | 1 | |
| Denaturation | 10 seg | 95 °C | 50 | |
| Data collection | 1 min | 60 °C | ||
| NoV (NoV I and NoV II) and HAV | Reverse Transcription | 10 min | 45 °C | 1 |
| Enzyme activation | 10 min | 95 °C | 1 | |
| Denaturation | 15 seg | 95 °C | 45 | |
| Data collection | 45 seg | 60 °C |
| Sampling | River | Dam Reservoir | WTP_R | WTP_D | Point in the Distribution Network | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Entero | NoV I | NoV II | HAV | Entero | NoV I | NoV II | HAV | Entero | NoV I | NoV II | HAV | Entero | NoV I | NoV II | HAV | Entero | NoV I | NoV II | HAV | |
| gc/L | gc/L | gc/L | gc/L | gc/L | ||||||||||||||||
| January | ND | ND | ND | ND | 0.2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 12.5 | ND |
| February (A) | ND | ND | 0.4 | ND | - | - | - | - | ND | ND | 2.6 | ND | - | - | - | - | - | - | - | - |
| February (B) | ND | 3.6 | 19.6 | ND | ND | ND | 2.0 | ND | ND | ND | 9.7 | ND | ND | ND | 7.9 | 0.1 | ND | ND | ND | ND |
| March (A) | 4.4 | 0.2 | ND | ND | - | - | - | - | ND | ND | ND | ND | - | - | - | - | - | - | - | - |
| March (B) | ND | 5.3 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | - | - | - | - |
| April | ND | 10.6 | 2.4 | ND | ND | ND | 0.8 | ND | ND | ND | ND | ND | ND | ND | 1.4 | ND | ND | ND | ND | ND |
| May | ND | ND | ND | ND | ND | ND | 1.4 | ND | ND | ND | 1.5 | ND | ND | ND | 4.5 | ND | ND | ND | 4.6 | ND |
| June | ND | ND | 0.5 | ND | ND | ND | 78.6 | ND | ND | ND | 0.6 | ND | ND | ND | 0.9 | ND | ND | ND | 0.1 | ND |
| July | ND | ND | ND | ND | - | - | - | - | ND | ND | 0.2 | ND | - | - | - | - | ND | ND | 0.7 | ND |
| August (A) | ND | ND | ND | ND | - | - | - | - | ND | ND | ND | ND | - | - | - | - | ND | ND | 0.2 | ND |
| August (B) | ND | ND | ND | ND | - | - | - | - | ND | ND | 0.2 | ND | - | - | - | - | - | - | - | - |
| September (A) | ND | 0.4 | ND | ND | - | - | - | - | ND | ND | 0.1 | ND | - | - | - | - | ND | ND | 0.1 | ND |
| September (B) | ND | 2.0 | 0.8 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 0.9 | ND | - | - | - | - |
| October (A) | ND | 137.0 | ND | ND | - | - | - | - | ND | ND | 0.2 | ND | - | - | - | - | - | - | - | - |
| October (B) | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 0.1 | ND | ND | ND | ND | ND | ND | ND | 0.1 | ND |
| November | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | - | - | - | - |
| December | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | - | - | - | - |
| Entero | NoV I | NoV II | HAV | Coliform Bacteria | Fecal Coliforms | E. coli | Intestinal Enterococci | Temp | pH | Total Chlorine | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Entero | 1 | 0.2 | −0.16 | −0.02 | 0.14 | 0.01 | 0.1 | 0.17 | −0.1 | −0.09 | −0.19 |
| NoV I | 0.2 | 1 | 0.03 | −0.05 | −0.01 | −0.1 | −0.16 | 0.48 ** | 0.1 | −0.11 | −0.08 |
| NoV II | −0.16 | 0.03 | 1 | 0.2 | −0.08 | −0.05 | −0.1 | −0.15 | −0.12 | 0.17 | 0.06 |
| HAV | −0.02 | −0.05 | 0.2 | 1 | −0.11 | −0.09 | −0.09 | −0.09 | −0.2 | 0.14 | 0.2 |
| Coliform bacteria | 0.14 | −0.01 | −0.08 | −0.11 | 1 | 0.92 *** | 0.92 *** | −0.04 | 0.30 * | −0.45 ** | −0.85 ** |
| Fecal coliforms | 0.01 | −0.1 | −0.05 | −0.09 | 0.92 *** | 1 | 0.94 *** | −0.17 | 0.16 | −0.39 * | −0.76 ** |
| E. coli | 0.1 | −0.16 | −0.1 | −0.09 | 0.92 *** | 0.94 *** | 1 | −0.16 | 0.22 | −0.41 ** | −0.73 ** |
| Intestinal enterococci | 0.17 | 0.48 ** | −0.15 | −0.09 | −0.04 | −0.17 | −0.16 | 1 | 0.19 | −0.16 | −0.16 |
| Temp | −0.1 | 0.1 | −0.12 | −0.2 | 0.30 * | 0.16 | 0.22 | 0.19 | 1 | −0.34 * | −0.29 |
| pH | −0.09 | −0.11 | 0.17 | 0.14 | −0.45 ** | −0.39 * | −0.41 ** | −0.16 | −0.34 * | 1 | 0.36 * |
| Total chlorine | −0.19 | −0.08 | 0.06 | 0.2 | −0.85 ** | −0.76 ** | −0.73 ** | −0.16 | −0.29 | 0.36 * | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salvador, D.; Caeiro, M.F.; Serejo, F.; Nogueira, P.; Carneiro, R.N.; Neto, C. Monitoring Waterborne Pathogens in Surface and Drinking Waters. Are Water Treatment Plants (WTPs) Simultaneously Efficient in the Elimination of Enteric Viruses and Fecal Indicator Bacteria (FIB)? Water 2020, 12, 2824. https://doi.org/10.3390/w12102824
Salvador D, Caeiro MF, Serejo F, Nogueira P, Carneiro RN, Neto C. Monitoring Waterborne Pathogens in Surface and Drinking Waters. Are Water Treatment Plants (WTPs) Simultaneously Efficient in the Elimination of Enteric Viruses and Fecal Indicator Bacteria (FIB)? Water. 2020; 12(10):2824. https://doi.org/10.3390/w12102824
Chicago/Turabian StyleSalvador, Daniel, Maria Filomena Caeiro, Fátima Serejo, Paulo Nogueira, Rui Neves Carneiro, and Célia Neto. 2020. "Monitoring Waterborne Pathogens in Surface and Drinking Waters. Are Water Treatment Plants (WTPs) Simultaneously Efficient in the Elimination of Enteric Viruses and Fecal Indicator Bacteria (FIB)?" Water 12, no. 10: 2824. https://doi.org/10.3390/w12102824
APA StyleSalvador, D., Caeiro, M. F., Serejo, F., Nogueira, P., Carneiro, R. N., & Neto, C. (2020). Monitoring Waterborne Pathogens in Surface and Drinking Waters. Are Water Treatment Plants (WTPs) Simultaneously Efficient in the Elimination of Enteric Viruses and Fecal Indicator Bacteria (FIB)? Water, 12(10), 2824. https://doi.org/10.3390/w12102824