Microbial Source-Tracking Reveals Origins of Fecal Contamination in a Recovering Watershed
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Sample Collection, Storage, and Processing
2.3. Physical–Chemical Parameters
2.4. Microbial Source-Tracking qPCR Assays
2.5. qPCR Quality Assurance and Quality Control
2.6. Statistical Methods
3. Results
3.1. Physical–Chemical Parameters
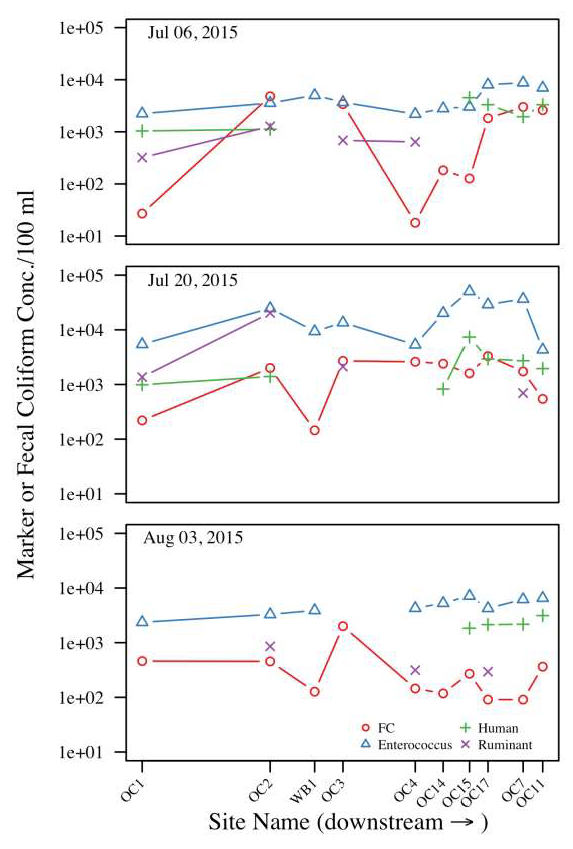
3.2. Fecal Indicator Bacteria Concentrations
3.3. Molecular Marker Concentrations
3.4. Associations Between General Fecal Indicators and MST Markers
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. Animal Waste, Water Quality, and Human Health; Dufour, A.P., Bos, R., Bartram, J., Eds.; IWA Publishing: London, UK, 2012. [Google Scholar]
- Paerl, H. Nutrient and other environmental controls of harmful cyanobacterial blooms along the freshwater-marine continuum. Adv. Exp. Med. Biol. 2008, 619, 217–237. [Google Scholar] [CrossRef] [PubMed]
- Onondaga Environmental Institute. Phase 1 Microbial Trackdown Study; A Report Prepared for NYSDEC and the Onondaga Lake Partnership; Onondaga Environmental Institute: Syracuse, NY, USA, 2013. [Google Scholar]
- Geldreich, E.E. Bacterial populations and indicator concepts in feces, sewage, stormwater and solid wastes. In Indicators of Viruses in Water and Food; Berg, G., Ed.; Ann Arbor Science: Ann Arbor, MI, USA, 1978. [Google Scholar]
- Silkie, S.S.; Nelson, K.L. Concentrations of host-specific and generic fecal markers measured by quantitative PCR in raw sewage and fresh animal feces. Water Res. 2009, 43, 4860–4871. [Google Scholar] [CrossRef] [PubMed]
- Bernhard, A.E.; Field, K.G. A PCR Assay To Discriminate Human and Ruminant Feces on the Basis of Host Differences in Bacteroides-Prevotella Genes Encoding 16S rRNA. Appl. Environ. Microbiol. 2000, 66, 4571–4574. [Google Scholar] [CrossRef] [PubMed]
- Reischer, G.H.; Kasper, D.C.; Steinborn, R.; Farnleitner, A.H.; Mach, R.L. A quantitative real-time PCR assay for the highly sensitive and specific detection of human faecal influence in spring water from a large alpine catchment area. Lett. Appl. Microbiol. 2007, 44, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Yampara-Iquise, H.; Zheng, G.; Jones, J.E.; Carson, C.A. Use of a Bacteroides thetaiotaomicron-specific alpha-1-6, mannanase quantitative PCR to detect human faecal pollution in water. J. Appl. Microbiol. 2008, 105, 1686–1693. [Google Scholar] [CrossRef] [PubMed]
- Stachler, E.; Kelty, C.; Sivaganesan, M.; Li, X.; Bibby, K.; Shanks, O.C. Quantitative CrAssphage PCR Assays for Human Fecal Pollution Measurement. Environ. Sci. Technol. 2017, 51, 9146–9154. [Google Scholar] [CrossRef] [PubMed]
- Green, H.C.; Haugland, R.A.; Varma, M.; Millen, H.T.; Borchardt, M.A.; Field, K.G.; Walters, W.A.; Knight, R.; Kelty, C.; Shanks, O. Improved HF183 quantitative real-time PCR assay for characterization of human fecal pollution in ambient surface water samples. Appl. Environ. Microbiol. 2014, 80, 3086–3094. [Google Scholar] [CrossRef] [PubMed]
- Reischer, G.H.; Kasper, D.C.; Steinborn, R.; Mach, R.L.; Farnleitner, A.H. Quantitative PCR method for sensitive detection of ruminant fecal pollution in freshwater and evaluation of this method in alpine karstic regions. Appl. Environ. Microbiol. 2006, 72, 5610–5614. [Google Scholar] [CrossRef] [PubMed]
- Mieszkin, S.; Yala, J.F.; Joubrel, R.; Gourmelon, M. Phylogenetic analysis of Bacteroidales 16S rRNA gene sequences from human and animal effluents and assessment of ruminant faecal pollution by real-time PCR. J. Appl. Microbiol. 2010, 108, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Santo Domingo, J.W.; Lamendella, R.; Edge, T.; Hill, S. Phylogenetic diversity and molecular detection of bacteria in gull feces. Appl. Environ. Microbiol. 2008, 74, 3969–3976. [Google Scholar] [CrossRef] [PubMed]
- Green, H.C.; Dick, L.K.; Gilpin, B.; Samadpour, M.; Field, K.G. Genetic markers for rapid PCR-based identification of gull, Canada goose, duck, and chicken fecal contamination in water. Appl. Environ. Microbiol. 2012, 78, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Green, H.C.; White, K.M.; Kelty, C.A.; Shanks, O.C. Development of rapid canine fecal source identification PCR-based assays. Environ. Sci. Technol. 2014, 48, 11453–11461. [Google Scholar] [CrossRef] [PubMed]
- Kildare, B.J.; Leutenegger, C.M.; McSwain, B.S.; Bambic, D.G.; Rajal, V.B.; Wuertz, S. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: A Bayesian approach. Water Res. 2007, 41, 3701–3715. [Google Scholar] [CrossRef] [PubMed]
- Coon, W.F.; Reddy, J.E. Hydrologic and Water-Quality Characterization and Modeling of the Onondaga Lake Basin, Onondaga County, New York; U.S. Geological Survey: Reston, VA, USA, 2008; p. 85.
- Rhea, J.R.; Russell, K.T.; Moran, E.; Glaser, D.; Ku, W.; Mastriano, J. Impacts of Advanced Tertiary Treatment on the Nitrogen Cycling of a Hypereutrophic Lake: A Case for 303(d) Delisting; Water Environment Foundation: Alexandria, VA, USA, 2006. [Google Scholar]
- Onondaga County Dept. of Water Environment Protection. Onondaga, NY ACJ Fourth Stipulation 2018 Annual Report; Onondaga County Dept. of Water Environment Protection: Syracuse, NY, USA, 2019; pp. 1–25. [Google Scholar]
- American Public Health Association; American Water Works Association; Water Environment Federation. Standard Methods for the Examination of Water and Wastewater, 23rd ed.; Baird, R., Eaton, A., Rice, E.W., Eds.; American Public Health Association: Washington, DC, USA, 2017. [Google Scholar]
- Ludwig, W.; Schleifer, K.H. How quantitative is quantitative PCR with respect to cell counts? Syst. Appl. Microbiol. 2000, 24, 556–562. [Google Scholar] [CrossRef]
- USEPA. Method 1611.1: Enterococci in Water by TaqMan® Quantitative Polymerase Chain Reaction (qPCR); USEPA: Washington, DC, USA, 2015.
- Green, H.C.; Field, K.G. Sensitive detection of sample interference in environmental qPCR. Water Res. 2012, 46, 3251–3260. [Google Scholar] [CrossRef] [PubMed]
- Tichopad, A.; Bar, T.; Pecen, L.; Kitchen, R.R.; Kubista, M.; Pfaffl, M.W. Quality control for quantitative PCR based on amplification compatibility test. Methods 2010, 50, 308–312. [Google Scholar] [CrossRef] [PubMed]
- R Core Team R. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Cheng, J.; Karambelkar, B.; Xie, Y. Leaflet: Create Interactive Web Maps with the JavaScript ‘Leaflet’ Library; R Package Version; Leaflet: London, UK, 2018. [Google Scholar]
- Bates, D.; Mäechler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Onondaga Environmental Institute. Phase 3 Microbial Trackdown Study; A Report Prepared for NYSDEC and the Onondaga Lake Partnership; Onondaga Environmental Institute: Syracuse, NY, USA, 2019. [Google Scholar]
- Onondaga Environmental Institute. Phase 2 Microbial Trackdown Study; A Report Prepared for NYSDEC and the Onondaga Lake Partnership; Onondaga Environmental Institute: Syracuse, NY, USA, 2015. [Google Scholar]
- Kappel, W.M. The Hydrogeology of the Tully Valley, Onondaga County, New York: An. Overview of Research, 1992–2012; 2014-1076; US Geological Survey: Reston, VA, USA, 2014; p. 28.
- Dillon, J. Improving Estimates of White-Tailed Deer Abundance Under Sub-Optimal Survey Conditions in Syracuse, New York; State University of New York: Syracuse, NY, USA, 2019. [Google Scholar]
- Onondaga Environmental Institute. OEI Analyses of CSO Capture and Pathogens in Onondaga Lake, and Recommendation for OEI Investigation of Pathogen Releases to Onondaga Lake Tributaries; Grannis, A.P., Pirro, N., Sage, S., Eds.; Onondaga Environmental Institute: Syracuse, NY, USA, 2007. [Google Scholar]
- Green, H.C.; Shanks, O.C.; Sivaganesan, M.; Haugland, R.A.; Field, K.G. Differential decay of human faecal Bacteroides in marine and freshwater. Environ. Microbiol. 2011, 13, 3235–3249. [Google Scholar] [CrossRef] [PubMed]

| Assay Name | Source Target | Primer Conc. (nM) | Probe Conc. (nM) | Reference |
|---|---|---|---|---|
| HF183 | Human | 1000 | 80 | [10] |
| Rum2Bac | Ruminant | 300 | 100 | [12] |
| DG3 | Canine | 1400 | 100 | [15] |
| Entero1 | Enterococcus | 1000 | 100 | [21,22] |
| Geometric Mean (Min.–Max.) 2 | Mean (Min.–Max.) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Site | Lat. | Land Use Class. | Class 1 | Fecal Coliforms (CFUs/100 mL) | Enterol1 (markers/100 mL) | HF183 (markers/100 mL) | Rum2Bac (markers/100 mL) | Conductivity (µS/cm) | Dissolved Oxygen (mg/L) | pH | Salinity (ppt) | Turbidity (NTUs) | Water Temperature (°C) |
| OC1 | 42.82 | Rural | C | 140 (27–460) | 3061 (2235–5437) | NA (990–1039) | NA (320–1366) | 996 (507–1484) | 10.2 (9.6–10.8) | 8.23 (8.18–8.28) | 0.5 (0.25–0.75) | 29.9 (8.2–51.6) | 18.3 (17.4–19.2) |
| OC2 | 42.90 | Rural | C | 1633 (454–4800) | 6611 (3286–24694) | NA (1118–1399) | 2807 (855–20326) | 1202 (951–1618) | 8.9 (8–9.5) | 8.13 (8.1–8.16) | 0.6 (0.47–0.82) | 50.2 (40.3–66.3) | 19.1 (15.9–21.6) |
| WB1 | 42.93 | Rural | C(T) | NA (127–145) | 5679 (3895–9363) | – | – | 2671 (645–6663) | 8.6 (8.2–8.9) | 8.06 (8.0–8.1) | 0.32 (0.31–0.34) | 25.3 (12.7–34.5) | 21.3 (19.3–22.6) |
| OC3 | 42.94 | Rural | C | 2638 (2000–3400) | NA (3687–13545) | – | NA (684–2123) | 933 (828–1144) | 8.7 (8.1–9.2) | 8.1 (8.06–8.12) | 0.46 (0.41–0.57) | 59 (25.6–110.4) | 20.2 (18.1–21.4) |
| OC4 | 42.99 | Urban | B | 189 (18–2600) | 3682 (2192–5327) | – | NA (314–640) | 1010 (848–1132) | 9.3 (8.5–9.9) | 8.12 (8.11–8.13) | 0.5 (0.42–0.56) | 29.7 (10.7–51.2) | 20.1 (18.5–21) |
| OC14 | 43.00 | Urban | B | 372 (118–2400) | 6689 (2807–20209) | NA (825–825) | – | 1030 (860–1174) | 8.9 (8.1–9.5) | 8.14 (8.13–8.16) | 0.51 (0.42–0.59) | 29.8 (8.4–55.2) | 19.9 (18.2–20.9) |
| OC15 | 43.07 | Urban | B | 380 (127–1600) | 10283 (3014–50474) | 3942 (1839–7364) | – | 1134 (954–1301) | 9.1 (7.9–9.7) | 7.94 (7.88–7.99) | 0.56 (0.47–0.65) | 34.9 (5.7–62.9) | 18.7 (17.4–19.9) |
| OC17 | 43.03 | Urban | B | 818 (91–3300) | 9970 (4249–29040) | 2748 (2141–3313) | NA (294–294) | 1194 (1026–1389) | 8.9 (7.8–9.7) | 7.93 (7.87–7.96) | 0.6 (0.51–0.7) | 36.2 (5.5–82.6) | 17.9 (16.5–19.1) |
| OC7 | 43.05 | Urban | C | 779 (91–3000) | 12571 (6199–36643) | 2259 (1949–2724) | NA (696–696) | 1225 (1050–1459) | 8.6 (7.8–9.4) | 7.9 (7.86–7.92) | 0.61 (0.52–0.74) | 42.9 (5.3–93.6) | 17.9 (16.4–19.4) |
| OC11 | 43.06 | Urban | C | 802 (364–2600) | 5832 (4335–6998) | 2728 (1955–3316) | – | 3013 (2200–4189) | 8.5 (7.8–9.3) | 7.84 (7.75–7.9) | 1.58 (1.13–2.24) | 18.3 (5.1–27.8) | 17.9 (16.3–19.6) |
| Outcome | Effect Estimate (95% Confidence Interval; p-Value) 1 | |||
|---|---|---|---|---|
| Latitude 2 | Land Use 3 | Sampling Visit 4 | ||
| 7/20/15 | 8/3/15 | |||
| Log10 Conductivity (µS/cm) | 1.16 (−0.14, 2.47; 0.082) | 0.08 (−0.12, 0.28; 0.407) | −0.12 (−0.31, 0.07; 0.211) | 0.03 (−0.16, 0.22; 0.759) |
| Dissolved Oxygen (mg/L) | −0.25 (−0.42, −0.07; 0.008) | −0.29 (−1.01, 0.42; 0.421) | 1.29 (1.00, 1.58; <0.001) | 1.10 (0.82, 1.38; <0.001) |
| pH | −1.46 (−2.11, −0.81; <0.001) | −0.15 (−0.29, −0.02; 0.030) | −0.02 (−0.6, 0.02; 0.362) | −0.04 (−0.08, 0.00; 0.033) |
| Water Temperature (°C) | −2.92 (0−14.82, 8.98; 0.631) | −0.79 (−2.42, −2.36; 0.341) | −3.04 (−3.73, −2.37; <0.001) | −0.76 (−142, −0.11; 0.024) |
| Log10 Salinity (ppt) | 1.51 (0.20, 2.81; 0.024) | 0.18 (−0.02, 0.38; 0.083) | −0.03 (−0.10, 0.04; 0.394) | 0.14 (0.07, 0.21; <0.001) |
| log10(Turbidity) (NTU) | −1.24 (−2.90, 0.42; 0.144) | −0.21 (−0.43, 0.01; 0.065) | −0.22 (−0.50, 0.05; 0.112) | −0.62 (−0.88, −0.35; <0.001) |
| Log10 Entero1/100 mL | 2.19 (−0.32, 4.70; 0.087) | 0.36 (0.02, 0.70; 0.041) | −0.55 (−0.96 −0.14; 0.009) | −0.73 (−1.14, −0.32; <0.001) |
| Log10 Fecal Coliforms/100 mL | 2.06 (−2.67, 6.78;0.394) | 0.08 (−0.62, 0.79; 0.823) | −0.56 (−1.12 −0.01; 0.048) | −0.69 (−1.24, −0.13; 0.016) |
| Human Marker Detection | − 5 | 55.98 (32.54, 79.41; <0.001) | −1.91 (−7.04, 3.23; 0.467) | −5.91 (−20.04, 8.22; p = 0.413) |
| Log10 HF183/100 mL | 2.17 (0.38, 3.96; 0.018) | 0.37 (0.04, 0.69; 0.030) | −0.01 (−0.18, 0.17; 0.947) | −0.12 (−0.32, 0.09; 0.271) |
| Ruminant Marker Detection | − 5 | −1.92 (−4.27, 0.43; 0.110) | 0.00 (−2.12, 2.12; 1.000) | −0.61 (−2.81, 1.59; 0.587) |
| Log10 Rum2Bac/100 mL | −1.35 (−6.00, 3.29; 0.569) | −0.28 (−0.95, 0.39; 0.416) | −0.72 (−1.05, −0.38; <0.001) | −1.01 (−1.43, −0.59; <0.001) |
| Outcome | Effect Estimate (95% Confidence Interval; p-Value) 2 | ||||
|---|---|---|---|---|---|
| Log10 Enterol 1 Concentration | Log10 Human Marker Concentration | Human Marker Detection | Log10 Ruminant Marker Concentration | Ruminant Marker Detection | |
| Log10 Fecal Coliform Concentration | 0.03 (−0.52, 0.59; 0.913) | 0.13 (−0.19, 0.45; 0.428) | 0.21 (−0.40, 0.82;0.503) | −0.23 (−0.58, 0.12; 0.197) | −0.42 (−0.97, 0.12; 0.127) |
| Log10 Entero1 Levels Concentration | − | 0.20 (0.02, 0.4; 0.03) | 0.35 (0.00, 0.70; 0.05) | 0.08 (−0.19, 0.32; 0.51) | 0.10 (−0.28, 0.48; 0.62) |
| Log10 Ruminant Marker Concentration | − | −0.02 (−0.323 0.29; 0.91) | −0.02 (−0.56, 0.53; 0.95) | − | − |
| Log10 Human Marker Concentration | − | − | − | − | −0.11 (−0.36, 0.14;0.38) |
| Human Marker Detection | − | − | − | − | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Green, H.; Weller, D.; Johnson, S.; Michalenko, E. Microbial Source-Tracking Reveals Origins of Fecal Contamination in a Recovering Watershed. Water 2019, 11, 2162. https://doi.org/10.3390/w11102162
Green H, Weller D, Johnson S, Michalenko E. Microbial Source-Tracking Reveals Origins of Fecal Contamination in a Recovering Watershed. Water. 2019; 11(10):2162. https://doi.org/10.3390/w11102162
Chicago/Turabian StyleGreen, Hyatt, Daniel Weller, Stephanie Johnson, and Edward Michalenko. 2019. "Microbial Source-Tracking Reveals Origins of Fecal Contamination in a Recovering Watershed" Water 11, no. 10: 2162. https://doi.org/10.3390/w11102162
APA StyleGreen, H., Weller, D., Johnson, S., & Michalenko, E. (2019). Microbial Source-Tracking Reveals Origins of Fecal Contamination in a Recovering Watershed. Water, 11(10), 2162. https://doi.org/10.3390/w11102162




