Identification of Human and Animal Fecal Contamination in Drinking Water Sources in the Kathmandu Valley, Nepal, Using Host-Associated Bacteroidales Quantitative PCR Assays
Abstract
1. Introduction
2. Materials and Methods
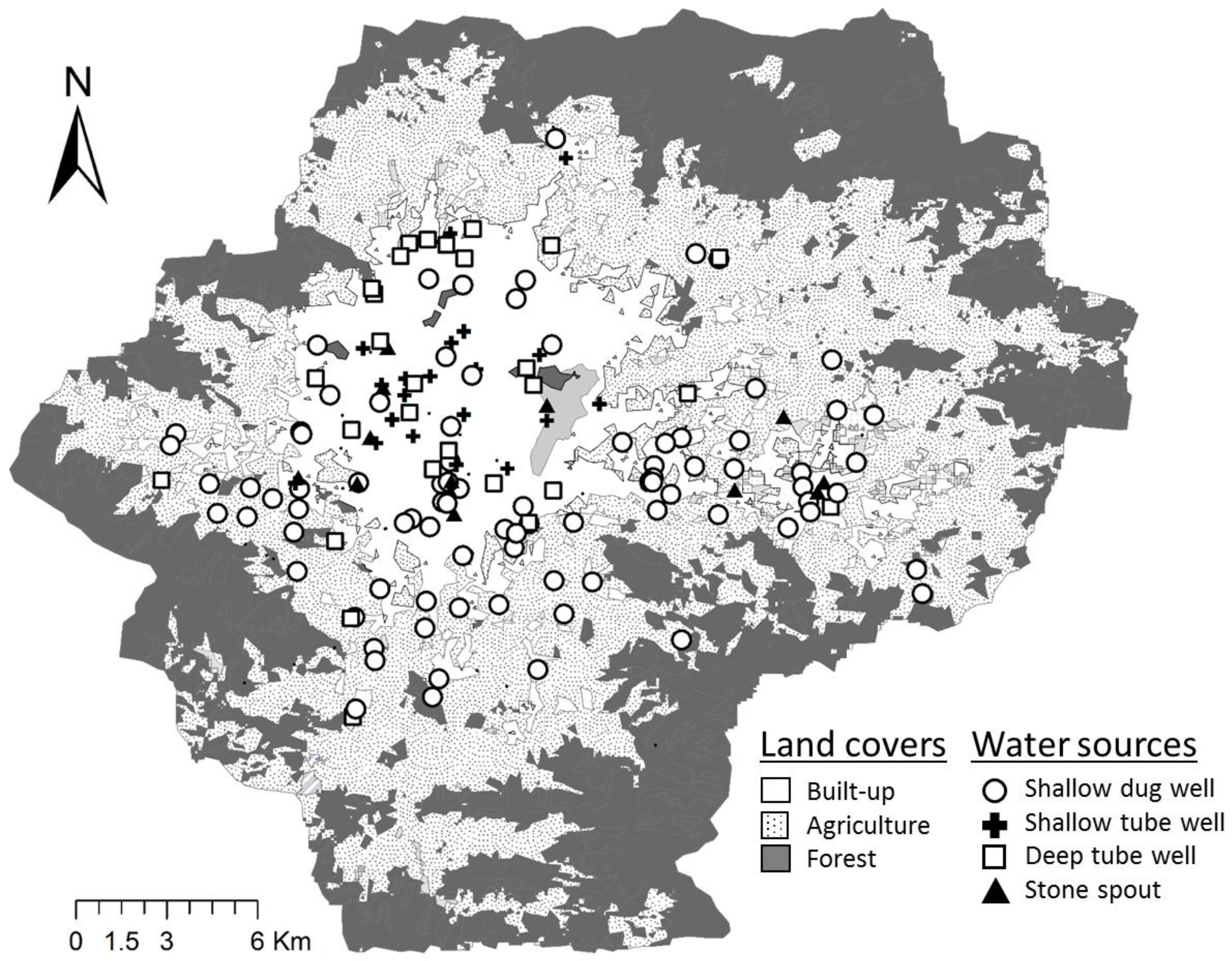
2.1. Study Site Description
2.2. Collection of Water Samples
2.3. Detection of E. coli and Total Coliforms
2.4. Bacterial DNA Extraction
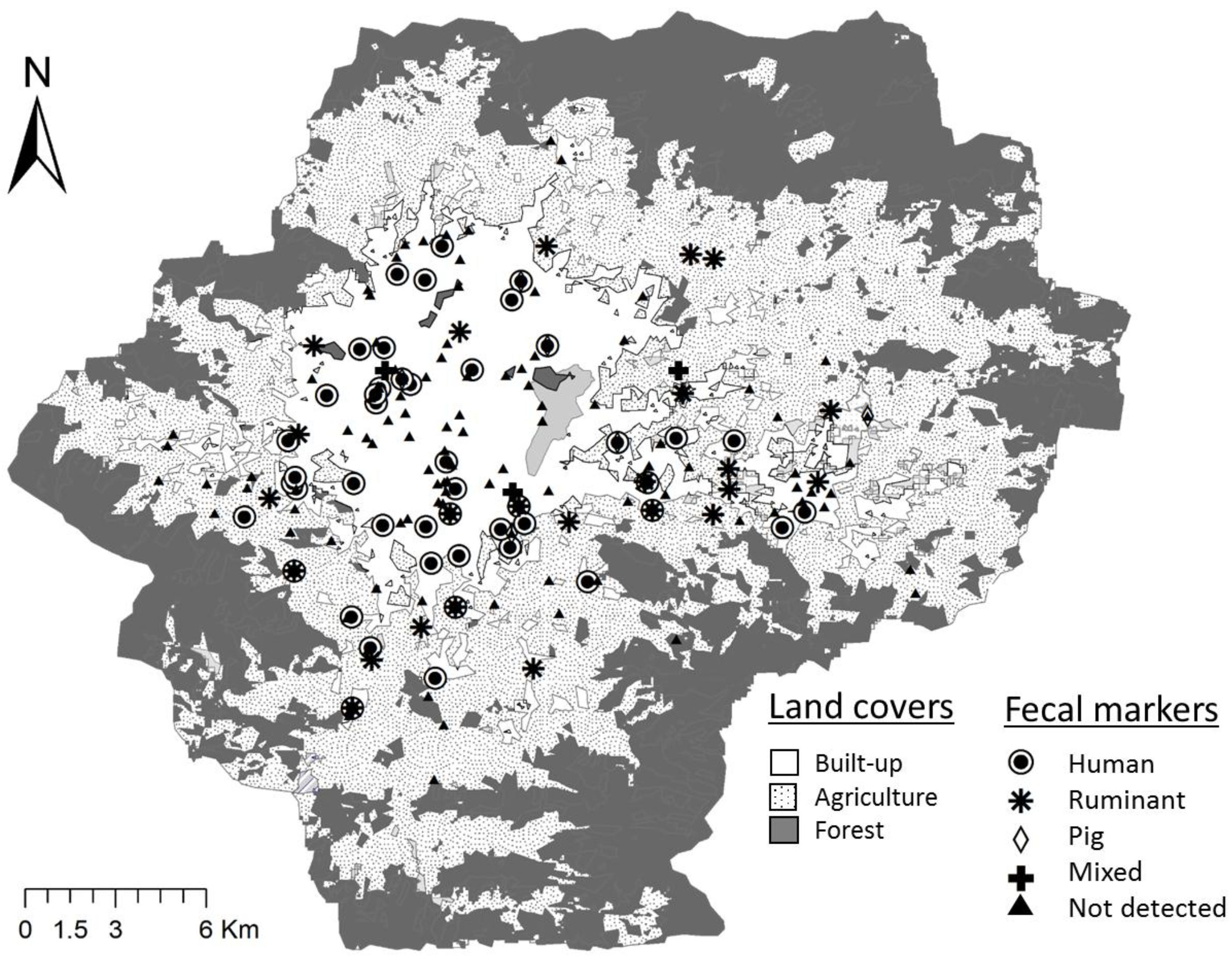
2.5. Application of the Selected Assays to the Fecal-Source Identification
2.6. Statistical Analysis
3. Results
3.1. Detection of FIB and Host-Associated Bacteroidales Genetic Markers
3.2. Comparison of FIB and Fecal Markers between Water Source Types
3.3. Comparison of FIB and Fecal Markers between Land Covers
3.4. Relationship between E. coli and Fecal Markers
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Beaudeau, P.; De Valk, H.; Vaillant, V.; Mannschott, C.; Tillier, C.; Mouly, D.; Ledrans, M. Lessons learned from ten investigations of waterborne gastroenteritis outbreaks, France, 1998–2006. J. Water Health 2008, 6, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Morris, B.L.; Lawrence, A.R.L.; Chilton, P.J.C.; Adams, B.; Calow, R.C.; Klinck, B.A. Groundwater and Its Susceptibility to Degradation: A Global Assessment of the Problem and Options for Management; Early warning and assessment report series, 03-3; United Nations Environment Programme: Nairobi, Kenya, 2003. [Google Scholar]
- Uy, D.; Haka, S.; Huya, C.; Srey, M.; Chunhieng, T.; Phoeurng, S.; Nasir, H.M.; Fredricks, D. Comparison of Tube-Well and Dug-Well Groundwater in the Arsenic Polluted Areas in Cambodia; Fukusi, K., Kurisu, F., Oguma, K., Furumai, H., Fontanos, P., Eds.; Southeast Asian Water Environment 4; International Water Association Publishing: London, UK, 2010. [Google Scholar]
- Ferguson, A.S.; Mailloux, B.J.; Ahmed, K.M.; Van Geen, A.; McKay, L.D.; Culligan, P.J. Hand-pumps as reservoirs for microbial contamination of well water. J. Water Health 2011, 9, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Haramoto, E.; Yamada, K.; Nishida, K. Prevalence of protozoa, viruses, coliphages and indicator bacteria in groundwater and river water in the Kathmandu Valley, Nepal. Trans. R. Soc. Trop. Med. Hyg. 2011, 105, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Barthiban, S.; Lloyd, B.J.; Maier, M. Applicability of a surveillance methodology for the microbiological safety of well water supplies, in a highly vulnerable hydrogeological setting—A case study based findings from the west coastal area of Sri Lanka. J. Water Resource Prot. 2012, 4, 899–912. [Google Scholar] [CrossRef]
- Shrestha, S.; Nakamura, T.; Malla, R.; Nishida, K. Seasonal variation in the microbial quality of shallow groundwater in the Kathmandu Valley, Nepal. Water Sci. Tech. Water Supply 2014, 14, 390–397. [Google Scholar] [CrossRef]
- Shrestha, S.; Haramoto, E.; Malla, R.; Nishida, K. Risk of diarrhoea from shallow groundwater contaminated with enteropathogens in the Kathmandu Valley, Nepal. J. Water Health 2015, 13, 259–269. [Google Scholar] [CrossRef]
- Shrestha, S.; Haramoto, E.; Shindo, J. Assessing the infection risk of enteropathogens from consumption of raw vegetables washed with contaminated water in Kathmandu Valley, Nepal. J. Appl. Microbiol. 2017, 123, 1321–1334. [Google Scholar] [CrossRef]
- Wu, J.; Yunus, M.; Islam, M.S.; Emch, M. Influence of climate extremes and land use on fecal contamination of shallow tubewells in Bangladesh. Environ. Sci. Technol. 2016, 50, 2669–2676. [Google Scholar] [CrossRef]
- Vernberg, W.; Scott, G.; Strozier, S.; Bemiss, J.; Daugomah, J. The Effects of Urbanization on Human and Ecosystem Health; Vernberg, F., Vernberg, W., Siewicki, T., Eds.; Sustainable Development in the Southeastern Coastal Zone; The University of South Carolina Press: Columbia, SC, USA, 1996; pp. 221–239. [Google Scholar]
- Paule, M.C.A.; Ventura, J.R.S.; Memon, S.; Lee, B.Y.; Jahng, D.; Kang, M.J.; Lee, C.H. Fecal contamination in Yongin watershed: Association to land use and land cover and stormwater quality. Desalin. Water Treat. 2015, 53, 3026–3038. [Google Scholar] [CrossRef]
- Okotto-Okotto, J.; Okotto, L.; Price, H.; Pedley, S.; Wright, J. A longitudinal study of long-term change in contamination hazards and shallow well quality in two neighbourhoods of Kisumu, Kenya. Int. J. Environ. Res. Public Health 2015, 12, 4275–4291. [Google Scholar] [CrossRef]
- Wardrop, N.A.; Hill, A.G.; Dzodzomenyo, M.; Aryeetey, G.; Wright, J.A. Livestock ownership and microbial contamination of drinking-water: Evidence from nationally representative household surveys in Ghana, Nepal and Bangladesh. Int. J. Hyg. Environ. Health 2018, 221, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Pilbeam, C.J.; Tripathi, B.P.; Sherchan, D.P.; Gregory, P.J.; Gaunt, J. Nitrogen balances for households in the mid-hills of Nepal. Agric. Ecosyst. Environ. 2000, 79, 61–72. [Google Scholar] [CrossRef]
- Schriewer, A.; Odagiri, M.; Wuertz, S.; Misra, P.R.; Panigrahi, P.; Clasen, T.; Jenkins, M.W. Human and animal fecal contamination of community water sources, stored drinking water and hands in rural India measured with validated microbial source tracking assays. Am. J. Trop. Med. Hyg. 2015, 93, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Harris, A.R.; Pickering, A.J.; Harris, M.; Doza, S.; Islam, M.S.; Unicomb, L.; Luby, S.; Davis, J.; Boehm, A.B. Ruminants contribute fecal contamination to the urban household environment in Dhaka, Bangladesh. Environ. Sci. Technol. 2016, 50, 4642–4649. [Google Scholar] [CrossRef]
- Odagiri, M.; Schriewer, A.; Daniels, M.E.; Wuertz, S.; Smith, W.A.; Clasen, T.; Schmidt, W.; Jin, Y.; Torondel, B.; Misra, P.R.; et al. Human fecal and pathogen exposure pathways in rural Indian villages and the effect of increased latrine coverage. Water Res. 2016, 100, 232–244. [Google Scholar] [CrossRef]
- US Environmental Protection Agency. Water Quality Standards for Coastal and Great Lakes Recreation Waters: Final Rule; Federal Register; US Environmental Protection Agency: Washington, DC, USA, 2004; Volume 69, pp. 67218–67243.
- US Environmental Protection Agency. Microbial Source Tracking Guide Document; EPA/600-R-05-064; US Environmental Protection Agency: Cincinnati, OH, USA, 2005.
- Bernhard, A.E.; Field, K.G. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 2000, 66, 4571–4574. [Google Scholar] [CrossRef]
- Bernhard, A.E.; Field, K.G. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 2000, 66, 1587–1594. [Google Scholar] [CrossRef]
- Layton, A.; McKay, L.; Williams, D.; Garrett, V.; Gentry, R.; Sayler, G. Development of Bacteroides 16S rRNA gene taqman-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl. Environ. Microbiol. 2006, 72, 4214–4224. [Google Scholar] [CrossRef] [PubMed]
- Haramoto, E. Detection of Waterborne Protozoa, Viruses, and Bacteria in Groundwater and Other Water Samples in the Kathmandu Valley, Nepal. IOP Conference Series: Earth and Environmental Science. 120 012004. 2018. Available online: https://doi.org/10.1088/1755-1315/120/1/012004 (accessed on 15 July 2018).
- Haramoto, E.; Osada, R. Assessment and application of host-specific Bacteroidales genetic markers for microbial source tracking of river water in Japan. PLoS ONE 2018, 13, e0207727. [Google Scholar] [CrossRef]
- Malla, B.; Ghaju Shrestha, R.; Tandukar, S.; Bhandari, D.; Inoue, D.; Sei, K.; Tanaka, Y.; Sherchand, J.B.; Haramoto, E. Validation of host-specific Bacteroidales quantitative PCR assays and their application to microbial source tracking of drinking water sources in the Kathmandu Valley, Nepal. J. Appl. Microbiol. 2018, 125, 609–619. [Google Scholar] [CrossRef]
- Wuertz, S.; Wang, D.; Reischer, G.; Farnleitner, A. Microbial Source Tracking: Methods, Applications, and Case Studies; Hagedorn, C., Blanch, A.R., Harwood, V.J., Eds.; Springer: New York, NY, USA, 2011; Chapter 4; pp. 90–91. [Google Scholar]
- Kildare, B.J.; Leutenegger, C.M.; McSwain, B.S.; Bambic, D.G.; Rajal, V.B.; Wuertz, S. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: A Bayesian approach. Water Res. 2007, 41, 3701–3715. [Google Scholar] [CrossRef] [PubMed]
- Reischer, G.H.; Kasper, D.C.; Steinborn, R.; Mach, R.L.; Farnleitner, A.H. Quantitative PCR method for sensitive detection of ruminant fecal pollution in freshwater and evaluation of this method in alpine karstic regions. Appl. Environ. Microbiol. 2006, 72, 5610–5614. [Google Scholar] [CrossRef] [PubMed]
- Mieszkin, S.; Furet, J.P.; Corthier, G.; Gourmelon, M. Estimation of pig fecal contamination in a river catchment by real-time PCR using two Pig-Specific Bacteroidales 16S rRNA genetic markers. Appl. Environ. Microbiol. 2009, 75, 3045–3054. [Google Scholar] [CrossRef] [PubMed]
- Central Bureau of Statistics. National Population and Housing Census 2011 (Village Development Committee/Municipality); Central Bureau of Statistics, Government of Nepal: Kathmandu, Nepal, 2012.
- Uddin, K.; Shrestha, H.L.; Murthy, M.S.R.; Bajracharya, B.; Shrestha, B.; Gilani, H.; Pradhan, S.; Dangol, B. Development of 2010 national land cover database for the Nepal. J. Environ. Manag. 2015, 148, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Regmi, R.K.; Mishra, B.K.; Masago, Y.; Luo, P.; Toyozumi-Kojima, A.; Jalilov, S.M. Applying a water quality index model to assess the water quality of the major rivers in the Kathmandu Valley, Nepal. Environ. Monit. Assess. 2017, 189–382. [Google Scholar] [CrossRef] [PubMed]
- Bhatta, G.D.; Doppler, W. Farming differentiation in the rural-urban interface of the middle mountains, Nepal: Application of analytic hierarchy process (AHP) modeling. J. Agric. Sci. 2010, 2, 37–51. [Google Scholar] [CrossRef]
- Kathmandu Upatyaka Khanepani Limited. KUKL 10th Annual Report; Kathmandu Upatyaka Khanepani Limited: Kathmandu, Nepal, 2018. [Google Scholar]
- Shrestha, S.; Aihara, Y.; Bhattarai, A.P.; Bista, N.; Rajbhandari, S.; Kondo, N.; Kazama, F.; Nishida, K.; Shindo, J. Dynamics of domestic water consumption in the urban area of the Kathmandu Valley: Situation analysis pre and post 2015 Gorkha Earthquake. Water 2017, 9, 222. [Google Scholar] [CrossRef]
- Ghaju Shrestha, R.; Tanaka, Y.; Malla, B.; Bhandari, D.; Tandukar, S.; Inoue, D.; Sei, K.; Sherchand, J.B.; Haramoto, E. Next-generation sequencing identification of pathogenic bacterial genes and their relationship with fecal indicator bacteria in different water sources in the Kathmandu Valley, Nepal. Sci. Total Environ. 2017, 601–602, 278–284. [Google Scholar] [CrossRef]
- Haramoto, E.; Kitajima, M. Quantification and genotyping of Aichi Virus 1 in water samples in the Kathmandu Valley, Nepal. Food Environ. Virol. 2017, 9, 350–353. [Google Scholar] [CrossRef]
- Ministry of Agricultural Development. Statistical Information on Nepalese Agriculture. Nepal; Government of Nepal: Kathmandu, Nepal, 2012.
- NPC & UN. Nepal MDG Acceleration Framework: Improving Access to Sanitation; National Planning Commission, Government of Nepal and United Nations in Nepal: Kathmandu, Nepal, 2013. Available online: http://www.undp.org/content/dam/undp/library/MDG/MDG%20Acceleration%20Framework/MAF%20Reports/RBAP/Nepal%20-%20WEB.pdf (accessed on 15 July 2018).
- Kishida, N.; Morita, H.; Haramoto, E.; Asami, M.; Akiba, M. One-year weekly survey of noroviruses and enteric adenoviruses in the Tone River water in Tokyo metropolitan area, Japan. Water Res. 2012, 46, 2905–2910. [Google Scholar] [CrossRef]
- Khush, R.S.; Arnold, B.F.; Srikanth, P.; Sudharsanam, S.; Ramaswamy, P.; Durairaj, N.; London, A.G.; Ramaprabha, P.; Rajkumar, P.; Balakrishnan, K.; et al. H2S as an indicator of water supply vulnerability and health risk in low-resource settings: A prospective cohort study. Am. J. Trop. Med. Hyg. 2013, 89, 251–259. [Google Scholar] [CrossRef]
- Pant, B.R. Groundwater quality in the Kathmandu valley of Nepal. Environ. Monit. Assess. 2011, 178, 477–485. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Guidelines for Drinking-Water Quality, 4th ed.; WHO: Geneva, Switzerland, 2011. [Google Scholar]
- Sobsey, M.D.; Khatib, L.A.; Hill, V.R.; Alocilja, E.; Pillai, S. Pathogens in Animal Wastes and the Impacts of Waste Management Practices on Their Survival, Transport and Fate; White papers on animal agriculture and the environment. MidWest Plan Service (MWPS); Iowa State University: Ames, IA, USA, 2001. [Google Scholar]
- Ercumen, A.; Pickering, A.J.; Kwong, L.H.; Arnold, B.F.; Parvez, S.M.; Alam, M.; Sen, D.; Islam, S.; Kullmann, C.; Chase, C.; et al. Animal feces contribute to domestic fecal contamination: evidence from E. coli measured in water, hands, food, flies, and soil in Bangladesh. Environ. Sci. Technol. 2017, 51, 8725–8734. [Google Scholar] [CrossRef] [PubMed]
- Dufour, A.; Bartram, J.; Bos, R.; Gannon, V.P.J. Animal Waste, Water Quality and Human Health, Emerging Issues in Water and Infectious Disease Series; International Water Association Publishing: London, UK, 2012. [Google Scholar]
- Boehm, A.B.; Soller, J.A.; Shanks, O.C. Human-associated fecal quantitative polymerase chain reaction measurements and simulated risk of gastrointestinal illness in recreational waters contaminated with raw sewage. Environ. Sci. Technol. Lett. 2015, 2, 270–275. [Google Scholar] [CrossRef]
- Ahmed, W.; Hamilton, K.A.; Lobos, A.; Hughes, B.; Staley, C.; Sadowsky, M.J.; Harwood, V.J. Quantitative microbial risk assessment of microbial source tracking markers in recreational water contaminated with fresh untreated and secondary treated sewage. Environ. Int. 2018, 117, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, R.R. Status and Strategy for Faecal Sludge Management in the Kathmandu Valley; HPCIDBC & UNHABITAT: Kathmandu, Nepal, 2011.
- Nyachhon, B.L. Economic Policy Network Policy Paper23–Service Enhancement and Development of Sanitary System in Urban and Semi-Urban Setting in Nepal; Ministry of Finance, Government of Nepal: Kathmandu, Nepal, 2006; 12p. [Google Scholar]
- Shukla, A. Wastewater Production, Treatment and Use in Nepal. 2012. Available online: https://www.ais.unwater.org/ais/pluginfile.php/232/mod_page/content/134/Nepal_CountryPaper.pdf (accessed on 15 July 2018).
- Pradhan, B.B.; Shrestha, B.R.; Punjali, K.R.; Lama, C.N. Kathmandu Valley Environmental Outlook; International Center for Integrated Mountain Development: Kathmandu, Nepal, 2007. [Google Scholar]
- Majorin, F.; Freeman, M.C.; Barnard, S.; Routray, P.; Boisson, S.; Clasen, T. Child feces disposal practices in rural Orissa: A cross sectional study. PLoS ONE 2014, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Howard, G.; Pedley, S.; Barrett, M.; Nalubega, M.; Johal, K. Risk factors contributing to microbiological contamination of shallow groundwater in Kampala, Uganda. Water Res. 2003, 37, 3421–3429. [Google Scholar] [CrossRef]
- Karn, S.K.; Harada, H. Surface water pollution in three urban territories of Nepal, India, and Bangladesh. Environ. Manag. 2001, 28, 483–496. [Google Scholar] [CrossRef]
- Devleesschauwer, B.; Aryal, A.; Joshi, D.D.; Rijal, S.; Sherchand, J.B.; Praet, N.; Speybroeck, N.; Duchateau, L.; Vercruysse, J.; Dorny, P. Epidemiology of Taenia solium in Nepal: Is it influenced by the social characteristics of the population and the presence of Taenia asiatica? Trop. Med. Int. Health 2012, 17, 1019–1022. [Google Scholar] [CrossRef]
- Shrestha, S.; Haramoto, E.; Sherchand, J.B.; Shindo, J. Detection of coliform bacteria in irrigation water and on vegetable surfaces in the Kathmandu valley of Nepal. JIOM 2016, 38, 43–47. [Google Scholar]
- Oun, A.; Kumar, A.; Harrigan, T.; Angelakis, A.; Xagoraraki, I. Effects of Biosolids and manure application on microbial water quality in rural areas in the US. Water 2014, 6, 3701–3723. [Google Scholar] [CrossRef]
- Jenkins, M.W.; Tiwari, S.; Lorente, M.; Gichaba, C.M.; Wuertz, S. Identifying human and livestock sources of fecal contamination in Kenya with host-specific Bacteroidales assays. Water Res. 2009, 43, 4956–4966. [Google Scholar] [CrossRef] [PubMed]
- Ervin, J.S.; Russell, T.L.; Layton, B.A.; Yamahara, K.M.; Wang, D.; Sassoubre, L.M.; Cao, Y.; Kelty, C.A.; Sivaganesan, M.; Boehm, A.B.; et al. Characterization of fecal concentrations in human and other animal sources by physical, culture-based, and quantitative real-time PCR Methods. Water Res. 2013, 47, 6873–6882. [Google Scholar] [CrossRef] [PubMed]
- Riedel, T.E.; Thulsiraj, V.; Zimmer-Faust, A.G.; Dagit, R.; Krug, J.; Hanley, K.T.; Adamek, K.; Ebentier, D.L.; Torres, R.; Cobian, U.; et al. Long-term monitoring of molecular markers can distinguish different seasonal patterns of fecal indicating bacteria sources. Water Res. 2015, 71, 227–243. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, A.; Sano, D.; Hatori, J.; Ishii, S.; Okabe, S. Chicken- and duck-associated Bacteroides-Prevotella genetic markers for detecting fecal contamination in environmental water. Appl. Microbiol. Biotechnol. 2013, 97, 7427–7437. [Google Scholar] [CrossRef] [PubMed]
- Sinigalliano, C.D.; Fleisher, J.M.; Gidley, M.L.; Solo-Gabriele, H.M.; Shibata, T.; Plano, L.R.; Elmir, S.M.; Wanless, D.; Bartkowiak, J.; Boiteau, R.; et al. Traditional and molecular analyses for fecal indicator bacteria in non-point source subtropical recreational marine waters. Water Res. 2010, 44, 3763–3772. [Google Scholar] [CrossRef] [PubMed]
| Water Sample | No. of Tested Samples | E. coli | Total Coliforms | ||||
|---|---|---|---|---|---|---|---|
| No. of Positive Samples (% Positive) | Concentration (log MPN/100 mL) | No. of Positive Samples (% Positive) | Concentration (log MPN/100 mL) | ||||
| Mean ± SD a | Max | Mean ± SD a | Max | ||||
| Shallow dug well | 181 | 149 (82) | 1.61 ± 1.59 | 5.38 | 178 (98) | 3.60 ± 1.27 | 7.08 |
| Shallow tube well | 45 | 13 (29) | −0.22 ± 1.40 | 4.46 | 36 (80) | 1.64 ± 1.77 | 5.38 |
| Deep tube well | 50 | 17 (34) | −0.17 ± 1.35 | 4.46 | 44 (88) | 1.79 ± 1.51 | 4.74 |
| Stone spout | 24 | 18 (75) | 1.34 ± 1.80 | 4.51 | 24 (100) | 2.69 ± 1.25 | 4.74 |
| Total | 300 | 197 (66) | 282 (94) | ||||
| Source | No. of Tested Samples | BacHum (Human) | BacR (Ruminant) | Pig2Bac (Pig) | No. of Samples Positive for at Least One Fecal Marker (% Positive) | |||
|---|---|---|---|---|---|---|---|---|
| No. of Positive Samples (% Positive) | Conc. a (log Copies/100 mL) | No. of Positive Samples (% Positive) | Conc. a (log Copies/100 mL) | No. of Positive Samples (% Positive) | Conc. a (log Copies/100 mL) | |||
| Shallow dug well | 166 | 40 (24) | 5.04–7.17 | 21 (13) | 4.25–7.81 | 5 (3) | 4.39–4.68 | 59 (36) |
| Shallow tube well | 33 | 6 (18) | 5.01–6.91 | 1 (3) | 4.47 | 2 (6) | 5.77 | 8 (24) |
| Deep tube well | 30 | 2 (7) | DNQ b | 3 (10) | 4.52–5.18 | 1 (3) | DNQ b | 5 (17) |
| Stone spout | 21 | 7 (33) | 5.50–6.88 | 3 (14) | 4.67–5.57 | 0 (0) | ND c | 10 (48) |
| Total | 250 | 55 (22) | 28 (11) | 8 (3) | 82 (33) | |||
| Factors | E. coli Concentration | BacHum Detection | BacR Detection | At Least One Marker Detection | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β a | 95% CI b | p | AOR c | 95% CI b | p | AORc | 95% CI b | p | AORc | 95% CI b | p | |||||
| Lower | Upper | Lower | Upper | Lower | Upper | Lower | Upper | |||||||||
| Multiple Linear Regression | Multiple Logistic Regression | |||||||||||||||
| Land cover | ||||||||||||||||
| Built-up | −0.18 | −0.57 | 0.20 | 0.345 | 3.60 | 1.63 | 7.92 | 0.002 | Ref. | 1.48 | 0.79 | 2.75 | 0.219 | |||
| Agricultural area | Ref. | 2.90 | 1.20 | 7.02 | 0.018 | Ref. | ||||||||||
| Source types | ||||||||||||||||
| Shallow dug well | 0.32 | −0.31 | 0.95 | 0.315 | Ref. | Ref. | Ref. | |||||||||
| Shallow tube well | −1.44 | −2.16 | −0.73 | <0.001 | 0.41 | 0.15 | 1.10 | 0.076 | 0.34 | 0.04 | 2.81 | 0.317 | 0.44 | 0.18 | 1.09 | 0.076 |
| Deep tube well | −1.43 | −2.13 | −0.73 | <0.001 | 0.14 | 0.03 | 0.63 | 0.010 | 0.86 | 0.23 | 3.27 | 0.828 | 0.26 | 0.09 | 0.75 | 0.012 |
| Stone spout | NA | 0.94 | 0.34 | 2.61 | 0.911 | 1.79 | 0.44 | 7.39 | 0.418 | 1.29 | 0.49 | 3.37 | 0.602 | |||
| Season | ||||||||||||||||
| Dry | Ref. | Ref. | Ref. | |||||||||||||
| Wet | 1.18 | 0.85 | 1.51 | <0.001 | 1.88 | 0.97 | 3.66 | 0.061 | 1.69 | 0.71 | 4.01 | 0.236 | 2.10 | 1.18 | 3.74 | 0.012 |
| Generalized Estimation Equation in Repeated Samples | ||||||||||||||||
| Land cover | ||||||||||||||||
| Built up | −0.02 | −0.43 | 0.39 | 0.914 | Ref. | Ref. | Ref. | |||||||||
| Agricultural area | 0.32 | 0.12 | 0.83 | 0.019 | 2.01 | 0.81 | 4.97 | 0.130 | 0.66 | 0.32 | 1.34 | 0.251 | ||||
| Source types | ||||||||||||||||
| Shallow dug well | 0.16 | −0.70 | 1.02 | 0.715 | Ref. | Ref. | Ref. | |||||||||
| Shallow tube well | −1.81 | −2.74 | 0.89 | <0.001 | 0.37 | 0.13 | 1.08 | 0.068 | 0.39 | 0.06 | 2.73 | 0.342 | 0.44 | 0.17 | 1.13 | 0.088 |
| Deep tube well | −1.64 | −2.58 | −0.71 | 0.001 | 0.18 | 0.04 | 0.85 | 0.030 | 0.42 | 0.05 | 3.53 | 0.424 | 0.20 | 0.05 | 0.74 | 0.016 |
| Stone spout | NA | 0.71 | 0.18 | 2.89 | 0.634 | 2.03 | 0.53 | 7.80 | 0.304 | 1.11 | 0.33 | 3.74 | 0.860 | |||
| Season | ||||||||||||||||
| Dry | Ref. | Ref. | Ref. | |||||||||||||
| Wet | 1.18 | 0.83 | 1.53 | <0.001 | 2.17 | 1.15 | 4.10 | 0.016 | 1.28 | 0.59 | 2.75 | 0.532 | 2.23 | 1.31 | 3.79 | 0.003 |
| Concentration of E. coli (log MPN/100 mL) | No. of Samples | No. of Positive Samples (% positive) | ||
|---|---|---|---|---|
| Human | Ruminant | At Least One Fecal Marker | ||
| <0.00 | 56 | 10 (18) | 2 (4) | 13 (23) |
| 0.00–1.50 | 64 | 5 (8) | 5 (8) | 10 (16) |
| 1.51–2.70 | 68 | 12 (18) | 9 (13) | 21 (31) |
| >2.70 | 62 | 28 (45) | 12 (19) | 38 (61) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malla, B.; Ghaju Shrestha, R.; Tandukar, S.; Bhandari, D.; Inoue, D.; Sei, K.; Tanaka, Y.; Sherchand, J.B.; Haramoto, E. Identification of Human and Animal Fecal Contamination in Drinking Water Sources in the Kathmandu Valley, Nepal, Using Host-Associated Bacteroidales Quantitative PCR Assays. Water 2018, 10, 1796. https://doi.org/10.3390/w10121796
Malla B, Ghaju Shrestha R, Tandukar S, Bhandari D, Inoue D, Sei K, Tanaka Y, Sherchand JB, Haramoto E. Identification of Human and Animal Fecal Contamination in Drinking Water Sources in the Kathmandu Valley, Nepal, Using Host-Associated Bacteroidales Quantitative PCR Assays. Water. 2018; 10(12):1796. https://doi.org/10.3390/w10121796
Chicago/Turabian StyleMalla, Bikash, Rajani Ghaju Shrestha, Sarmila Tandukar, Dinesh Bhandari, Daisuke Inoue, Kazunari Sei, Yasuhiro Tanaka, Jeevan B. Sherchand, and Eiji Haramoto. 2018. "Identification of Human and Animal Fecal Contamination in Drinking Water Sources in the Kathmandu Valley, Nepal, Using Host-Associated Bacteroidales Quantitative PCR Assays" Water 10, no. 12: 1796. https://doi.org/10.3390/w10121796
APA StyleMalla, B., Ghaju Shrestha, R., Tandukar, S., Bhandari, D., Inoue, D., Sei, K., Tanaka, Y., Sherchand, J. B., & Haramoto, E. (2018). Identification of Human and Animal Fecal Contamination in Drinking Water Sources in the Kathmandu Valley, Nepal, Using Host-Associated Bacteroidales Quantitative PCR Assays. Water, 10(12), 1796. https://doi.org/10.3390/w10121796






