Abstract
Atmospheric air is a microbial habitat of pathogenic bioaerosols that may pose serious risks to humans. A commonly laboratory-based approach for the diagnosis of such infections in the bloodstream is the blood culture analysis. Its clinical relevance is attributed to the fact that these infections are characterized by high rates of morbidity and mortality, requiring the need for efficient methods for rapid diagnosis. For this reason, our study aimed to develop a method of manometric monitoring for the rapid detection of viable microorganisms in blood culture vials. A methodology was developed to detect pressure variation in intra-vials through a manometric instrument that was coupled to vials of blood culture containing culture broth that allowed microbial growth. This device allowed the early detection of microbial activity based on the production or use of intra-flask gases as a result of microbial metabolic activity. The analyzed variables were the pressure as a function of time, microbial species, and culture medium. The highest pressure found in the flasks without microorganisms was 40 mmHg between 2 and 6 h, and the lowest pressure was −42 mmHg between 21 and 24 h. The variation of the internal pressure in blood culture flasks according to different groups of microorganisms as a function of time demonstrated that the fermentative gram-negative bacilli and gram-positive cocci exhibited a significant increase in relation to their respective control groups (p < 0.001). The non-fermenting gram-negative bacilli showed expected results in relation to the pressure variation in which the production of negative pressures was noticed during the period of analysis, with a significant difference with respect to their control groups (p < 0.001). The developed methodology for the early detection of microorganisms responsible for bloodstream infection was demonstrated to be effective.
1. Introduction
Sepsis is considered a public health concern of worldwide dimension due to high rates of morbidity and mortality [1]. It is estimated that about 18 million cases occur and more than 27% are responsible for the incidence of mortality [2,3]. The latest key-facts available from the World Health Organization (WHO) date back to 2017, reporting a total of 48.9 million cases and 11 million sepsis-related deaths, reaching ca. 20% of all deaths worldwide [4]. In Brazil, the incidence of sepsis increased by 50.5% during the period 2006 to 2015 [5].
Due to the severity of sepsis, the use of empirical treatment with broad-spectrum antimicrobials is necessary whenever suspicion occurs [6,7]. Besides, frequency of nosocomial infections, resulting from microbiological contamination of surfaces and/or air of inpatient treatment facilities [8] such as hospitals, pharmacy shops and other clinical facilities, increases the need for an efficient and fast diagnostic tool. Indeed, the quality of air in public facilities is instrumental to ensure global healthcare, as air often contains microbial pathogens that may reduce immunity. A statistically significant correlation between the concentration of bacteria and the season of the year, size and location of pharmacy shops has recently been reported to be under study [9]. Medical protocols strongly indicate the need for blood collection for blood culture before antimicrobials are administered due to the imminent need to know the etiologic agent, as well as their susceptibility profile to antimicrobials [10]. The need for information provided by blood culture becomes even more evident due to the increasing rates of multiple-resistant microorganisms [11,12,13].
Blood culture is a widely used test for the investigation of bloodstream infections (BSI), being considered the best laboratory test for the diagnosis of sepsis [14,15]. Different methodologies—automated, semi-automated and manual methods—are available on the market and all have in common the need to inoculate the blood sample in specific flasks, containing culture media capable of favoring the growth of microorganisms.
The automated blood culture methodologies, considered the most efficient and used worldwide, demonstrate advantages in terms of specificity, sensitivity and an average positivity time of 24 h [16,17,18]. However, microbiology laboratories with no financial resources are at a disadvantage because they need monitoring equipment that has a high cost of acquisition and maintenance [18,19,20]. Manual methodologies are thus used as an alternative. Although they are efficient and economically viable, they do not have positivity indicators—requiring subcultures at 48 h intervals for detection—and the time to positivity of blood culture usually varies between 36 and 72 h [19,21,22]. The implementation of this methodology contributes to increased turnaround time and, consequently, compromises patient care [23]. In addition, the delayed diagnosis may reflect the high health costs related to length of stay and use of antimicrobials [24]. It is noted that the development of fast and accurate methodologies that identify specific pathogens and provide information on sensitivity to antimicrobial agents is essential for the improvement of detection of BSI and sepsis [25,26]. As the growth of microorganisms is always followed by a series of metabolic changes, the intensity of these latter may increase or decrease depending on the concentration of microorganisms. The physiological indicators available include the detection of DNA, RNA, ATP, carbon dioxide production and oxygen consumption [27,28].
A suggestive alternative are techniques that aim to detect microorganisms through microbial metabolism in closed blood culture systems [29]. Current methods available on the market are based on the pressure variation in blood culture vials through the production or consumption of gases [29,30,31]. Some of them use indicating devices attached to the bottles [19], while others use automated detection equipment [31,32].
Finally, there is a need to seek methods that combine advantages, such as low cost, high sensitivity and rapid diagnosis, aiming not only at the diagnostic improvement, but also affordable acquisition by the laboratories. Therefore, the ultimate aim of this work was the development of devices capable of detecting the smallest variations in pressure behavior in blood culture flasks, followed by the production or metabolization of biogas by the microorganisms present in the sample, as an effective, simple, fast and inexpensive alternative for laboratories that do not have the benefits of automation. This study aimed to develop a methodology based on a diagnostic kit for the detection of viable microorganisms in blood culture vials more quickly.
2. Materials and Methods
2.1. Selection of Microorganisms
The choice of the microorganisms used in this study was based on two different factors: their relationship as an etiological agent of sepsis and the prevalence described in the literature from the data presented by Baykara et al. (2018) [33], Yusef et al. (2018) [34] and WHO, (2020) [35]. The bacteria selected for this work are described in Table 1. The ATCC bacterial strains were obtained from the Tropical Culture Collection of the Foundation André Tosello (Campinas, SP, Brazil) and Controllab (Rio de Janeiro, RJ, Brazil). The bacterial strains of clinical specimens were kindly provided by Monera Microbiology Laboratory (Aracaju, SE, Brazil).

Table 1.
ATCC organisms and clinical isolates applied in this study.
2.2. Isolation and Identification of Microorganism
The lyophilized ATCC bacterial strains were suspended for reactivation in BHI (brain heart infusion) enrichment medium (Oxoid LTD, Basingstoke, UK) inside 5 mL test tubes which were subsequently incubated in a bacteriological oven (Quimis® q316m) with electronic thermostat set at 35 °C for 24 h, following the manufacturer’s recommendations. After incubation period, the bacteria were isolated in blood agar using streak plate technique (Oxoid LTD, Basingstoke, UK) with the aid of a platinum inoculation loop. Additionally, a new fresh isolate was performed for the clinical isolates from their primary culture into blood agar plates. The plates were then incubated under the same conditions previously described. After 24 h incubation, the bacteria were phenotypically identified using the automated Vitek® 2 (Biomérieux) system. For each experiment 3 different strains: ATCC isolate (i-ATCC); Clinical isolate 1 (CI-1); Clinical isolate 2 (CI-2), were obtained under the same growth conditions.
2.3. Preparation of Broth Culture
The culture media used in this work were Brain Heart Infusion-BHI broths (MD-media Master Diagnostica, SP, Brazil), Thioglycolate-THIO (Kasvi, PR, Brazil) and Trypticase Soy Broth-TSB (MD-media Master Diagnostica, SP, Brazil). For the broth cultures preparation, 18.5 g, 14.9 g and 15 g of BHI, THIO and TSB powders were weighed, respectively, in 500 mL flasks with the aid of a semi-analytical balance (bl-3200 h Shimadzu). After weighing, 500 mL of distilled water was added in each flask to dissolve the powder, followed by homogenization with the aid of a glass stick for complete dissolution. Afterwards, 45 mL of the broth were distributed in glass flasks (80 mL). The vials were hermetically sealed with a rubber stopper and sterilized in moist heat for 15 min at 121 °C in a vertical autoclave (AV Phoenix Luferco, Araraquara, SP, Brazil). After sterilization and cooling to room temperature, the flasks were incubated at 35 ± 2 °C/24 h in a bacteriological oven for sterility test. Lastly, culture media that did not exhibit turbidity or changes in color and pH recommended by the manufacturer were used. The color changes were observed visually in an environment with light and a light background. The pH was measured using reactive pH tapes (Macherey-nagel®) with sensitivity from 0 to 14.
2.4. Device for Detecting Pressure in Blood Culture Vials
Twelve aneroid manometers (Wenzhou Medical Instruments Co. Ltd., Wenzhou, China, with Inmetro certification) were used to measure intra-bottle pressure from production or consumption of gases by bacteria and other control tests contained in the headspace of the flasks. The pressure gauges employed have detection sensitivities between 0 to 300 mmHg (clockwise) and 0 to −300 (counterclockwise), where 0.00135951 kgf/cm2 is equivalent to the force required to move the pressure marker to 1 mmHg (Figure 1).
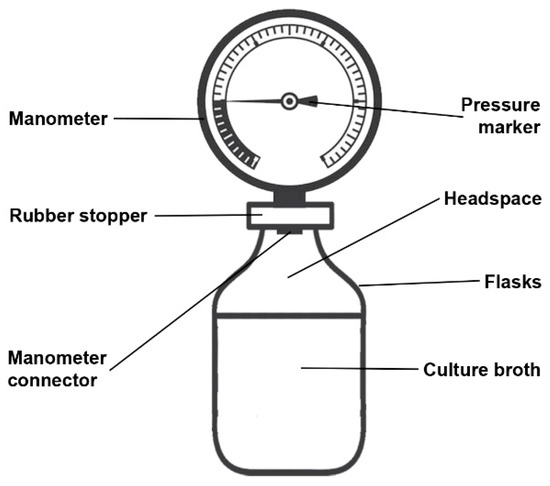
Figure 1.
Scheme of the intra-bottle pressure measurement device.
The pressure gauges were connected to the flasks through the rubber stopper. With the aid of a tweezers previously sterilized in dry heat, the sterile rubber caps were punctured (1 to 2 mm) in their central region and later attached to the manometer connector (5 mm). The glass vials containing the previously prepared culture media were uncapped next to a lamp flame in order to avoid contamination, and immediately the cap attached to the prepared manometer was connected to the mouth vial. After setting up each instrument, the leak test was performed. Using a 10 mL syringe attached to a needle, 10 mL of air was introduced into the syringe plunger in order to observe the pressure increase in the system. This pressure was maintained for 60 s to analyze possible air leaks and consequently the pressure loss. Instruments that did not show pressure loss during the tests were used.
2.5. Selection and Evaluation of Different Culture Media for the Detection of Microorganisms
Culture media were prepared in BHI, THIO and TSB culture media, as well as the manometers coupled according to the previously described recommendations. Three bottles of each culture medium were used for each bacterium used (triplicate test). The bacteria used were Enterobacter cloacae ATCC 13,047 and Staphylococcus aureus ATCC 25,923. The bacterial inoculates were prepared in test tubes with 4 mL of sterile 0.9% saline. Initially, with the aid of the bacteriological loop, 2 to 3 colonies were transferred from the culture media to saline solution, where it was homogenized until its complete dissolution. The concentration of bacteria corresponded to the McFarland 0.5 scale (≅1.5 × 108 CFU/mL). A 5 mL syringe and sterile needle-diameter 0.70 mm and length 25 mm were used to aspirate the inoculum from the test tube and transfer it to the flask through the rubber stopper. The aspiration needle attached to the syringe was submerged in the saline to obtain 1 mL of the suspension, where the plunger of the syringe was pulled negatively. Subsequently, a sterile needle was bent at 40° and inserted into the rubber cap of the vial. The syringe was detached from the suction needle and it was attached to the needle previously inserted in the cap. The plunger was pushed positively in order to transfer the inoculum into the bottle. The syringe was first uncoupled to let the needle generate a free bridge between the external environment and allow the intra-vial pressure to be zero. The pressure gauges remained attached to the flasks for 24 h, incubated in a bacteriological oven at 35 ± 2 °C. The readings were performed with an interval of 1 h for 6 h from the zero time and for 24 h observing the pointer marking on the graduation of the manometer.
2.6. Determination of Maximum and Minimum Pressure in Blood Culture Flasks Containing Selected Culture Medium and Determination of Statistical Time-to-Positivity (Sttp) and Time-to-Positivity (TTP)
To evaluate the maximum and minimum pressure, a group of 63 bottles of BHI broth were used. Then, the aneroid gauges were reconnected to the vials as previously described. Subsequently, 1 mL of sterile 0.9% saline solution was added with a syringe and needle through the rubber stopper of the bottle. The flasks were then incubated in a microbiological incubator at a temperature of 35 ± 2 °C for 24 h. The reading times took place at intervals of 5 min during the 24 h with the aid of a 13.0 mp camera from a cell phone (Motorola® model xt1600). The analysis of the images allowed to analyze the pressure changes every 1 h from time 0 (time of incubation). To determine the TTP and STTP, the pressure values obtained from the results of the experiments described in Section 2.5 were used. TTP was determined by calculating the mean and standard deviation of pressures over time (0 to 24 h). The determination of the STTP was performed with the analysis of the interquartile range of the results of the pressures obtained over time (0 to 24 h). Subsequently, the module value of the highest and lowest pressures observed were calculated using a safety margin of 25%.
2.7. Analysis of the Internal Pressure Variation Profile in Blood Culture Flasks According to Different Groups of Microorganisms
For this analysis, the bacteria described in Table 1 were used. The assays were carried out with four groups: control group (flask with 45 mL of BHI broth + 1 mL of sterile 0.9% saline); isolated group 1 (flask with 45 mL of BHI broth + bacterial suspension corresponding to 0.5 McFarland scale [≅1.5 × 108 CFU/mL] of clinical isolate 1); isolated group 2 (flask with 45 mL of BHI broth + bacterial suspension corresponding to 0.5 McFarland scale [≅1.5 × 108 CFU/mL] of clinical isolate 2) and isolated ATCC group (flask with 45 mL of BHI broth + corresponding bacterial suspension the McFarland 0.5 scale [≅1.5 × 108 CFU/mL of the ATCC isolate). The tests continued with the coupling of the aneroid manometers to the flasks and the addition of bacterial suspensions and sterile 0.9% saline solution in the flask contents, followed by incubation in a bacteriological oven and reading as previously described.
2.8. Statistical Analysis
Initially, the data were submitted to an analysis of normal distribution using Shapiro-wilk’s method. Then, the homogeneity of variances or heteroscedasticity were performed using Bartlett’s test (for gaussian data) or Levene’s test (for non-gaussian data). Gaussian data were expressed as mean ± standard error of the mean. Non-gaussian data were expressed as median and interquartile range (iqr). Differences between gaussian data means and or with homoscedasticity were analyzed using the analysis of variance (ANOVA) test, followed by the Tukey and Dunnett multiple comparisons test. Differences between non-gaussian and/or heteroscedasticity data means were analyzed using the Kruskal–Wallis test, followed by the Tukey multiple comparison test. Differences between means were considered significant when the p-value is less than 0.05. Graph pad prism version 8.0.2 was used to perform the calculations.
3. Results and Discussion
3.1. Performance and Selection of Different Culture Media
To determine the best culture broth and detect the presence of bacteria by pressure variation, preliminary tests were necessary. The results depicted in Figure 2, using Enterobacter cloacae ATCC 13,047 revealed no significant differences in pressures between the culture media used during the first 4 h (p > 0.05). Between 5 and 6 h, a significant pressure increase was observed in BHI broth in response to the production of gases inside the flasks when compared to the other evaluated broths (TSB and THIO) (p < 0.05). Moreover, a significant increase in pressure was observed in BHI and TSB, and both presented pressures of 300 ± 0 mmHg (maximum pressure that the device can detect) after 24 h, when compared with THIO broth (p < 0.05). These results demonstrate that, although all the broth media tested are excellent culture media for the recovery of microorganisms, BHI and TSB were considered the most efficient in terms of gas production.
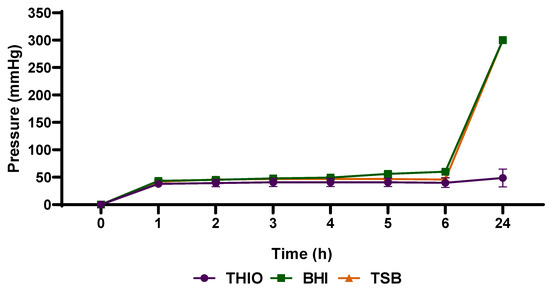
Figure 2.
Analysis of different culture media for the detection of increased pressure after gas production by Enterobacter cloacae ATCC 13047. Data are expressed as mean ± standard deviation from the mean. Significant difference between groups: p < 0.05 (ANOVA followed by Tukey’s test).
As shown in Figure 3, the cultivation of Staphylococcus aureus ATCC 25,923 in blood culture flasks containing BHI broth was also efficient to allow the detection of significant pressure increase after 5 h. Therefore, because of its excellent performance in allowing the bacteria to produce gases in sufficient quantity to determine significant pressure variations inside the flasks, BHI broth was the medium chosen for carrying out the following tests. As there is no ideal culture medium for the isolation of microorganisms derived from the bloodstream [36,37], this study sought to evaluate the ability of bacteria to grow and simultaneously produce or consume enough gases to generate pressure variations in different media culture routinely applied for blood culture. Culture broths support the growth of a wide range of bacterial species such as tryptone soy broth (TSB), which favors the growth of yeasts, fungi and aerobic bacteria-and brain heart infusion (BHI), which promotes growth of streptococci, pneumococci, meningococci, enterobacteria, non-fermenters, yeasts and fungi [38]. Thioglycolate is another commonly used broth that favors the growth of aerobic, facultative and mainly anaerobic microorganisms, due to the O2 reducing capacity [36,39]. The growth of these microorganisms will depend on the constituents of the media as well as the presence of oxygen in the flasks.
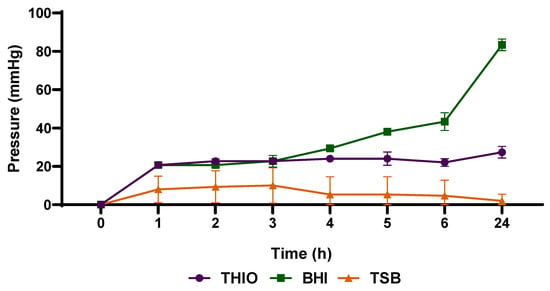
Figure 3.
Analysis of different culture media in detecting the increase in pressure after gas production by Staphylococcus aureus ATCC 25923. Data are expressed as mean ± standard deviation from the mean. Significant difference between groups: p < 0.05 (ANOVA followed by Tukey’s test).
The majority of laboratories have commonly used a set of two bottles, including aerobic and anaerobic atmospheres, to perform routine blood cultures [40]. The use of the two atmospheres, combined with the same culture medium, aims to maximize the yield of obligately aerobic microorganisms, obligately anaerobes and facultative anaerobes. It is worth mentioning that the BHI broth in an aerobic atmosphere favored the growth and production of gases by the bacteria used here. TSB and THIO broths, despite allowing the growth of microorganisms, were not efficient for the production of gases when compared to BHI.
The choice of bacteria Enterobacter cloacae ATCC 13,047 and Staphylococcus aureus ATCC 25,923 for this study is justified because they are facultative anaerobic capable of using fermentation and respiration as an energy source and consequently gas production. In addition, these species represent the two major groups of pathogenic bacteria, gram negative and gram positive, respectively [41].
3.2. Definition of the Maximum and Minimum Pressure for the Detection Time of Microbial Activity
According to the data depicted in Figure 4, pressures varied substantially over time during analyzes using BHI flasks. The highest pressure found in bottles without microorganisms was 40 mmHg between 2 and 6 h and the lowest pressure was 42 mmHg between 21 and 24 h. These results were already expected due to physicochemical changes related to temperature, pressure and gas solubility. The positive pressures observed over time are related to the events of vaporization of the contents of the culture medium. The flasks, before being incubated in a bacteriological oven, exhibited a pressure of 0 mmHg shortly after preparation at room temperature. Inside the microbiological oven at a temperature of 35 ± 2 °C, the constituent molecules of the broth cause agitation as a result of thermal energy transfer [42]. As a result, the vaporized molecules in the headspace collide with the walls of the flasks, generating force evidenced by the increase in intra-flask pressure. More volatile substances present in the intra-vial content can also flow quickly, from liquid to gaseous [43].
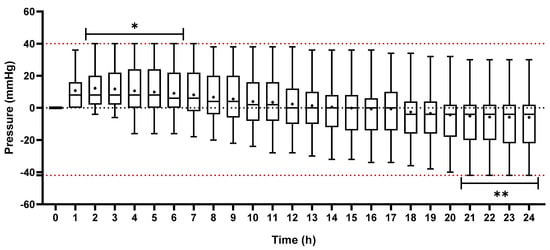
Figure 4.
Analysis of the pressure distribution in blood culture flasks containing only BHI broth and 0.9% NaCl solution for 24 h. The data are presented in mean (+) and interquartile range. The lower and upper box-plot rods represent the minimum and maximum pressures, respectively. *: maximum positive pressure measured during the experiment. **: maximum negative pressure measured during the experiment.
The vapor pressure of a liquid can be described as a colligative property related to its molecular characteristics and temperature. The lower the forces of attraction between the molecules in the liquid phase, the less difficult it is for them to flow into the vapor phase and, consequently, the greater the vapor pressure of this liquid and its volatility [44]. Additionally, each molecule has its own colligative properties. These physical phenomena explain the pressure increase in flasks containing culture medium without microorganisms. Furthermore, it is possible to observe the formation of a vacuum, evidenced by the negative pressures. Over time, a dynamic equilibrium occurs between vaporization and condensation of the vapor gas and the molecules return to the liquid phase, along with the dissolution of the pre-existing gases in the headspace of the flask. Finally, the diffusion of gases and the condensation of substances promote a decrease in pressure at a constant temperature [43,45,46].
The pressure variations in BHI culture medium without microorganism will depend on the amount of substances present in the medium. It is important to note that BHI broth is a culture medium rich in nutrients with a complex formulation derived from the infusion of brain and heart, peptone and glucose [47]. Therefore, in the current investigation it was noticed the pressure variations of BHI broth without microorganisms submitted under the same conditions as with microorganisms. Subsequently, cutoff points (intervals) for the detection of microorganisms (TTP) were also determined.
3.3. Profile of Internal Pressure Variation in Blood Culture Flasks According to Different Groups of Microorganisms X Time
As illustrated in Figure 5, Figure 6 and Figure 7, the internal pressure in blood culture bottles containing fermentative gram-negative bacilli and gram-positive cocci demonstrated a relevant increase in relation to their respective control groups (p < 0.001). These data suggest the progressive formation of gases by such microorganisms as a result of their metabolic activity. Bacteria can use different substrates and metabolic pathways to produce essential substances for their survival. Nevertheless, the production of gases and volatile substances at different levels is a common feature for all [48].
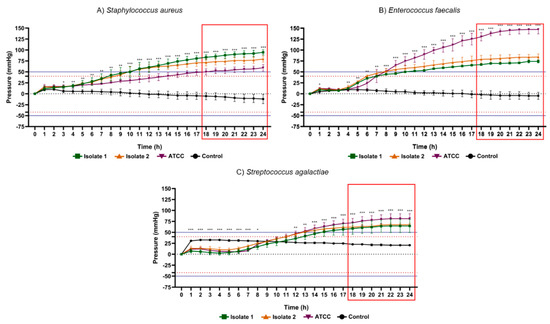
Figure 5.
Results of 24 h pressure gauge curves of Staphylococcus aureus (A), Enterococcus faecalis (B) and Streptococcus agalactiae (C) in culture flasks with BHI broth compared to the control (BHI culture medium). Data are expressed as mean ± standard deviation of the mean. Significant difference in relation to the control: * p < 0.05, ** p < 0.01 and *** p < 0.001 (ANOVA and Tukey’s test). Black dotted line: pressure 0 mmHg on the y axis. Dotted lines in red: maximum positive pressure (40 mmHg) and maximum negative pressure (−42 mmHg). Solid blue lines: (50 mmHg) and minimum (−52.5 mmHg) of pressure for positive test. Red rectangle: time-to-positivity (TTP).
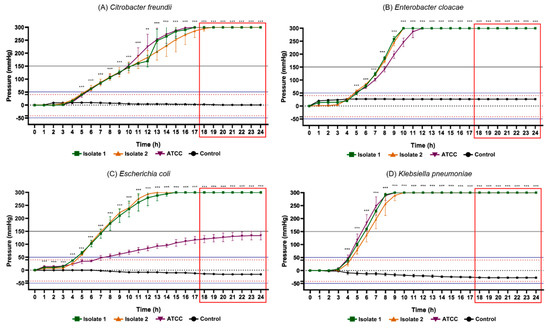
Figure 6.
Results of the 24 h pressure gauge curves of Citrobacter freundii (A), Enterobacter cloacae (B), Escherichia coli (C) and Klebsiella pneumoniae (D) in culture flasks with BHI broth compared to the control (BHI culture medium). Data are expressed as mean ± standard deviation of the mean. Significant difference in relation to the control: ** p < 0.01 and *** p < 0.001 (ANOVA and Tukey’s test). Black dotted line: pressure 0 mmHg on the y axis. Dotted lines in red: maximum positive pressure (40 mmHg) and maximum negative pressure (−42 mmHg). Solid blue lines: (50 mmHg) and minimum (−52.5 mmHg) of pressure for positive test. Red rectangle: time-to-positivity (TTP).
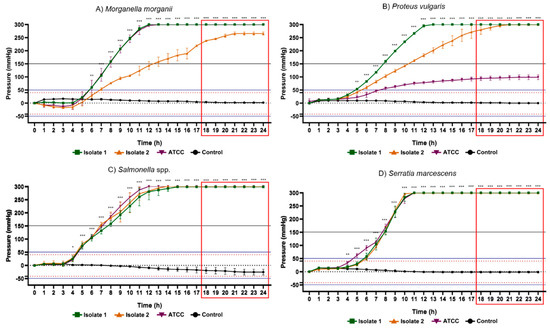
Figure 7.
Results of the 24 h pressure gauge curves of Morganella morganii (A), Proteus vulgaris (B), Salmonella spp. (C) and Serratia marcescens (D) in culture flasks with BHI broth compared with the control (BHI culture medium). Data are expressed as mean ± standard deviation of the mean. Significant difference in relation to the control: * p < 0.05, ** p < 0.01 and *** p < 0.001 (ANOVA and Tukey’s test). Black dotted line: pressure 0 mmHg on the y axis. Dotted lines in red: maximum positive pressure (40 mmHg) and maximum negative pressure (−42 mmHg). Solid blue lines: (50 mmHg) and minimum (−52.5 mmHg) of pressure for positive test. Red rectangle: time-to-positivity (TTP).
Oxidative cell respiration, or fermentation, is a metabolic pathway performed by all bacteria for energy acquisition. These are processes that involve the use of oxygen and the production of CO2. The quantity of gases used or produced will depend on the metabolic rate, availability of nutrients and number of viable bacteria. Therefore, the yield of the oxidative pathway for ATP production, despite being higher than the fermentative pathway, produces less biogas production because it occurs at a lower speed [49,50,51]. In this context, it is known that bacteria perform the form of metabolism that most benefits them energetically. The evaluated glucose-nonfermenting gram-negative rods, and gram-positive bacteria have the ability to breathe anaerobically even in the presence of oxygen. In summary, it is possible to suggest that fermentation was the pathway responsible for the highest gas production and, therefore, the results of the present study were expected [49].
Among the glucose-nonfermenting gram-negative rods evaluated in this study, all isolates of Citrobacter freundii, Enterobacter cloacae, Klebsiella pneumoniae, Salmonella spp. and Serratia marcescens exhibited the highest peaks of positive internal pressure in the flasks (300 ± 0 mmHg), while the lowest values were presented by Escherichia coli ATCC (133.3 ± 16.29 mmHg), Morganella morganii isolate 2 (265.3 ± 6.11 mmHg) and Proteus vulgaris ATCC (98.67 ± 9.01 mmHg). Concerning gram-positive cocci, Staphylococcus aureus isolate 1 (94.67 ± 5.77 mmHg), Enterococcus faecalis ATCC (147.33 ± 9.86 mmHg) and Streptococcus agalactiae ATCC (81.33 ± 11.02 mmHg) demonstrated greatest pressure increase, while Staphylococcus aureus ATCC (59.33 ± 7.57 mmHg), Enterococcus faecalis isolate 1 (74.00 ± 3.46 mmHg) and Streptococcus agalactiae isolate 1 (64.67 ± 15.11 mmHg) the lowest pressure. Accordingly, in all vials containing these bacteria, the pressure values obtained were notoriously higher than the maximum records obtained in all control groups. These data support the hypothesis that microorganisms are capable of significantly changing the internal pressure of blood culture bottles in a closed system due to their metabolic products. Therefore, the data collected in this study are relevant for the subsequent elaboration of a device capable of detecting the presence of microorganisms through pressure variations.
Glucose metabolism can lead to different products through the glycolytic pathway and these substances depends on the availability of nutrients in the culture medium used. Among those final products are lactic acid, ethanol, hydrogen, carbon dioxide and volatile fatty acids [52,53]. In this study, the BHI broth used to recover the known microorganisms proved to be effective for the production of gases by fermentative gram-negative bacilli and gram-positive bacteria.
The fermentative gram-negative bacilli studied are from the order Enterobacterales [54]. These organisms are known to be efficient in fermentation processes using organic substances as a carbon source and their main gaseous products are CO2 and H2 [55,56,57,58]. For instance, the genus Enterobacter is known for its effectiveness in the production of gases, especially hydrogen. Studies report the widespread use of this genus in fermentation processes due to its metabolic versatility and adaptive capacity to the different atmospheres imposed [59,60,61].
The gram-positive bacteria applied in this study also have facultative anaerobic metabolism [62]. A study reported by Ferreira et al. (2013) [63] demonstrated that Staphylococcus aureus can grow aerobically or anaerobically using glucose as a carbon source. The authors also reported that, in the presence of O2, S. aureus uses the oxidative pathway whereas in its absence the fermentative pathway is used producing acetate, lactate, ethanol and 2,3-butanediol as products. The Streptococcus and Enterococcus genera are also able to form products from their metabolism process [64,65,66]. The substances produced by the metabolic pathways appear to be involved in the increase of intra-bottle pressure.
Non-fermenting gram-negative bacilli reported expected results in regard to pressure variation besides negative pressure induction during the analysis time being noted. The investigated bacteria do not use the fermentative metabolic pathway; therefore, they use the oxidative respiratory pathway for glucose degradation and energy generation. Furthermore, bacteria can use oxygen and nitrogen-reducing molecules contained in the [51]. It is assumed that negative pressures are generated as a result of the diffusion of pre-existing gases in the headspace of the flasks as the bacteria use the reducing molecules [43].
As illustrated in Figure 8, glucose non-fermenting bacteria such as Acinetobacter baumannii ATCC (−142.70 ± 3.05 mmHg), Pseudomonas aeruginosa ATCC (−98.67 ± 16.29 mmHg) and Stenotrophomonas maltophilia isolate 2 (−100. 00 ± 3.46 mmHg) provided highest negative pressure peaks while the lowest values were represented by Acinetobacter baumannii isolate 2 (−114.70 ± 11.02 mmHg), Pseudomonas aeruginosa isolate 1 (−78.00 ± 23.07 mmHg) and Stenotrophomonas maltophilia ATCC (−96.00 ± 7.21 mmHg). The maximum pressures of the tested bacteria were higher than the maximum records obtained in the control groups.
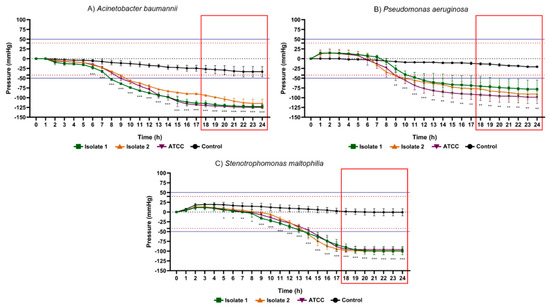
Figure 8.
Results of the 24 h pressure gauge curves of Acinetobacter baumannii (A), Pseudomonas aeruginosa (B) and Stenotrophomonas maltophilia (C) in culture flasks with BHI broth compared to the control (BHI culture medium). Data are expressed as mean ± standard deviation of the mean. Significant difference in relation to the control: * p < 0.05, ** p < 0.01 and *** p < 0.001 (ANOVA and Tukey’s test). Black dotted line: pressure 0 mmHg on the y axis. Dotted lines in red: maximum positive pressure (40 mmHg) and maximum negative pressure (−42 mmHg). Solid blue lines: (50 mmHg) and minimum (−52.5 mmHg) of pressure for positive test. Red rectangle: time-to-positivity (TTP).
Glucose metabolism in nonfermenting gram-negative bacilli occurs via phosphorylation to glucose-6-phosphate or through direct oxidation to gluconate, catalyzed by membrane-bound glucose dehydrogenase. Glucose dehydrogenase in turn transfers electrons from carbohydrates directly to the electron transport chain [67]. The final electron acceptors of most species in this group are O2, although other molecules such as nitrate can be used [68,69,70]. Therefore, we can correlate the negative pressures to the vacuum generated during the condensation or diffusion of the gaseous substances present in the headspace of the blood culture flask.
3.4. Determination of Statistical Time-to-Positivity (STTP) and Time-to-Positivity (TTP)
Another aspect of great relevance was the time taken to determine the positivity rate of bacterial metabolic activity in the different microorganisms studied. Table 2 displays the statistical time-to-positivity (STTP), representing the period in which the flasks with microorganisms revealed mean values of internal pressure significantly different from the control group, and time-to-positivity (TTP), constituting the moment from of which the pressure values, in module, would be above the limits adopted for positivity of the test (~|50| mmHg) with a safety margin of 25%. The fermentative gram-negative bacilli showed STTP between 5 and 6 h, and maximum TTP (highest TTP in the group) was 14 h besides that this was the group of bacteria which mainly produced gases through their optional aerobic metabolism. Gram positive cocci exhibited STTP between 3 and 12 h and maximum TTP of 18 h. Glucose non-fermenting gram-negative rods, on the contrary, exhibited an STTP value between 6 and 10 h and a maximum TTP of 21 h.

Table 2.
Time of positivity of microorganisms under investigation. STTP: statistical time-to-positivity; TTP: time-to-positivity.
In addition, the results obtained in this current study evidence that the time of positivity of all microorganisms corroborates a study reported by Puerta-alcalde et al. (2018) [71], in which they suggest most of the microorganisms that cause BSI are positive over the first 24 h from the incubation time. Therefore, the present methodology employed demonstrated effectiveness to detect the presence of microorganisms using the BHI broth. On top of that, several aspects should be taken into account when comes to standard positive time for the detection of microorganisms in the blood. These factors include methodology, incubation time, bacterial load, volume of blood in the bottles and culture conditions [72,73,74]. Therefore, the TTP obtained with the methodology developed in this study can be considered significantly satisfactory when compared to the STTP found in some recent studies. The mean tpp found was lower (11 am) than the values reported by Guerti et al. (2011) [75] (21.33 h), Abdelhamid (2017) [76] (21.10 h) and by Ning et al. (2016) [17] (24.91 h), who used automated blood culture methodologies. In contrast, some microorganisms such as S. aureus, Streptococcus spp., Enterococcus and Acinetobacter spp. Can take from 48 h to 72 h to positive [76]. This information corroborates the findings found in this study, where after 24 h these bacteria presented the highest TTP.
In summary, automated methodologies manifest several advantages regarding the STTP when compared to manual methods [73]. Therefore, despite the manual methodology being applied in this work, the positivity rate is similar to the times resulting from automated methodologies [20,29].
In the present analysis, the behavior of the internal pressure in blood culture vials over 24 h also revealed a different profile among the microorganisms evaluated. Gram positive cocci indicated pressures between 50.0 mmHg and 150.0 mmHg, whereas fermenting gram-negative rods expressed pressures greater than 150 mmHg. On the other hand, non-fermenting gram-negative rods showed negative pressures below 50.00 mmHg. Thus, the intensity of the increase or decrease in internal pressure in blood culture bottles after 24 h also seems to provide subsidies to suggest which group of microorganisms is present in the blood culture sample.
Preliminary data are crucial and contribute to the diagnostic reasoning during the identification of the etiological agent of sepsis. Therefore, the time-to-positivity can indicate the paths by which the microbiologist will take to identify and provide preliminary information about the microorganism under analysis [77].
It is also important to note that, when we point out the bacterial load in the blood sample and the sensitivity of the methodology used, we refer to the time-to-positivity which, therefore, will be correlated with the patient’s prognosis [78]. Studies indicate that the time-to-positivity serves as an indicator of sepsis prognosis [78,79,80,81] and has been associated with increase in hospitalization rates, length of stay and hospital costs. For this reason, methodologies that provide sensitivity and specificity in the short-term diagnosis directly affect early intervention mostly regarding antimicrobial therapy.
4. Conclusions
In summary, this study demonstrated the relevance of early detection bacteremia through a new methodology system, which proved to be an efficient rapid diagnostic technology. According to the methodologies applied and within the experimental conditions implemented in the present study, it was concluded that the BHI medium proved to be the most suitable for the analysis of blood pressure behavior in blood culture vials containing different bacterial species associated with sepsis. We also found that variations in blood pressure greater than |50| mmHg are indicative of the presence of microorganisms in blood culture flasks. Gram positive cocci and glucose non-fermenting gram-negative rods promote an increase in blood pressure, whereas non-fermenting gram-negative rods cause a progressive reduction in blood pressure values in blood culture vials over time. Pressure behavior (increase or decrease in intra-bottle pressure), as well as the magnitude of the increase in pressure values, may suggest the group of bacteria present in blood culture vials. The maximum time required to guarantee the positivity of the tested method is 18 h for positive pressures and 21 for negative pressures. The methodology based on the detection of variations in pressure behavior in blood culture bottles developed in this study for the early detection of microorganisms responsible for BSI was effective. To summarize, the findings of our study suggest that microorganisms that cause BSI can be detected in blood culture vials by varying intra-vial pressure, and can be a straight-forward diagnostic kit for the early detection of pathogenic bioaerosols in the blood. Subsequent studies to determine the minimum amount of detectable microorganism and tests to evaluate the effectiveness compared to standard methods are suggested towards the improvement of the methodology in relation to its sensitivity.
Author Contributions
A.G.d.S.-N., M.S.P., M.C.d.S., L.L.A. and R.R.S.P. have written and formatted the review. J.C.C., P.S. and R.L.C.d.A.-J. have reviewed and edited. A.G.d.S.-N., M.S.P., M.C.d.S., L.L.A. and R.R.S.P. have contributed to the writing and preparation of figures and tables. A.G.d.S.-N., M.S.P., L.L.A., J.C.C., E.B.S. and R.L.C.d.A.-J. have conceptualized, structured, reviewed and supervised the literature research. All authors have read and agreed to the published version of the manuscript.
Funding
This work has been funded by the Fundação de Apoio à Pesquisa e à Inovação Tecnológica do Estado de Sergipe-Program Centelha (FAPITEC/SE) and by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Portuguese Science and Technology Foundation (FCT) through the project UIDB/04469/2020 (strategic fund) funded by national funds, and co-financed Education (FCT/MEC) from national funds and FEDER, under the Partnership Agreement PT2020.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available from corresponding authors upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Deisz, R.; Rademacher, S.; Gilger, K.; Jegen, R.; Sauerzapfe, B.; Fitzner, C.; Stoppe, C.; Benstoem, C.; Marx, G. Additional Telemedicine Rounds as a Successful Performance-Improvement Strategy for Sepsis Management: Observational Multicenter Study. J. Med. Internet Res. 2019, 21, e11161. [Google Scholar] [CrossRef] [PubMed]
- Tillmann, B.; Wunsch, H. Epidemiology and Outcomes. Crit. Care Clin. 2018, 34, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, G.O.; Castell, C.D. Epidemiologia das infecções graves nas unidades de terapia intensiva latino-americanas. Rev. Bras. Ter. Intensiva 2016, 28, 261–263. [Google Scholar]
- World Health Organization. Sepsis. Available online: https://www.who.int/news-room/fact-sheets/detail/sepsis (accessed on 28 April 2021).
- Quintano Neira, R.A.; Hamacher, S.; Japiassú, A.M. Epidemiology of sepsis in Brazil: Incidence, lethality, costs, and other indicators for Brazilian Unified Health System hospitalizations from 2006 to 2015. PLoS ONE 2018, 13, e0195873. [Google Scholar] [CrossRef] [PubMed]
- Buckman, S.A.; Turnbull, I.R.; Mazuski, J.E. Empiric Antibiotics for Sepsis. Surg. Infect. 2018, 19, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Fouks, Y.; Samueloff, O.; Levin, I.; Many, A.; Amit, S.; Cohen, A. Assessing the effectiveness of empiric antimicrobial regimens in cases of septic/infected abortions. Am. J. Emerg. Med. 2020, 38, 1123–1128. [Google Scholar] [CrossRef]
- Robakowska, M.; Bronk, M.; Tyrańska-Fobke, A.; Ślęzak, D.; Kraszewski, J.; Balwicki, Ł. Patient Safety Related to Microbiological Contamination of the Environment of a Multi-Profile Clinical Hospital. Int. J. Environ. Res. Public Health 2021, 18, 3844. [Google Scholar] [CrossRef]
- Jankowiak, E.; Kubera, Ł.; Małecka-Adamowicz, M.; Dembowska, E. Microbiological air quality in pharmacies and an antibiotic resistance profile of staphylococci species. Aerobiologia 2020, 36, 551–563. [Google Scholar] [CrossRef]
- Rhodes, A.; Evans, L.E.; Alhazzani, W.; Levy, M.M.; Antonelli, M.; Ferrer, R.; Kumar, A.; Sevransky, J.E.; Sprung, C.L.; Nunnally, M.E.; et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock: 2016. Intensive Care Med. 2017, 43, 304–377. [Google Scholar] [CrossRef]
- Opota, O.; Croxatto, A.; Prod’hom, G.; Greub, G. Blood culture-based diagnosis of bacteraemia: State of the art. Clin. Microbiol. Infect. 2015, 21, 313–322. [Google Scholar] [CrossRef]
- Rivera-Izquierdo, M.; Benavente-Fernández, A.; López-Gómez, J.; Láinez-Ramos-Bossini, A.J.; Rodríguez-Camacho, M.; Valero-Ubierna, M.D.C.; Martín-delosReyes, L.M.; Jiménez-Mejías, E.; Moreno-Roldán, E.; Lardelli-Claret, P.; et al. Prevalence of Multi-Resistant Microorganisms and Antibiotic Stewardship among Hospitalized Patients Living in Residential Care Homes in Spain: A Cross-Sectional Study. Antibiotics 2020, 9, 324. [Google Scholar] [CrossRef]
- Tassinari, M.; Zannoli, S.; Farabegoli, P.; Pedna, M.F.; Pierro, A.; Mastroianni, A.; Fontan, R.; Luongo, L.; Sarnataro, G.; Menegatti, E.; et al. Rapid diagnosis of bloodstream infections in the critically ill: Evaluation of the broad-range PCR/ESI-MS technology. PLoS ONE 2018, 13, e0197436. [Google Scholar] [CrossRef]
- Ghosh, S.; Basu, G. A hospital based study on clinico microbiological profile of neonatal septicemia. Asian J. Med. Sci. 2018, 9, 25–30. [Google Scholar] [CrossRef]
- Timsit, J.-F.; Ruppé, E.; Barbier, F.; Tabah, A.; Bassetti, M. Bloodstream infections in critically ill patients: An expert statement. Intensive Care Med. 2020, 46, 266–284. [Google Scholar] [CrossRef]
- Chung, Y.; Kim, I.H.; Han, M.; Kim, H.S.; Kim, H.S.; Song, W.; Kim, J.S. A comparative evaluation of BACT/ALERT FA PLUS and FN PLUS blood culture bottles and BD BACTEC Plus Aerobic and Anaerobic blood culture bottles for antimicrobial neutralization. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 2229–2233. [Google Scholar] [CrossRef]
- Ning, Y.; Hu, R.; Yao, G.; Bo, S. Time to positivity of blood culture and its prognostic value in bloodstream infection. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 619–624. [Google Scholar] [CrossRef]
- Somily, A.M.; Habib, H.A.; Torchyan, A.A.; Sayyed, S.B.; Absar, M.; Al-Aqeel, R.; Binkhamis, A.K. Time-to-detection of bacteria and yeast with the BACTEC FX versus BacT/Alert Virtuo blood culture systems. Ann. Saudi Med. 2018, 38, 194–199. [Google Scholar] [CrossRef]
- Atkinson-Dunn, R.; Michael Dunne, W., Jr. Conventional Blood Culture Methods. In The Dark Art of Blood Cultures; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2017; pp. 21–38. [Google Scholar] [CrossRef]
- Li, Z.; Liu, S.; Chen, H.; Zhang, X.; Ling, Y.; Zhang, N.; Hou, T. Comparative evaluation of BACTEC FX, BacT/ALERT 3D, and BacT/ALERT VIRTUO-automated blood culture systems using simulated blood cultures. Acta Clin. Belg. 2020, 7, 1–8. [Google Scholar] [CrossRef]
- Ahmad, A.; Iram, S.; Hussain, S.; Yusuf, N.W. Diagnosis of paediatric sepsis by automated blood culture system and conventional blood culture. J. Pak. Med. Assoc. 2017, 67, 192–195. [Google Scholar] [PubMed]
- Wilson, M.L.; Weinstein, M.P.; Reller, L.B. Commercial Blood Culture Systems and Methods. In Manual of Commercial Methods in Clinical Microbiology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; pp. 11–20. [Google Scholar] [CrossRef]
- Palmer, H.R.; Palavecino, E.L.; Johnson, J.W.; Ohl, C.A.; Williamson, J.C. Clinical and microbiological implications of time-to-positivity of blood cultures in patients with Gram-negative bacilli bacteremia. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 955–959. [Google Scholar] [CrossRef] [PubMed]
- Paoli, C.J.; Reynolds, M.A.; Sinha, M.; Gitlin, M.; Crouser, E. Epidemiology and Costs of Sepsis in the United States-An Analysis Based on Timing of Diagnosis and Severity Level. Crit. Care Med. 2018, 46, 1889–1897. [Google Scholar] [CrossRef]
- Blaschke, A.J.; Hersh, A.L.; Beekmann, S.E.; Ince, D.; Polgreen, P.M.; Hanson, K.E. Unmet diagnostic needs in infectious disease. Diagn. Microbiol. Infect. Dis. 2015, 81, 57–59. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Caliendo, A.M.; Gilbert, D.N.; Ginocchio, C.C.; Hanson, K.E.; May, L.; Quinn, T.C.; Tenover, F.C.; Alland, D.; Blaschke, A.J.; Bonomo, R.A.; et al. Better tests, better care: Improved diagnostics for infectious diseases. Clin. Infect. Dis. 2013, 57 (Suppl. S3), S139–S170. [Google Scholar] [CrossRef] [PubMed]
- Kocincová, A.S.; Nagl, S.; Arain, S.; Krause, C.; Borisov, S.M.; Arnold, M.; Wolfbeis, O.S. Multiplex bacterial growth monitoring in 24-well microplates using a dual optical sensor for dissolved oxygen and pH. Biotechnol. Bioeng. 2008, 100, 430–438. [Google Scholar] [CrossRef]
- Lazcka, O.; Del Campo, F.J.; Muñoz, F.X. Pathogen detection: A perspective of traditional methods and biosensors. Biosens. Bioelectron. 2007, 22, 1205–1217. [Google Scholar] [CrossRef]
- Menchinelli, G.; Liotti, F.M.; Fiori, B.; De Angelis, G.; D’Inzeo, T.; Giordano, L.; Posteraro, B.; Sabbatucci, M.; Sanguinetti, M.; Spanu, T. In vitro Evaluation of BACT/ALERT® VIRTUO®, BACT/ALERT 3D®, and BACTEC™ FX Automated Blood Culture Systems for Detection of Microbial Pathogens Using Simulated Human Blood Samples. Front. Microbiol. 2019, 10, 221. [Google Scholar] [CrossRef]
- Dreyer, A.W.; Ismail, N.A.; Nkosi, D.; Lindeque, K.; Matthews, M.; van Zyl, D.G.; Hoosen, A.A. Comparison of the VersaTREK blood culture system against the Bactec9240 system in patients with suspected bloodstream infections. Ann. Clin. Microbiol. Antimicrob. 2011, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Yue, P.; Zhou, M.; Kudinha, T.; Xie, X.; Du, J.; Song, H.; Zhang, L.; Ma, X.; Weng, L.; Chai, W.; et al. Clinical Performance Evaluation of VersaTrek 528 Blood Culture System in a Chinese Tertiary Hospital. Front. Microbiol. 2018, 9, 2027. [Google Scholar] [CrossRef] [PubMed]
- Samuel, L.P.; Pimentel, J.D.; Tibbetts, R.J.; Martin, R.; Hensley, R.; Meier, F.A. Comparison of time to positivity of the VersaTREK® REDOX 80-mL and the REDOX EZ draw 40-mL blood culture bottles for common bacterial bloodstream pathogens. Diagn. Microbiol. Infect. Dis. 2011, 71, 101–105. [Google Scholar] [CrossRef]
- Baykara, N.; Akalın, H.; Arslantaş, M.K.; Hancı, V.; Çağlayan, Ç.; Kahveci, F.; Demirağ, K.; Baydemir, C.; Ünal, N. Epidemiology of sepsis in intensive care units in Turkey: A multicenter, point-prevalence study. Crit. Care 2018, 22, 93. [Google Scholar] [CrossRef]
- Yusef, D.; Shalakhti, T.; Awad, S.; Algharaibeh, H.; Khasawneh, W. Clinical characteristics and epidemiology of sepsis in the neonatal intensive care unit in the era of multi-drug resistant organisms: A retrospective review. Pediatr. Neonatol. 2018, 59, 35–41. [Google Scholar] [CrossRef]
- World Health Organization. Global Report on the Epidemiology and Burden of Sepsis: Current Evidence, Identifying Gaps and Future Directions; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Hansen, G.T. Laboratory Blood Cultures: Past, Present, and Future. Clin. Microbiol. Newsl. 2016, 38, 119–128. [Google Scholar] [CrossRef]
- Pashirova, T.N.; Lukashenko, S.S.; Zakharov, S.V.; Voloshina, A.D.; Zhiltsova, E.P.; Zobov, V.V.; Souto, E.B.; Zakharova, L.Y. Self-assembling systems based on quaternized derivatives of 1,4-diazabicyclo[2.2.2]octane in nutrient broth as antimicrobial agents and carriers for hydrophobic drugs. Colloids Surf. B Biointerfaces 2015, 127, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Reimer, L.G.; Reller, L.B.; Wang, W.L.; Mirrett, S. Controlled evaluation of trypticase soy broth with and without gelatin and yeast extract in the detection of bacteremia and fungemia. Diagn. Microbiol. Infect. Dis. 1987, 8, 19–24. [Google Scholar] [CrossRef]
- Ganguli, L.A.; Turton, L.J.; Tillotson, G.S. Evaluation of Fastidious Anaerobe Broth as a blood culture medium. J. Clin. Pathol. 1982, 35, 458–461. [Google Scholar] [CrossRef]
- Riley, J.A.; Heiter, B.J.; Bourbeau, P.P. Comparison of recovery of blood culture isolates from two BacT/ALERT FAN aerobic blood culture bottles with recovery from one FAN aerobic bottle and one FAN anaerobic bottle. J. Clin. Microbiol. 2003, 41, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Shoji, K.; Komuro, H.; Watanabe, Y.; Miyairi, I. The utility of anaerobic blood culture in detecting facultative anaerobic bacteremia in children. Diagn. Microbiol. Infect. Dis. 2013, 76, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Camel, T.d.O.; Filgueiras, C.A.L. A importância da lei de Gay-Lussac para a classificação dos compostos orgânicos. Química Nova 2013, 36, 738–747. [Google Scholar] [CrossRef]
- Kast, W.; Hohenthanner, C.R. Mass transfer within the gas-phase of porous media. Int. J. Heat Mass Transf. 2000, 43, 807–823. [Google Scholar] [CrossRef]
- Yeon, J.-W.; Jung, S.-H. Effects of temperature and solution composition on evaporation of iodine as a part of estimating volatility of iodine under gamma irradiation. Nucl. Eng. Technol. 2017, 49, 1689–1695. [Google Scholar] [CrossRef]
- Hashim, F.A.; Houssein, E.H.; Mabrouk, M.S.; Al-Atabany, W.; Mirjalili, S. Henry gas solubility optimization: A novel physics-based algorithm. Future Gener. Comput. Syst. 2019, 101, 646–667. [Google Scholar] [CrossRef]
- Petropoulos, J.H.; Havredaki, V.I. On the fundamental concepts underlying Henry-law adsorption and adsorbed gas transport in porous solids. J. Chem. Soc. Faraday Trans. 1986, 82, 2531–2545. [Google Scholar] [CrossRef]
- Jawan, R.; Abbasiliasi, S.; Tan, J.S.; Mustafa, S.; Halim, M.; Ariff, A.B. Influence of Culture Conditions and Medium Compositions on the Production of Bacteriocin-Like Inhibitory Substances by Lactococcus lactis Gh1. Microorganisms 2020, 8, 1454. [Google Scholar] [CrossRef]
- Ladygina, N.; Dedyukhina, E.G.; Vainshtein, M.B. A review on microbial synthesis of hydrocarbons. Process. Biochem. 2006, 41, 1001–1014. [Google Scholar] [CrossRef]
- Bouvet, O.M.M.; Lenormand, P.; Ageron, E.; Grimont, P.A.D. Taxonomic diversity of anaerobic glycerol dissimilation in the Enterobacteriaceae. Res. Microbiol. 1995, 146, 279–290. [Google Scholar] [CrossRef]
- Freude, C.; Blaser, M. Carbon Isotope Fractionation during Catabolism and Anabolism in Acetogenic Bacteria Growing on Different Substrates. Appl. Environ. Microbiol. 2016, 82, 2728–2737. [Google Scholar] [CrossRef] [PubMed]
- Qadri, S.M.; Carr, D.L.; Narayanan, R.; Ottman, J. Accuracy of a rapid carbohydrate oxidation microtube method for identification of nonfermentative gram-negative bacilli. J. Clin. Microbiol. 1982, 15, 43–47. [Google Scholar] [CrossRef] [PubMed]
- de Kok, S.; Meijer, J.; van Loosdrecht, M.C.; Kleerebezem, R. Impact of dissolved hydrogen partial pressure on mixed culture fermentations. Appl. Microbiol. Biotechnol. 2013, 97, 2617–2625. [Google Scholar] [CrossRef]
- Temudo, M.F.; Kleerebezem, R.; van Loosdrecht, M. Influence of the pH on (open) mixed culture fermentation of glucose: A chemostat study. Biotechnol. Bioeng. 2007, 98, 69–79. [Google Scholar] [CrossRef]
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar] [CrossRef]
- Wang, J.; Yin, Y. Fermentative hydrogen production using pretreated microalgal biomass as feedstock. Microb. Cell Fact. 2018, 17, 22. [Google Scholar] [CrossRef] [PubMed]
- Bott, M. Anaerobic citrate metabolism and its regulation in enterobacteria. Arch. Microbiol. 1997, 167, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Song, W.; Cheng, J.; Liu, M.; Zhang, C.; Cen, K. Improvement of fermentative hydrogen production using genetically modified Enterobacter aerogenes. Int. J. Hydrog. Energy 2017, 42, 3676–3681. [Google Scholar] [CrossRef]
- Yazdani, S.S.; Gonzalez, R. Anaerobic fermentation of glycerol: A path to economic viability for the biofuels industry. Curr. Opin. Biotechnol. 2007, 18, 213–219. [Google Scholar] [CrossRef]
- Barbirato, F.; Soucaille, P.; Bories, A. Physiologic Mechanisms Involved in Accumulation of 3-Hydroxypropionaldehyde during Fermentation of Glycerol by Enterobacter agglomerans. Appl. Environ. Microbiol. 1996, 62, 4405–4409. [Google Scholar] [CrossRef] [PubMed]
- Converti, A.; Perego, P. Use of carbon and energy balances in the study of the anaerobic metabolism of Enterobacter aerogenes at variable starting glucose concentrations. Appl. Microbiol. Biotechnol. 2002, 59, 303–309. [Google Scholar] [CrossRef]
- Pachapur, V.L.; Sarma, S.J.; Brar, S.K.; Le Bihan, Y.; Buelna, G.; Verma, M. Biohydrogen production by co-fermentation of crude glycerol and apple pomace hydrolysate using co-culture of Enterobacter aerogenes and Clostridium butyricum. Bioresour. Technol. 2015, 193, 297–306. [Google Scholar] [CrossRef]
- Post, K.W. Overview of Bacteria. In Diseases of Swine; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2019; pp. 743–748. [Google Scholar] [CrossRef]
- Ferreira, M.T.; Manso, A.S.; Gaspar, P.; Pinho, M.G.; Neves, A.R. Effect of oxygen on glucose metabolism: Utilization of lactate in Staphylococcus aureus as revealed by in vivo NMR studies. PLoS ONE 2013, 8, e58277. [Google Scholar] [CrossRef]
- Doi, Y. Lactic acid fermentation is the main aerobic metabolic pathway in Enterococcus faecalis metabolizing a high concentration of glycerol. Appl. Microbiol. Biotechnol. 2018, 102, 10183–10192. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y. L-lactate production from biodiesel-derived crude glycerol by metabolically engineered Enterococcus faecalis: Cytotoxic evaluation of biodiesel waste and development of a glycerol-inducible gene expression system. Appl. Environ. Microbiol. 2015, 81, 2082–2089. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Poyart, C.; Trieu-Cuot, P.; Lamberet, G.; Gruss, A.; Gaudu, P. Respiration metabolism of Group B Streptococcus is activated by environmental haem and quinone and contributes to virulence. Mol. Microbiol. 2005, 56, 525–534. [Google Scholar] [CrossRef] [PubMed]
- van Schie, B.J.; Hellingwerf, K.J.; van Dijken, J.P.; Elferink, M.G.; van Dijl, J.M.; Kuenen, J.G.; Konings, W.N. Energy transduction by electron transfer via a pyrrolo-quinoline quinone-dependent glucose dehydrogenase in Escherichia coli, Pseudomonas aeruginosa, and Acinetobacter calcoaceticus (var. lwoffi). J. Bacteriol. 1985, 163, 493–499. [Google Scholar] [CrossRef]
- Jia, Y.; Zhou, M.; Chen, Y.; Luo, J.; Hu, Y. Carbon selection for nitrogen degradation pathway by Stenotrophomonas maltophilia: Based on the balances of nitrogen, carbon and electron. Bioresour. Technol. 2019, 294, 122114. [Google Scholar] [CrossRef] [PubMed]
- Peleg, A.Y.; de Breij, A.; Adams, M.D.; Cerqueira, G.M.; Mocali, S.; Galardini, M.; Nibbering, P.H.; Earl, A.M.; Ward, D.V.; Paterson, D.L.; et al. The success of acinetobacter species; genetic, metabolic and virulence attributes. PLoS ONE 2012, 7, e46984. [Google Scholar] [CrossRef]
- Su, W.; Zhang, L.; Li, D.; Zhan, G.; Qian, J.; Tao, Y. Dissimilatory nitrate reduction by Pseudomonas alcaliphila with an electrode as the sole electron donor. Biotechnol. Bioeng. 2012, 109, 2904–2910. [Google Scholar] [CrossRef] [PubMed]
- Puerta-Alcalde, P.; Cardozo, C.; Suárez-Lledó, M.; Rodríguez-Núñez, O.; Morata, L.; Fehér, C.; Marco, F.; Del Río, A.; Martínez, J.A.; Mensa, J.; et al. Current time-to-positivity of blood cultures in febrile neutropenia: A tool to be used in stewardship de-escalation strategies. Clin. Microbiol. Infect. 2019, 25, 447–453. [Google Scholar] [CrossRef]
- Jones, R.L.; Sayles, H.R.; Fey, P.D.; Rupp, M.E. Effect of Clinical Variables on the Volume of Blood Collected for Blood Cultures in an Adult Patient Population. Infect. Control. Hosp. Epidemiol. 2017, 38, 1493–1497. [Google Scholar] [CrossRef] [PubMed]
- Pongsachareonnont, P.; Honglertnapakul, W.; Chatsuwan, T. Comparison of methods for identifying causative bacterial microorganisms in presumed acute endophthalmitis: Conventional culture, blood culture, and PCR. BMC Infect. Dis. 2017, 17, 165. [Google Scholar] [CrossRef]
- Schwarzenbacher, J.; Kuhn, S.O.; Vollmer, M.; Scheer, C.; Fuchs, C.; Rehberg, S.; Balau, V.; Hahnenkamp, K.; Bohnert, J.A.; Gründling, M. On-site blood culture incubation shortens the time to knowledge of positivity and microbiological results in septic patients. PLoS ONE 2019, 14, e0225999. [Google Scholar] [CrossRef]
- Guerti, K.; Devos, H.; Ieven, M.M.; Mahieu, L.M. Time to positivity of neonatal blood cultures: Fast and furious? J. Med. Microbiol. 2011, 60, 446–453. [Google Scholar] [CrossRef]
- Abdelhamid, S.M. Time to Positivity and Antibiotic Sensitivity of Neonatal Blood Cultures. J. Glob. Infect. Dis. 2017, 9, 102–107. [Google Scholar] [CrossRef]
- Pence, M.A.; McElvania TeKippe, E.; Burnham, C.A. Diagnostic assays for identification of microorganisms and antimicrobial resistance determinants directly from positive blood culture broth. Clin. Lab. Med. 2013, 33, 651–684. [Google Scholar] [CrossRef]
- Xu, H.; Cheng, J.; Yu, Q.; Li, Q.; Yi, Q.; Luo, S.; Li, Y.; Zhang, G.; Tian, X.; Cheng, D.; et al. Prognostic role of time to positivity of blood culture in children with Pseudomonas aeruginosa bacteremia. BMC Infect. Dis. 2020, 20, 665. [Google Scholar] [CrossRef]
- Li, Y.; Li, Q.; Zhang, G.; Ma, H.; Wu, Y.; Yi, Q.; Jiang, L.; Wan, J.; Suo, F.; Luo, Z. Time to positivity of blood culture is a risk factor for clinical outcomes in Staphylococcus aureus bacteremia children: A retrospective study. BMC Infect. Dis. 2019, 19, 437. [Google Scholar] [CrossRef] [PubMed]
- Peralta, G.; Rodríguez-Lera, M.J.; Garrido, J.C.; Ansorena, L.; Roiz, M.P. Time to positivity in blood cultures of adults with Streptococcus pneumoniae bacteremia. BMC Infect. Dis. 2006, 6, 79. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.C.; Lee, C.C.; Li, C.W.; Li, M.C.; Ko, W.C.; Lee, N.Y. Time-to-positivity of blood culture: An independent prognostic factor of monomicrobial Pseudomonas aeruginosa bacteremia. J. Microbiol. Immunol. Infect. 2017, 50, 486–493. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).