The GIS2 Gene Is Repressed by a Zinc-Regulated Bicistronic RNA in Saccharomyces cerevisiae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Plasmid Constructs
2.3. Quantitative RT-PCR Analysis
2.4. Northern Blot Analysis
2.5. Protein Extraction and Immunoblotting
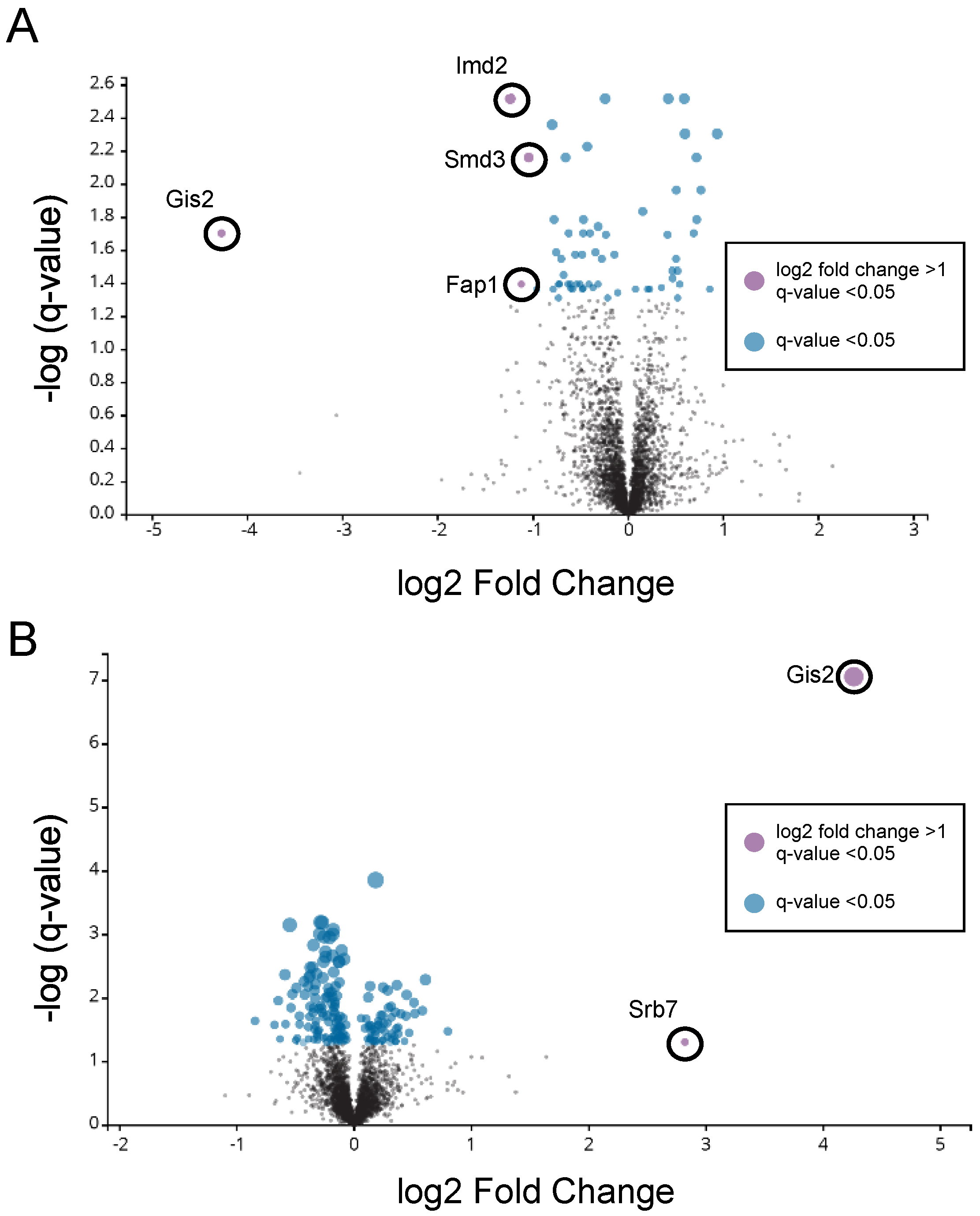
2.6. Mass Spectrometry Analysis
2.7. Competitive Growth Assays
3. Results
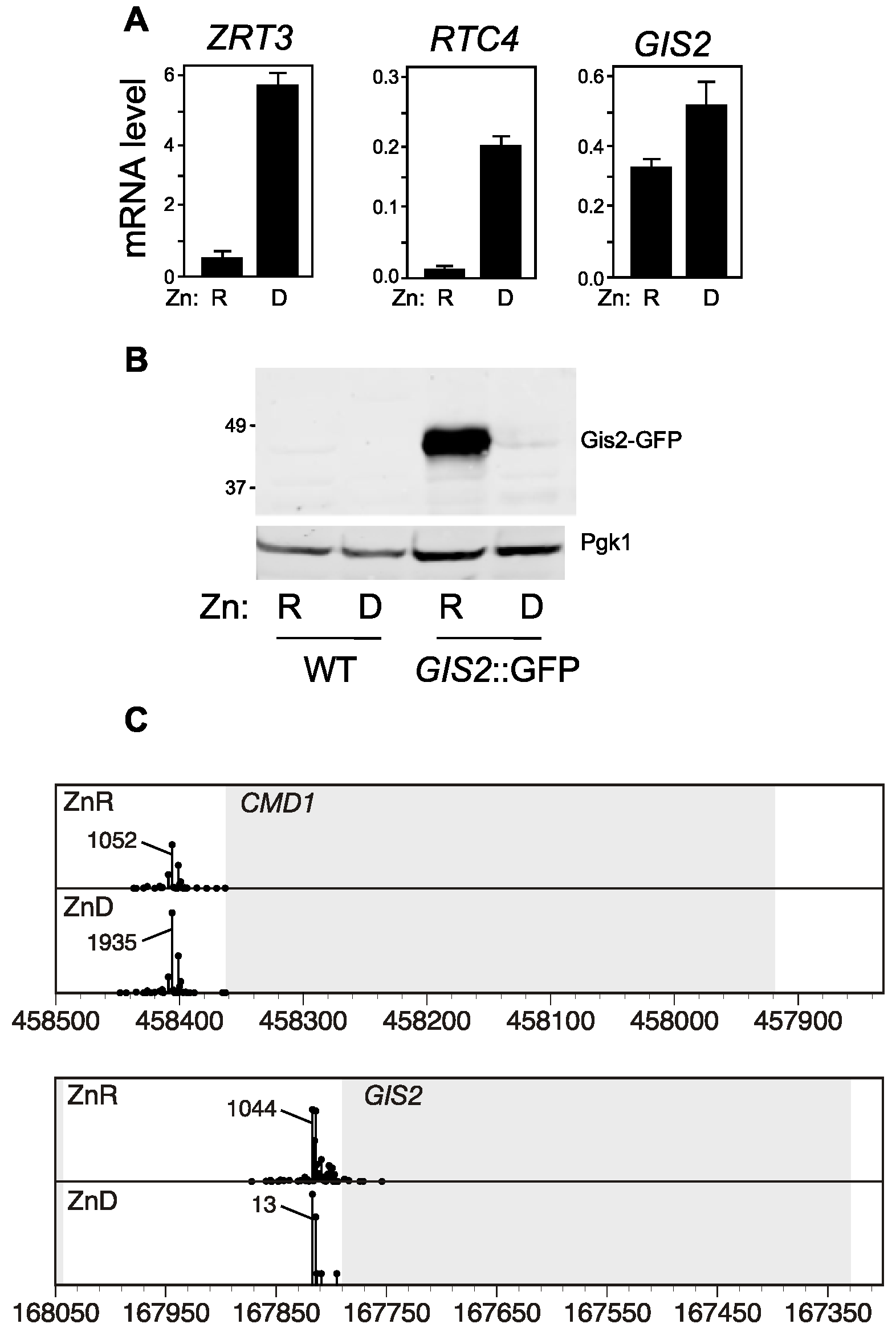
3.1. Opposite Regulation of GIS2 mRNA and Gis2 Protein Levels in Response to Zinc
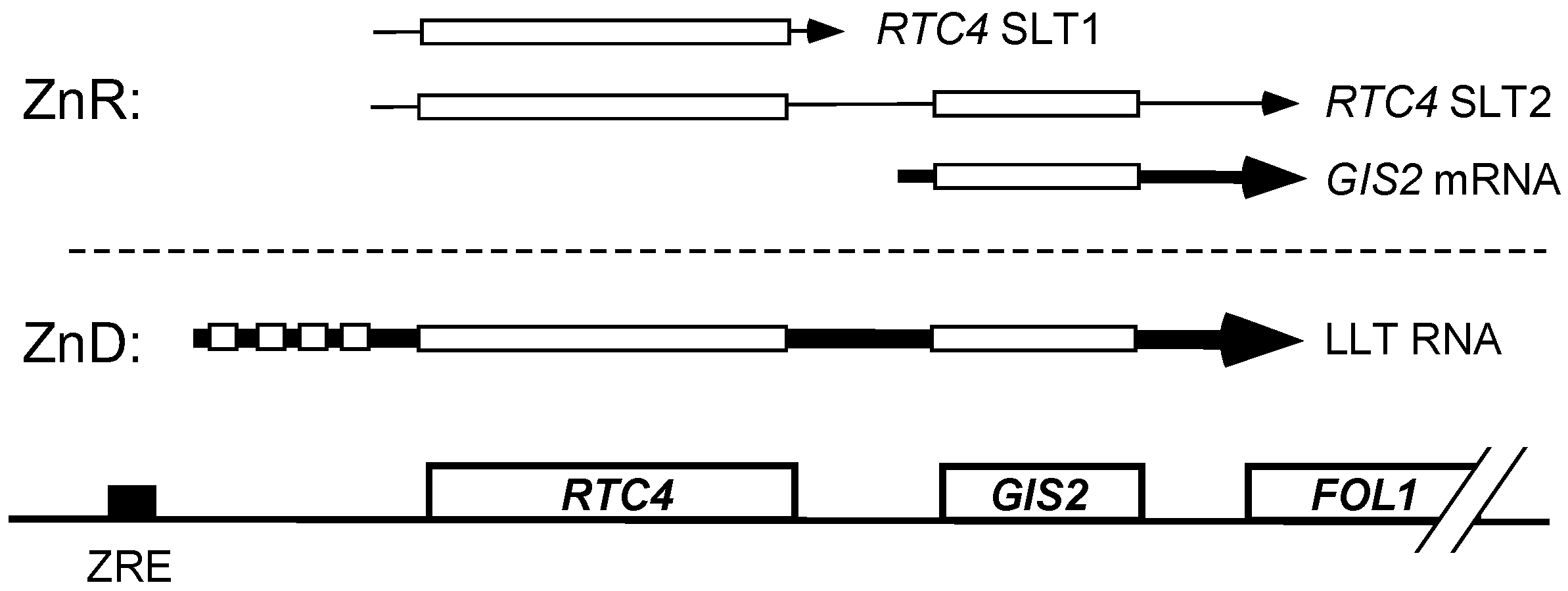
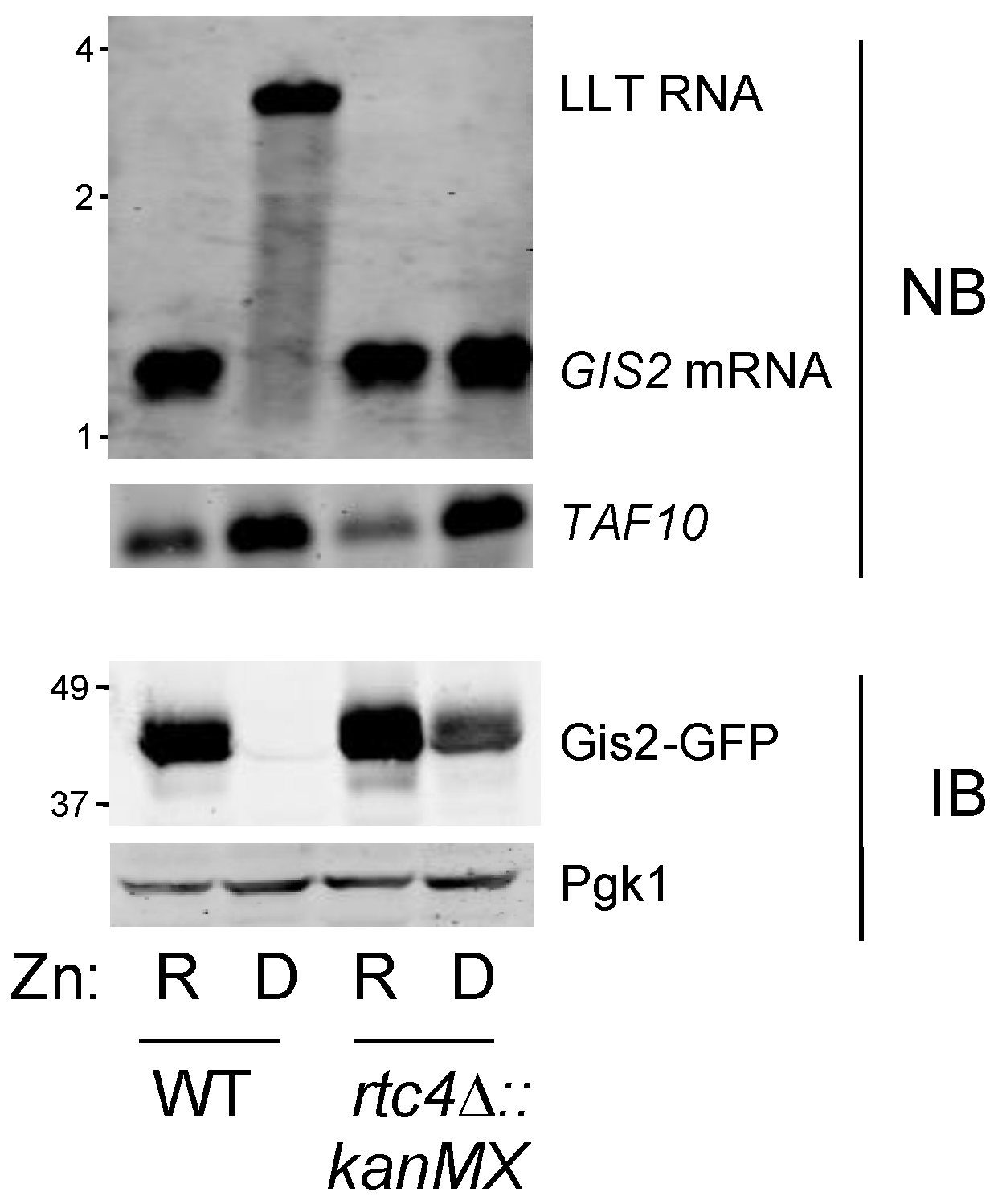
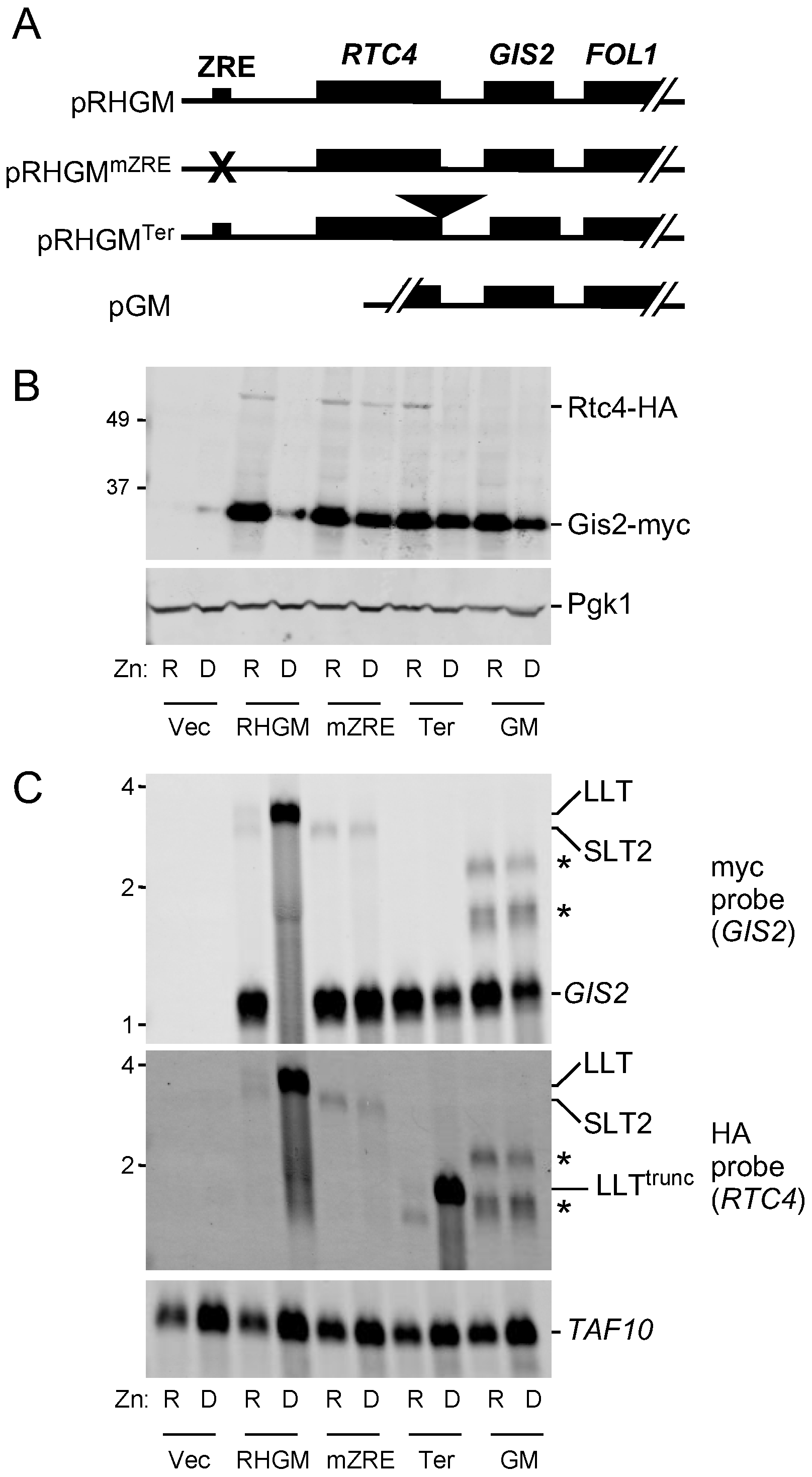
3.2. Co-Regulation of RTC4 and GIS2 by a Bicistronic Regulatory RNA
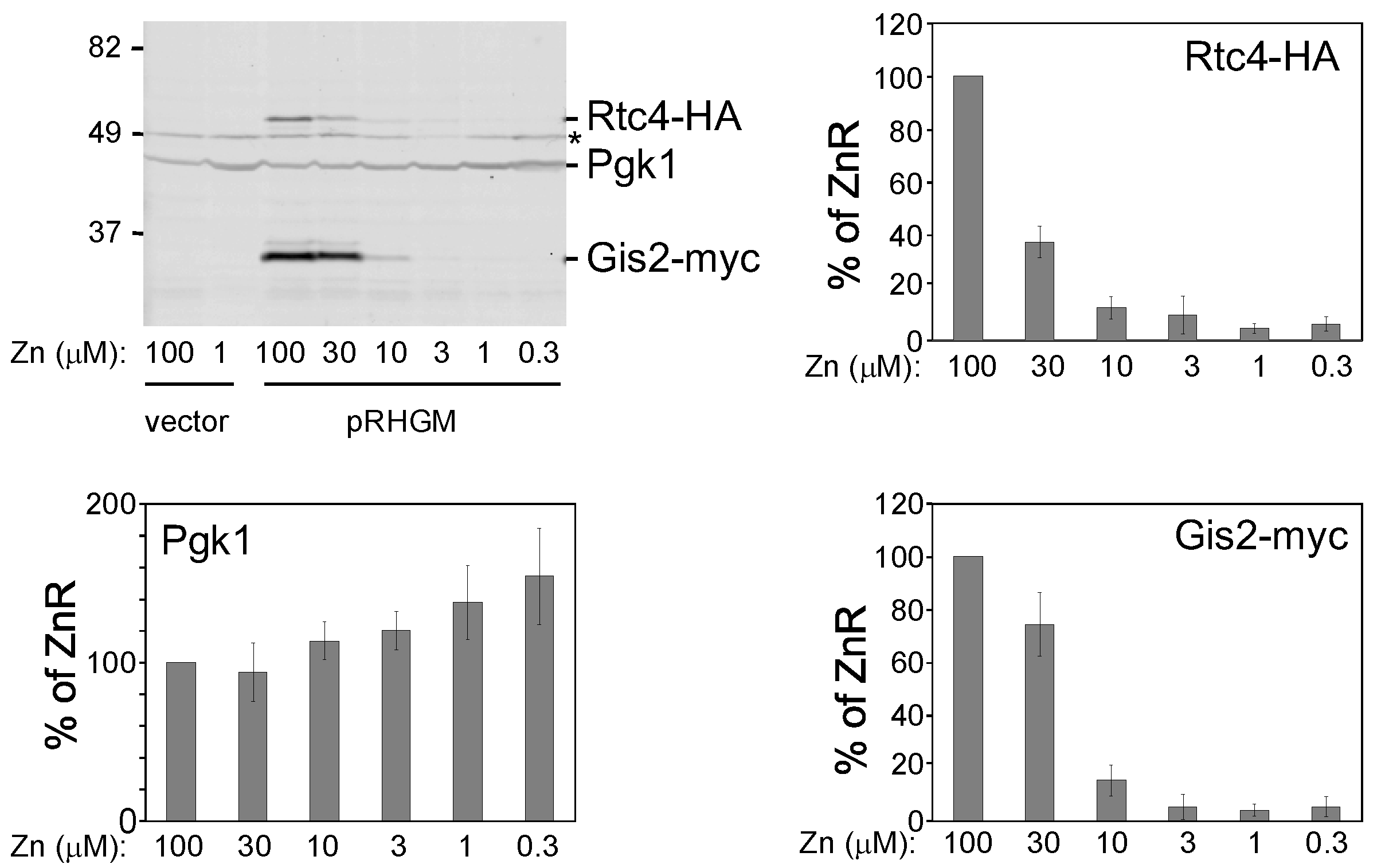
3.3. Probing the Functional Significance of GIS2 Regulation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Johnston, M.; Davis, R.W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol. Cell. Biol. 1984, 4, 1440–1448. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wei, W.; Gagneur, J.; Perocchi, F.; Clauder-Munster, S.; Camblong, J.; Guffanti, E.; Stutz, F.; Huber, W.; Steinmetz, L.M. Bidirectional promoters generate pervasive transcription in yeast. Nature 2009, 457, 1033–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnone, J.T.; Robbins-Pianka, A.; Arace, J.R.; Kass-Gergi, S.; McAlear, M.A. The adjacent positioning of co-regulated gene pairs is widely conserved across eukaryotes. BMC Genom. 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- Karginov, T.A.; Pastor, D.P.H.; Semler, B.L.; Gomez, C.M. Mammalian polycistronic mRNAs and disease. Trends Genet. 2017, 33, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Merchant, S.S.; Helmann, J.D. Elemental economy: Microbial strategies for optimizing growth in the face of nutrient limitation. Adv. Microb. Phys. 2012, 60, 91–210. [Google Scholar]
- Eide, D.J. Homeostatic and adaptive responses to zinc deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 2009, 284, 18565–18569. [Google Scholar] [CrossRef] [PubMed]
- Bird, A.J.; Gordon, M.; Eide, D.J.; Winge, D.R. Repression of ADH1 and ADH3 during zinc deficiency by Zap1-induced intergenic RNA transcripts. EMBO J. 2006, 25, 5726–5734. [Google Scholar] [CrossRef] [PubMed]
- Taggart, J.; MacDiarmid, C.W.; Haws, S.; Eide, D.J. Zap1-dependent transcription from an alternative upstream promoter controls translation of RTC4 mRNA in zinc-deficient Saccharomyces cerevisiae. Mol. Microbiol. 2017, 106, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Ard, R.; Allshire, R.C.; Marquardt, S. Emerging properties and functional consequences of noncoding transcription. Genetics 2017, 207, 357–367. [Google Scholar] [PubMed]
- Zhao, H.; Eide, D. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. USA 1996, 93, 2454–2458. [Google Scholar] [CrossRef] [PubMed]
- Brachmann, C.B.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains of Saccharomyces cerevisiae 288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- North, M.; Steffen, J.; Loguinov, A.V.; Zimmerman, G.R.; Vulpe, C.D.; Eide, D.J. Genome-wide functional profiling identifies genes and processes important for zinc-limited growth of Saccharomyces cerevisiae. PLoS Genet. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Huh, W.K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Pisat, N.P.; Pandey, A.; Macdiarmid, C.W. MNR2 regulates intracellular magnesium storage in Saccharomyces cerevisiae. Genetics 2009, 183, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Kunes, S.; Schatz, P.J.; Botstein, D. Plasmid construction by homologous recombination in yeast. Gene 1987, 58, 201–216. [Google Scholar] [CrossRef]
- Wach, A.; Brachat, A.; Pohlmann, R.; Philippsen, P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 1994, 10, 1793–1808. [Google Scholar] [CrossRef] [PubMed]
- Sikorski, R.S.; Hieter, P. A system of shuttle vectors and host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989, 122, 19–27. [Google Scholar] [PubMed]
- MacDiarmid, C.W.; Taggart, J.; Jeong, J.; Kerdsomboon, K.; Eide, D.J. Activation of the yeast UBI4 polyubiquitin gene by Zap1 via an intragenic promoter is critical for zinc-deficient growth. J. Biol. Chem. 2016, 291, 18880–18896. [Google Scholar] [CrossRef] [PubMed]
- MacDiarmid, C.W.; Taggart, J.; Kerdsomboon, K.; Kubisiak, M.; Panascharoen, S.; Schelble, K.; Eide, D.J. Peroxiredoxin chaperone activity is critical for protein homeostasis in zinc-deficient yeast. J. Biol. Chem. 2013, 288, 31313–31327. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Matic, I.; Hilger, M.; Nagaraj, N.; Selbach, M.; Olsen, J.V.; Mann, M. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 2009, 4, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Bairoch, A.; Apweiler, R.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. The Universal Protein Resource (UniProt). Nucleic Acids Res. 2005, 33, D154–D159. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Weisenhorn, E.; MacDiarmid, C.W.; Andreini, C.; Bucci, M.; Taggart, J.; Banci, L.; Russell, J.; Coon, J.J.; Eide, D.J. The cellular economy of the Saccharomyces cerevisiae zinc proteome. Metallomics. under review.
- Tatip, S.; Taggart, J.; Eide, D.J. Regulation of the yeast HNT1 gene by zinc-responsive transcription start site changes. Curr. Genet. to be submitted.
- Scherrer, T.; Femmer, C.; Schiess, R.; Aebersold, R.; Gerber, A.P. Defining potentially conserved RNA regulons of homologous zinc-finger RNA-binding proteins. Genome Biol. 2011, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.H.; Taggart, J.; Song, P.X.; MacDiarmid, C.; Eide, D.J. An MSC2 promoter-lacZ fusion gene reveals zinc-responsive changes in sites of transcription initiation that occur across the yeast genome. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; University of Wisconsin-Madison, Madison, WI, USA. Results of immunoblot analyses. 2018. [Google Scholar]
- Breslow, D.K.; Cameron, D.M.; Collins, S.R.; Schuldiner, M.; Stewart-Ornstein, J.; Newman, H.W.; Braun, S.; Madhani, H.D.; Krogan, N.J.; Weissman, J.S. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 2008, 5, 711–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, W.; Ma, D.; Xiao, C.; Wang, Z.; Zhang, J. The genomic landscape and evolutionary resolution of antagonistic pleiotropy in yeast. Cell Rep. 2012, 2, 1399–1410. [Google Scholar] [CrossRef] [PubMed]
- Pelechano, V.; Wei, W.; Steinmetz, L.M. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 2013, 497, 127–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blumenthal, T.; Davis, P.; Garrido-Lecca, A. Operon and non-operon gene clusters in the C. elegans genome. WormBook 2015, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Balvay, L.; Soto Rifo, R.; Ricci, E.P.; Decimo, D.; Ohlmann, T. Structural and functional diversity of viral IRESes. Biochim. Biophys. Acta 2009, 1789, 542–557. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Unbehaun, A.; Spahn, C.M.T. Ribosomal chamber music: Toward an understanding of IRES mechanisms. Trends Biochem. Sci. 2017, 42, 655–668. [Google Scholar] [CrossRef] [PubMed]
- Carrozza, M.J.; Li, B.; Florens, L.; Suganuma, T.; Swanson, S.K.; Lee, K.K.; Shia, W.J.; Andersaon, S.; Yates, J.; Washburn, M.P.; et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 2005, 123, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Michalak, P. Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics 2008, 91, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Addinall, S.G.; Downey, M.; Yu, M.; Zubko, M.K.; Dewar, J.; Leake, A.; Hallinan, J.; Shaw, O.; James, K.; Wilkinson, D.J.; et al. A genomewide suppressor and enhancer analysis of cdc13-1 reveals varied cellular processes influencing telomere capping in Saccharomyces cerevisiae. Genetics 2008, 180, 2251–2266. [Google Scholar] [CrossRef] [PubMed]
- Balciunas, D.; Ronne, H. Yeast genes GIS1-4: Multicopy suppressors of the Gal− phenotype of snf1 mig1 srb8/10/11 cells. Mol. Gen. Genet. 1999, 262, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.; Farr, G.W.; Fernandez, C.F.; Lauden, L.; McCormack, J.C.; Wolin, S.L. Yeast Gis2 and its human ortholog CNBP are novel components of stress-induced RNP granules. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Leipheimer, J.; Bloom, A.L.M.; Baumstark, T.; Panepinto, J.C. CNBP Homologues Gis2 and Znf9 interact with a putative G-quadruplex-forming 3′ untranslated region, altering polysome association and stress tolerance in Cryptococcus neoformans. mSphere 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Liang, Y.; Deng, W.; Shimizu, K.; Ashique, A.M.; Li, E.; Li, Y.P. The zinc-finger protein CNBP is required for forebrain formation in the mouse. Development 2003, 130, 1367–1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liquori, C.L.; Ricker, K.; Moseley, M.L.; Jacobsen, J.F.; Kress, W.; Naylor, S.L.; Day, J.W.; Ranum, L.P. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science 2001, 293, 864–867. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wang, Y.; Abe, Y.; Cheney, L.; Udd, B.; Li, Y.P. Haploinsuffciency for Znf9 in Znf9+/− mice is associated with multiorgan abnormalities resembling myotonic dystrophy. J. Mol. Biol. 2007, 368, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Benhalevy, D.; Gupta, S.K.; Danan, C.H.; Ghosal, S.; Sun, H.W.; Kazemier, H.G.; Paeschke, K.; Hafner, M.; Juranek, S.A. The human CCHC-type zinc finger nucleic acid-binding protein binds G-rich elements in target mRNA coding sequences and promotes translation. Cell Rep. 2017, 18, 2979–2990. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.U.; Bartel, D.P. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science 2016, 353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, E.; Lee, T.; Kim, J.H.; Park., A.; Ra, E.A.; Kang, S.; Choi, H.; Choi, J.; Huh, H.; Lee, J.E.; et al. CNBP acts as a key transcriptional regulator of sustained expression of interleukin-6. Nucleic Acids Res. 2017, 45, 3280–3296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gromoller, A.; Lehming, N. Srb7p is a physical and physiological target of Tup1p. EMBO J. 2000, 19, 6845–6852. [Google Scholar] [CrossRef] [PubMed]
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 2014, 11, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Ghaemmaghami, S.; Huh, W.K.; Bower, K.; Howson, R.W.; Belle, A.; Dephoure, N.; O’shea, E.K.; Weissman, J.S. Global analysis of protein expression in yeast. Nature 2003, 425, 737–741. [Google Scholar] [CrossRef] [PubMed]
- MacDiarmid, C.W.; Gaither, L.A.; Eide, D. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 2000, 19, 2845–2855. [Google Scholar] [CrossRef] [PubMed]
- Qiao, W.; Mooney, M.; Bird, A.J.; Winge, D.R.; Eide, D.J. Zinc binding to a regulatory zinc-sensing domain monitored in vivo by using FRET. Proc. Natl. Acad. Sci. USA 2006, 103, 8674–8679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Zinc Replete | Zinc Deficient | ||||||
|---|---|---|---|---|---|---|---|
| Strain | % in To Inoculum | % after 15 Generations | Exp/Cont Ratio | p-Value a | % after 15 Generations | Exp/Cont Ratio | p-Value a |
| WT | 53.9 | 52.7 ± 0.2 | - | - | 49.8 ± 2.6 | - | - |
| gis2∆ | 49.9 | 46.1 ± 1.4 | 0.9 | 0.02 | 49.9 ± 1.3 | 1.0 | NS |
| tsa1∆ | 50.1 | 28.9 ± 1.2 | 0.6 | 0.001 | 1.4 ± 0.1 | 0.03 | 0.001 |
| WT vector | 51.7 | 49.0 ± 0.6 | - | - | 45.2 ± 0.6 | - | - |
| WT pRGmZRE | 50.9 | 49.5 ± 0.7 | 1.0 | NS | 53.2 ± 0.9 | 1.2 | 0.006 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taggart, J.; Wang, Y.; Weisenhorn, E.; MacDiarmid, C.W.; Russell, J.; Coon, J.J.; Eide, D.J. The GIS2 Gene Is Repressed by a Zinc-Regulated Bicistronic RNA in Saccharomyces cerevisiae. Genes 2018, 9, 462. https://doi.org/10.3390/genes9090462
Taggart J, Wang Y, Weisenhorn E, MacDiarmid CW, Russell J, Coon JJ, Eide DJ. The GIS2 Gene Is Repressed by a Zinc-Regulated Bicistronic RNA in Saccharomyces cerevisiae. Genes. 2018; 9(9):462. https://doi.org/10.3390/genes9090462
Chicago/Turabian StyleTaggart, Janet, Yirong Wang, Erin Weisenhorn, Colin W. MacDiarmid, Jason Russell, Joshua J. Coon, and David J. Eide. 2018. "The GIS2 Gene Is Repressed by a Zinc-Regulated Bicistronic RNA in Saccharomyces cerevisiae" Genes 9, no. 9: 462. https://doi.org/10.3390/genes9090462
APA StyleTaggart, J., Wang, Y., Weisenhorn, E., MacDiarmid, C. W., Russell, J., Coon, J. J., & Eide, D. J. (2018). The GIS2 Gene Is Repressed by a Zinc-Regulated Bicistronic RNA in Saccharomyces cerevisiae. Genes, 9(9), 462. https://doi.org/10.3390/genes9090462




