FDHE-IW: A Fast Approach for Detecting High-Order Epistasis in Genome-Wide Case-Control Studies
Abstract
:1. Introduction
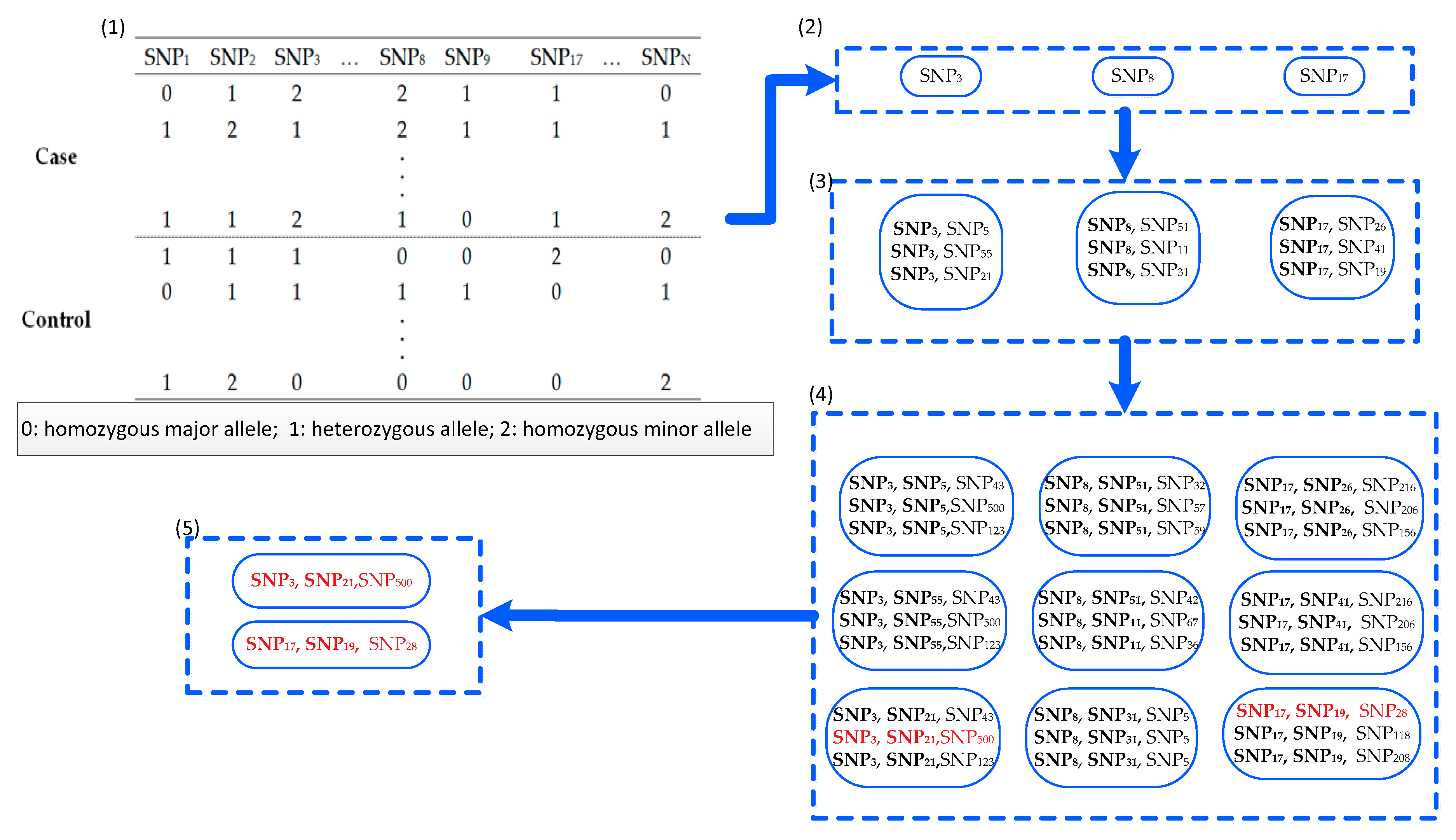
2. Materials and Methods
2.1. Definitions
- (1)
- 0 ≤ IWF(X, Y) ≤ 2
- (2)
- 1 ≤ IWF(X, Y) ≤ 2 if X interacts with Y.
- (3)
- 0 ≤ IWF(X, Y) ≤ 1 if X is redundant to Y.
| Algorithm 1: FDHE-IW |
| Inputs: D (s1, s2, …, sN, C)—the given data set with N + 1 columns; si denotes the values of the ith SNP locus for all samples. T—the candidate size; θ—the threshold of the G-test p-value; k—the number of SNPs in a k-way SNP combination; and K—the number to find the SNP combinations based on a seed SNP. Outputs: SNP combinations (SC)—the k-way SNP combinations that are associated with disease status. |
|
|
2.2. Performance Evaluation and Simulation Data Sets
2.2.1. Performance Evaluation
2.2.2. Simulation Data Sets and Case Study
2.2.3. Paremeters Setting
3. Results
3.1. Simulated Models
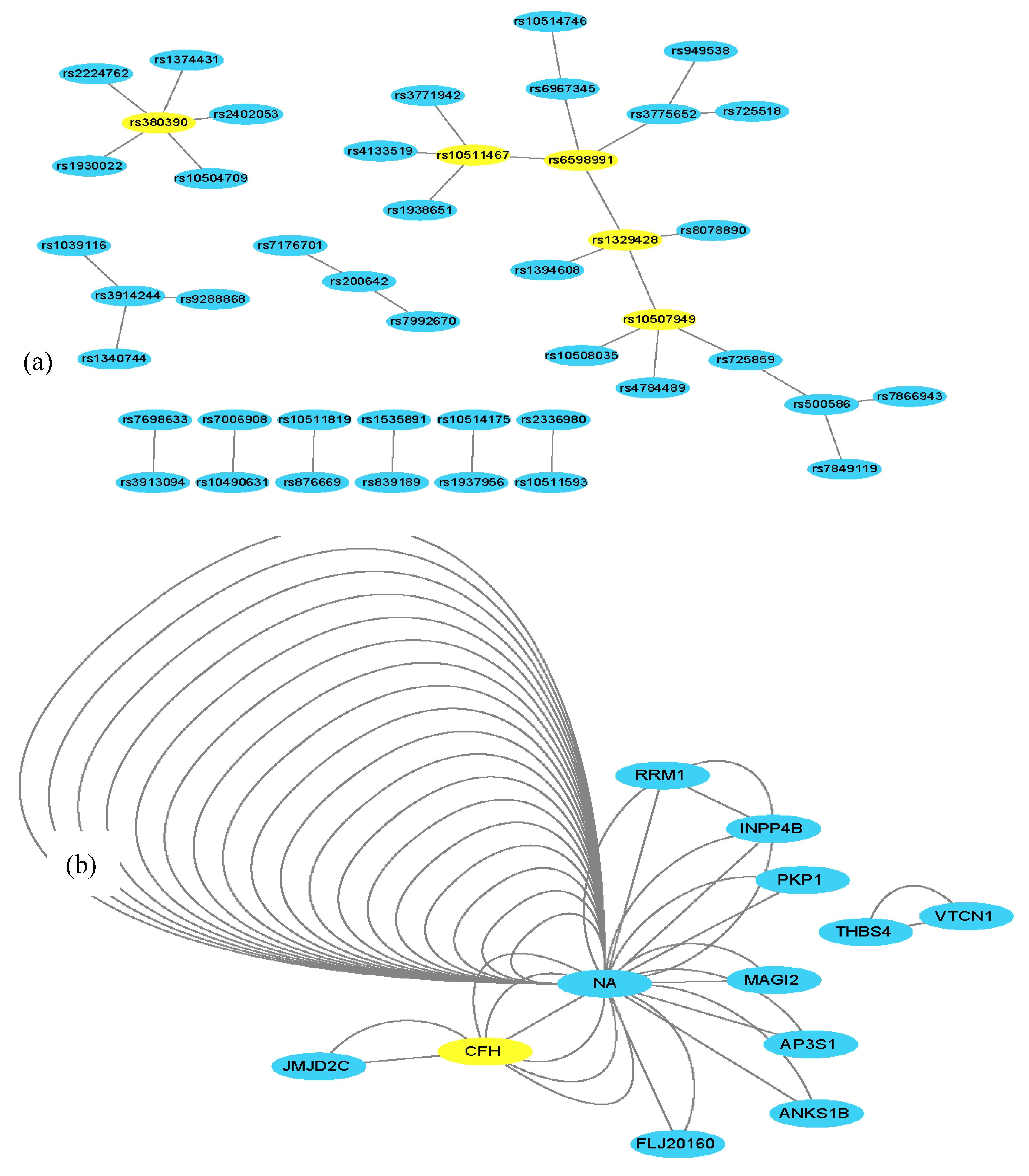
3.2. Experimental Results Using an AMD Dataset
4. Discussion
5. Conclusions
5.1. Advantage
5.2. Limitations
5.3. Future Work
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Manolio, T.A. Genomewide association studies and assessment of the risk of disease. N. Engl. J. Med. 2010, 363, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.J.; Zeiss, C.; Chew, E.Y.; Tsai, J.Y.; Sackler, R.S.; Haynes, C.; Henning, A.K.; SanGiovanni, J.P.; Mane, S.M.; Mayne, S.T.; et al. Complement factor H polymorphism in age-related macular degeneration. Science 2005, 308, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Upton, A.; Trelles, O.; Cornejo-García, J.A.; Perkins, J.R. Review: High-performance computing to detect epistasis in genome scale data sets. Brief. Bioinform. 2016, 17, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R. Gene-gene interaction. In Encyclopedia of Behavioral Medicine; Gellman, M.D., Turner, J.R., Eds.; Springer: New York, NY, USA, 2013; pp. 841–842. [Google Scholar]
- Stanfill, A.G.; Starlarddavenport, A. Primer in Genetics and Genomics, Article 7-Multifactorial Concepts: Gene-Gene Interactions. Biol. Res. Nurs. 2018, 20, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.H.; Asselbergs, F.W.; Williams, S.M. Bioinformatics challenges for genome-wide association studies. Bioinformatics 2010, 26, 445–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, X.; Yang, C.; Yang, Q.; Xue, H.; Fan, X.; Tang, N.L.; Yu, W. BOOST: A fast approach to detecting gene–gene interactions in genome-wide case–control studies. Am. J. Hum. Genet. 2010, 87, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.Y.; Yang, C.; Xiang, W.; Yu, W. Gboost: A gpu-based tool for detecting gene–gene interactions in genome–wide case control studies. Bioinformatics 2011, 27, 1309. [Google Scholar]
- Yang, G.; Jiang, W.; Yang, Q.; Yu, W. PBOOST: A GPU based tool for parallel permutation tests in genome-wide association studies. Bioinformatics 2015, 31, 1460–1462. [Google Scholar] [CrossRef] [PubMed]
- Li, X. A fast and exhaustive method for heterogeneity and epistasis analysis based on multi-objective optimization. Bioinformatics 2017, 33, 2829–2836. [Google Scholar] [CrossRef] [PubMed]
- Hahn, L.W.; Ritchie, M.D.; Moore, J.H. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics 2003, 19, 376–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; He, Z.; Wan, X.; Yang, Q.; Xue, H.; Yu, W. SNPHarvester: A filtering-based approach for detecting epistatic interactions in genome-wide association studies. Bioinformatics 2009, 25, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Crawford, L.; Ping, Z.; Mukherjee, S. Detecting epistasis with the marginal epistasis test in genetic mapping studies of quantitative traits. PLoS Genet. 2017, 13, e1006869. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, J.S. Bayesian inference of epistatic interactions in case–control studies. Nat. Genet. 2007, 39, 1167–1173. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, X.; Robbins, K.; Rekaya, R. AntEpiSeeker: Detecting epistatic interactions for case-control studies using a two-stage ant colony optimization algorithm. BMC Res. Notes 2010, 3, 117. [Google Scholar] [CrossRef] [PubMed]
- Aflakparast, M.; Salimi, H.; Gerami, A.; Dubé, M.P.; Visweswaran, S.; Masoudi-Nejad, A. Cuckoo search epitasis: A new method for exploring significant genetic interactions. Heredity 2014, 112, 666–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, P.-J.; Shen, H.-B. MACOED: A multi-objective ant colony optimization algorithm for SNP epistasis detection in genome-wide association studies. Bioinformatics 2015, 31, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Tuo, S.; Zhang, J.; Yuan, X.; Zhang, Y.; Liu, Z. FHSA-SED: Two-locus model detection for genome-wide association study with harmony search algorithm. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Tuo, S.; Zhang, J.; Yuan, X.; He, Z.; Liu, Y.; Liu, Z. Niche harmony search algorithm for detecting complex disease associated high-order SNP combinations. Sci. Rep. 2017, 7, 11529. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Sun, Y.; Liu, J.X.; Xia, J.; Zhang, J.; Zheng, C.H. CINOEDV: A co-information based method for detecting and visualizing n -order epistatic interactions. BMC Bioinform. 2016, 17, 214. [Google Scholar] [CrossRef] [PubMed]
- Sinoquet, C.; Niel, C. Enhancement of a stochastic Markov blanket framework with ant colony optimization, to uncover epistasis in genetic association studies. Bioinformatics 2018, 15, 673–6780. [Google Scholar]
- Liu, J.; Yu, G.; Jiang, Y.; Wang, J. HiSeeker: Detecting high-order SNP interactions based on pair-wise SNP combinations. Genes 2017, 8, 153. [Google Scholar] [CrossRef] [PubMed]
- Tuba, M. Plenary lecture 3: Swarm Intelligence Algorithms Parameter Tuning. In Proceedings of the WSEAS International Conference on Computer Engineering and Applications, and Proceedings of the 2012 American Conference on Applied Mathematics, Cambridge, UK, 25–27 January 2012. [Google Scholar]
- Menezes, B.A.M.; Wrede, F.; Kuchen, H.; de Lima Neto, F.B. Parameter Selection for Swarm Intelligence Algorithms—Case Study on Parallel Implementation of FSS. In Proceedings of the 2017 IEEE Latin American Conference on Computational Intelligence, Arequipa, Peru, 8–10 November 2017. [Google Scholar]
- Vinh, N.X.; Zhou, S.; Chan, J.; Bailey, J. Can high-order dependencies improve mutual information based feature selection? Pattern Recognit. 2016, 53, 46–58. [Google Scholar] [CrossRef] [Green Version]
- Shishkin, A.; Bezzubtseva, A.; Drutsa, A.; Shishkov, I.; Gladkikh, E.; Gusev, G.; Serdyukov, P. Efficient High-Order Interaction-Aware Feature Selection Based on Conditional Mutual Information. In Proceedings of the 30th Conference on Neural Information Processing Systems, Barcelona, Spain, 5–10 December 2016. [Google Scholar]
- Song, Q.; Ni, J.; Wang, G. a fast clustering-based feature subset selection algorithm for high-dimensional data. IEEE Trans. Knowl. Data Eng. 2012, 25, 1–14. [Google Scholar] [CrossRef]
- Claude, E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Jakulin, A.; Bratko, I. Testing the Significance of Attribute Interactions. In Proceedings of the Twenty-First International Conference on Machine Learning, Banff, AB, Canada, 4–8 July 2004; pp. 409–416. [Google Scholar]
- Zeng, Z.; Zhang, H.; Zhang, R.; Yin, C. A novel feature selection method considering feature interaction. Pattern Recognit. 2015, 48, 2656–2666. [Google Scholar] [CrossRef]
- Niel, C.; Sinoquet, C.; Dina, C.; Rocheleau, G.; Kelso, J. SMMB—A stochastic Markov-blanket framework strategy for epistasis detection in GWAS. Bioinformatics 2018, 34, 2773–2780. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.H. G–Test Goodness-of-Fit. In Handbook of Biological Statistics, 3rd ed.; Sparky House Publishing: Baltimore, MD, USA, 2014; pp. 53–58. [Google Scholar]
- Harremoës, P.; Tusnády, G. Information divergence is more chi squared distributed than the chi squared statistic. arXiv, 2012; arXiv:1202.1125. [Google Scholar]
- Crow, J.H. Weinberg and language impediments. Genetics 1999, 152, 821–825. [Google Scholar] [PubMed]
- Urbanowicz, R.J.; Kiralis, J.; Sinnott-Armstrong, N.A.; Heberling, T.; Fisher, J.M.; Moore, J.H. GAMETES: A fast, direct algorithm for generating pure, strict, epistatic models with random architectures. BioData Min. 2012, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Vélez, J.I.; Chandrasekharappa, S.C.; Henao, E.; Martinez, A.F.; Harper, U.; Jones, M.; Solomon, B.D.; Lopez, L.; Garcia, G.; Aguirre-Acevedo, D.C.; et al. Pooling/bootstrap-based GWAS (pbGWAS) identifies new loci modifying the age of onset in PSEN1 p.Glu280Ala Alzheimer’s disease. Mol. Psychiatr. 2013, 18, 568–575. [Google Scholar] [CrossRef] [PubMed]
- Tutz, G.; Ramzan, S. Improved methods for the imputation of missing data by nearest neighbor methods. Comput. Stat. Data Anal. 2015, 90, 84–99. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, Z.; Ji, Q.; Li, Q. Histone demethylase JMJD2C: Epigenetic regulators in tumors. Oncotarget 2017, 8, 91723–91733. [Google Scholar] [CrossRef] [PubMed]
- Hong, Q.; Yu, S.; Yang, Y.; Liu, G.; Shao, Z. A polymorphism in JMJD2C alters the cleavage by caspase-3 and the prognosis of human breast cancer. Oncotarget 2014, 5, 4779–4787. [Google Scholar] [CrossRef] [PubMed]
- Burton, A.; Azevedo, C.; Andreassi, C.; Riccio, A.; Saiardi, A. Inositol pyrophosphates regulate JMJD2C-dependent histone demethylation. Proc. Natl. Acad. Sci. USA 2013, 110, 18970–18975. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Sun, Y.; Li, S.; Liu, J.; Zheng, C.; Zhang, J. An improved opposition-based learning particle swarm optimization for the detection of SNP-SNP interactions. BioMed Res. Int. 2015, 2015, 524821. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Shang, J.; Liu, J.; Li, S.; Zheng, C. epiACO—A method for identifying epistasis based on ant Colony optimization algorithm. BioData Min. 2017, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Tang, W.; Wu, X.; Fu, W. A random forest approach to the detection of epistatic interactions in case-control studies. BMC Bioinform. 2009, 10, S65. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Meng, Y.; Yu, N.; Pan, Y. Cloud computing for detecting high-order genome-wide epistatic interaction via dynamic clustering. BMC Bioinform. 2014, 15, 102. [Google Scholar] [CrossRef] [PubMed]
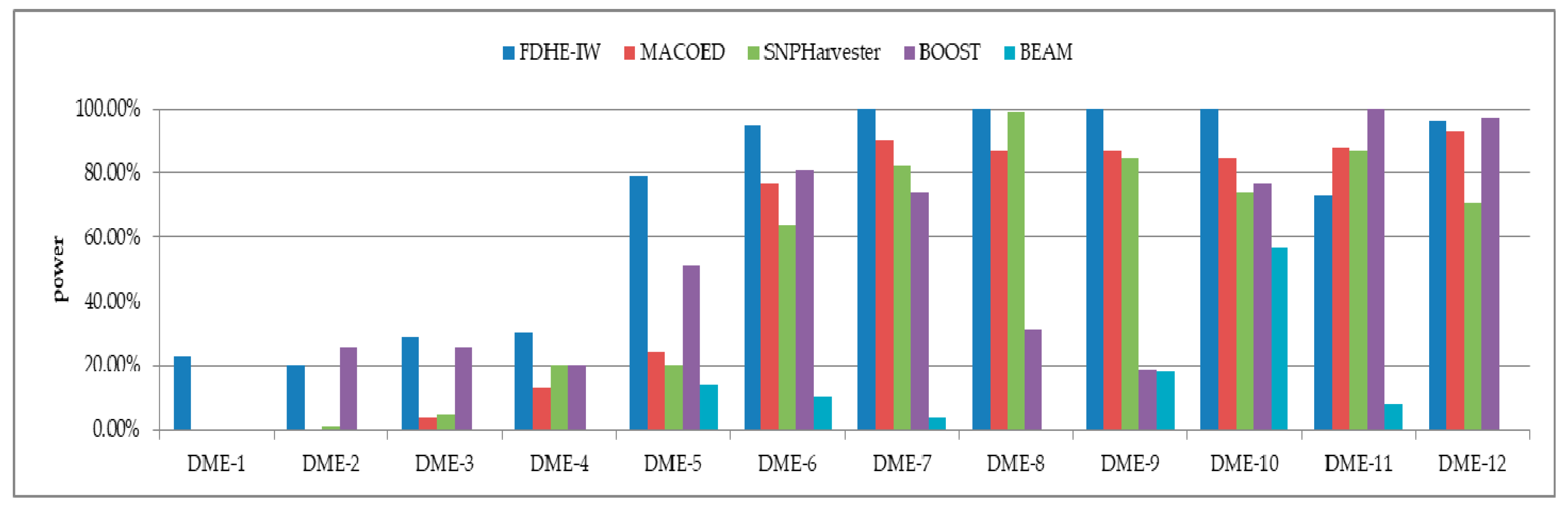
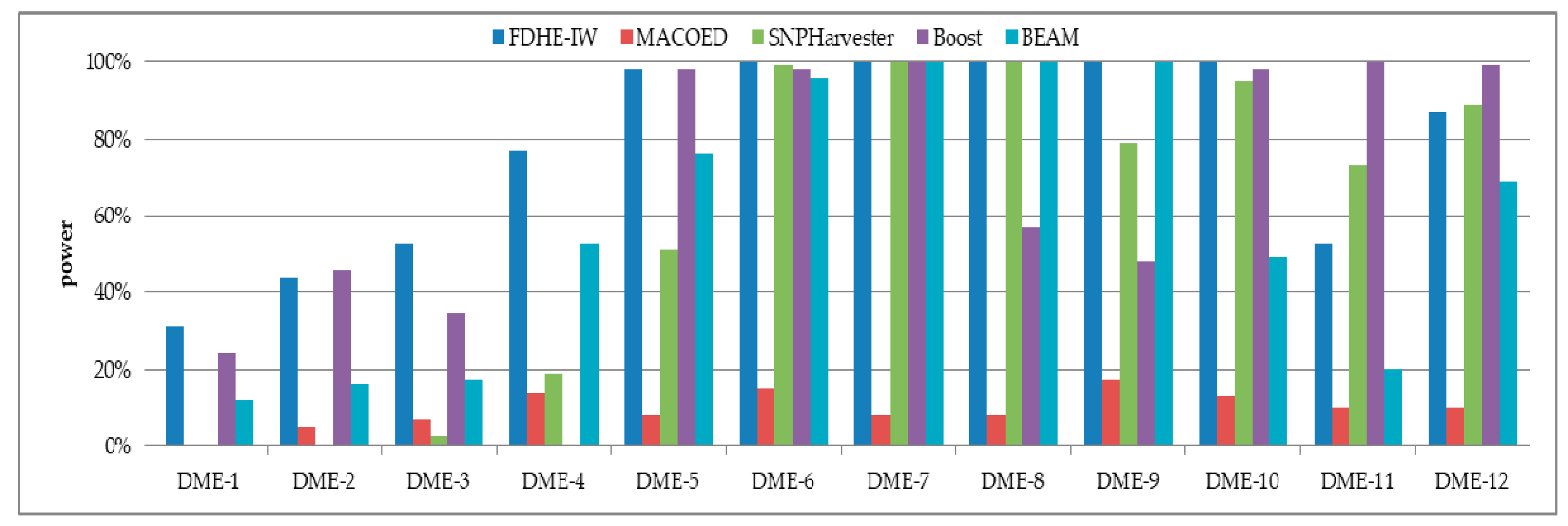
| Models | 100 SNPs (1600 Sample Size) | 1000 SNPs (4000 Sample Size) | ||||
|---|---|---|---|---|---|---|
| Recall | Precision | F-Measure | Recall | Precision | F-Measure | |
| DME-1 | 26.09% | 54.55% | 35.29% | 67.74% | 84.00% | 75.00% |
| DME-2 | 15.00% | 27.27% | 19.35% | 75.00% | 78.57% | 76.74% |
| DME-3 | 44.83% | 44.83% | 44.83% | 79.25% | 55.26% | 65.12% |
| DME-4 | 60.00% | 33.96% | 43.37% | 92.21% | 43.56% | 59.17% |
| DME-5 | 69.62% | 91.67% | 79.14% | 100.00% | 85.22% | 92.02% |
| DME-6 | 82.11% | 78.00% | 80.00% | 100.00% | 53.48% | 69.69% |
| DME-7 | 96.00% | 64.43% | 77.11% | 100.00% | 35.46% | 52.36% |
| DME-8 | 100.00% | 31.35% | 47.73% | 100.00% | 33.33% | 50.00% |
| DME-9 | 95.00% | 73.08% | 82.61% | 99.00% | 73.88% | 84.62% |
| DME-10 | 95.00% | 93.14% | 94.06% | 99.00% | 86.84% | 92.52% |
| DME-11 | 98.41% | 100.00% | 99.20% | 100.00% | 100.00% | 100.00% |
| DME-12 | 96.88% | 97.89% | 97.38% | 98.85% | 98.85% | 98.85% |
| Models | Algorithms | Recall | Precision | F-Measure | Runtime(s) |
|---|---|---|---|---|---|
| Multiplicative model | FDHE-IW | 36.5% | 40.2% | 35.7% | 2.95 |
| MACOED | 68.5% | 90.8% | 71.7% | 10.8 | |
| BEAM | 14.8% | 10.3% | 7.7% | 8.52 | |
| BOOST | 0.5% | 62.5% | 0.5% | 0.6 | |
| SNPHarvester | 0.3% | 50.0% | 0.5% | 2.97 | |
| Threshold model | FDHE-IW | 86.9% | 66.4% | 71.0% | 2.95 |
| MACOED | 98.0% | 83.0% | 89.3% | 11.40 | |
| BEAM | 84.5% | 68.3% | 60.5% | 8.52 | |
| BOOST | 34.8% | 99.0% | 37.1% | 0.95 | |
| SNPHarvester | 51.8% | 47.0% | 29.0% | 2.92 | |
| Concrete model | FDHE-IW | 96.3% | 91.0% | 93.3% | 2.95 |
| MACOED | 98.8% | 84.8% | 91.0% | 11.59 | |
| BEAM | 81.3% | 62.3% | 70.1% | 8.52 | |
| BOOST | 66.3% | 87.3% | 69.7% | 0.66 | |
| SNPHarvester | 91.3% | 57.3% | 67.7% | 2.90 |
| Models | Algorithms | Recall | Precision | F-Measure | Runtime(s) |
|---|---|---|---|---|---|
| Multiplicative model | FDHE-IW | 78.55% | 65.35% | 69.01% | 65.4 |
| MACOED | 75.00% | 32.85% | 20.82% | 440 | |
| BEAM | 35.12% | 37.84% | 17.65% | 308 | |
| BOOST | 18.50% | 22.95% | 9.40% | 4 | |
| SNPHarvester | 5.55% | 32.20% | 3.61% | 130 | |
| Threshold model | FDHE-IW | 100.00% | 51.87% | 66.02% | 66.6 |
| MACOED | 100.00% | 14.31% | 11.08% | 450 | |
| BEAM | 98.46% | 75.27% | 42.64% | 199 | |
| BOOST | 88.50% | 62.50% | 33.70% | 9 | |
| SNPHarvester | 87.50% | 29.45% | 12.24% | 143 | |
| Concrete model | FDHE-IW | 99.21% | 89.89% | 94.00% | 35.3 |
| MACOED | 100.00% | 38.60% | 23.89% | 303 | |
| BEAM | 66.94% | 63.05% | 31.68% | 133 | |
| BOOST | 72.50% | 20.83% | 15.82% | 6 | |
| SNPHarvester | 84.00% | 92.45% | 43.80% | 62.3 |
| SNP1 | Gene1 | SNP2 | Gene2 | SNP3 | Gene3 | G-test p-Value |
|---|---|---|---|---|---|---|
| rs380390 | CFH | rs1930022 | NA | rs3913094 | NA | 2.22 × 10−16 |
| rs380390 | CFH | rs10504709 | NA | rs2402053 | NA | 1.11 × 10−16 |
| rs380390 | CFH | rs10504709 | NA | rs2380684 | NA | 1.11 × 10−16 |
| rs380390 | CFH | rs2380684 | NA | rs2224762 | JMJD2C | 0 |
| rs380390 | CFH | rs10504548 | NA | rs2224762 | JMJD2C | 0 |
| rs380390 | CFH | rs2402053 | NA | rs2224762 | JMJD2C | 0 |
| rs380390 | CFH | rs718263 | NCALD | rs2224762 | JMJD2C | 0 |
| rs1329428 | CFH | rs3775652 | INPP4B | rs6598991 | NA | 2.22 × 10−16 |
| rs725518 | RRM1 | rs3775652 | INPP4B | rs1002979 | NA | 2.22 × 10−16 |
| snp1 | Gen1 | SNP2 | Gen2 | SNP3 | Gen3 | SNP4 | Gen4 | G-test p-Value |
|---|---|---|---|---|---|---|---|---|
| rs1740752 | PCCA | rs4772270 | NA | rs7044653 | NA | rs6598991 | NA | 0 |
| rs4772270 | NA | rs7044653 | NA | rs1329428 | CFH | rs6598991 | NA | 0 |
| FDHE-IW | BEAM | epi Forest | DCHE | FHSA-SED | NHSA-DHSC | epiACO | |
|---|---|---|---|---|---|---|---|
| Relevant SNPs or genes identified | SNPs: rs380390 rs1329428 rs10511467 rs6598991 rs10507949 rs3776652 Genes: CFH JMJD2C INPP4B | SNPs: rs380390 rs1329428 Gene: CFH | SNPs: rs380390 rs1329428 rs1394608 rs7104698 Gene: CFH | SNPs: rs380390 rs1329428 rs1394608 rs1740752 rs1363688 rs10512174 rs618499 rs1926489 Genes: CFH ZNF25 SGCD LRIG3 DRD1 ISCA1 | SNPs: rs380390 rs1329428 rs10272438 rs1740752 rs3775652 rs1394608 rs1363688 rs10511467 Genes: CFH BBS9 SGCDINPP4B | SNPs: rs380390 rs1329428 rs10272438 rs1363688 rs1394608 rs3775652 rs7104698 rs10511467 rs10512413 Genes: CFH INPP4B BBS9 ABL1ANKS1B | SNPs: rs380390 rs1329428 rs1363688 rs1394608 rs2224762 rs9328536 rs943008 rs718263 Genes: CFH MED27 KDM4C NCALD NEDD9 |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tuo, S. FDHE-IW: A Fast Approach for Detecting High-Order Epistasis in Genome-Wide Case-Control Studies. Genes 2018, 9, 435. https://doi.org/10.3390/genes9090435
Tuo S. FDHE-IW: A Fast Approach for Detecting High-Order Epistasis in Genome-Wide Case-Control Studies. Genes. 2018; 9(9):435. https://doi.org/10.3390/genes9090435
Chicago/Turabian StyleTuo, Shouheng. 2018. "FDHE-IW: A Fast Approach for Detecting High-Order Epistasis in Genome-Wide Case-Control Studies" Genes 9, no. 9: 435. https://doi.org/10.3390/genes9090435
APA StyleTuo, S. (2018). FDHE-IW: A Fast Approach for Detecting High-Order Epistasis in Genome-Wide Case-Control Studies. Genes, 9(9), 435. https://doi.org/10.3390/genes9090435




