Comparative Transcriptomics of Root Development in Wild and Cultivated Carrots
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. RNA Extraction and RNAseq
2.3. RNAseq Data Analysis
3. Results and Discussion
3.1. RNAseq Summary and Identification of Novel Transcripts
3.2. Differential Expression
3.2.1. cDEG.1.2: Genes Differentially Expressed in the Developing Storage Root (T2), as Compared with the Young Plant (T1)
3.2.2. cDEG.1.3—Genes Differentially Expressed in the Mature Storage Root (T3), as Compared with the Young Plant (T1)
3.2.3. cDEG.2.3—Genes Differentially Expressed in the Mature Storage Root (T3), as Compared with the Developing Storage Root (T2)
3.2.4. DEGs between Wild in Cultivated D. carota at the Three Timepoints (wcDEG.1, wcDEG.2, and wcDEG.3)
3.3. Genetic Determinants of the Carrot Storage Root Development
3.3.1. Regulation of the Carrot Storage Root Development
3.3.1.1. Transcription Factors
3.3.1.2. Post-Translational Protein Modifications
3.3.1.3. Hormonal Signaling
3.3.1.4. Redox Signaling
3.3.1.5. Regulation Complexity
3.3.2. DEGs Involved in the Carrot Storage Root Development
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. The Statistics Division of FAO. Available online: http://faostat3.fao.org/ (accessed on 18 April 2018).
- Simon, P.W. Plant breeding for human nutritional quality. Plant Breed. Rev. 2009, 31, 325–392. [Google Scholar]
- Iorizzo, M.; Senalik, D.A.; Ellison, S.L.; Grzebelus, D.; Cavagnaro, P.F.; Allender, C.; Brunet, J.; Spooner, D.M.; Van Deynze, A.; Simon, P.W. Genetic structure and domestication of carrot (Daucus carota subsp. sativus) (Apiaceae) 1. Am. J. Bot. 2013, 100, 930–938. [Google Scholar] [CrossRef] [PubMed]
- Banga, O. Origin and distribution of the western cultivated carrot. Genet. Agrar. 1963, 17, 357–370. [Google Scholar]
- Stolarczyk, J.; Janick, J. Carrot: History and Iconography. Chron. Horticult. 2011, 51, 12–18. [Google Scholar]
- Macko-Podgórni, A.; Machaj, G.; Stelmach, K.; Senalik, D.; Grzebelus, E.; Iorizzo, M.; Simon, P.W.; Grzebelus, D.; Macko-Podgorni, A.; Machaj, G.; et al. Characterization of a genomic region under selection in cultivated carrot (Daucus carota subsp. sativus) reveals a candidate domestication gene. Front. Plant Sci. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Rong, J.; Lammers, Y.; Strasburg, J.L.; Schidlo, N.S.; Ariyurek, Y.; de Jong, T.J.; Klinkhamer, P.G.L.; Smulders, M.J.M.; Vrieling, K. New insights into domestication of carrot from root transcriptome analyses. BMC Genom. 2014, 15, 895. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-L.L.; Jia, X.-L.L.; Xu, Z.-S.S.; Wang, F.; Xiong, A.-S.S. Sequencing, assembly, annotation, and gene expression: Novel insights into the hormonal control of carrot root development revealed by a high-throughput transcriptome. Mol. Genet. Genom. 2015, 290, 1379–1391. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Ellison, S.; Senalik, D.; Zeng, P.; Satapoomin, P.; Huang, J.; Bowman, M.; Iovene, M.; Sanseverino, W.; Cavagnaro, P.; et al. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 2016, 48, 657–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S.; Lönnstedt, I.; Speed, T.; Robinson, M.; Smyth, G.; McCarthy, D.; Chen, Y.; Smyth, G.; et al. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Leng, N.; Dawson, J.; Kendziorski, C. EBSeq: An R Package for Differential Expression Analysis Using RNA-seq Data. 2018. Available online: http://www.bioconductor.org/packages/release/bioc/vignettes/EBSeq/inst/doc/EBSeq_Vignette.pdf (accessed on 16 April 2018).
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- MacQueen, J.B. Some Methods for classification and Analysis of Multivariate Observations. In Proceedings of the 5th Berkeley Symposium on Mathematical Statistics and Probability, Berkeley, CA, USA, 21 June–18 July 1965 and 27 December 1965–7 January 1966; University of California Press: Berkeley, CA, USA, 1967. [Google Scholar]
- Lloyd, S.P. Least Squares Quantization in PCM. IEEE Trans. Inf. Theory 1982. [Google Scholar] [CrossRef]
- Tao, X.; Gu, Y.-H.; Wang, H.-Y.; Zheng, W.; Li, X.; Zhao, C.-W.; Zhang, Y.-Z. Digital Gene Expression Analysis Based on Integrated De Novo Transcriptome Assembly of Sweet Potato [Ipomoea batatas (L.) Lam.]. PLoS ONE 2012, 7, e36234. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, X.; He, Q.; Liu, X.; Xu, W.; Li, L.; Gao, J.; Wang, F. Transcriptome analysis of the roots at early and late seedling stages using Illumina paired-end sequencing and development of EST-SSR markers in radish. Plant Cell Rep. 2012, 31, 1437–1447. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; Senalik, D.A.; Grzebelus, D.; Bowman, M.; Cavagnaro, P.F.; Matvienko, M.; Ashrafi, H.; Van Deynze, A.; Simon, P.W. De novo assembly and characterization of the carrot transcriptome reveals novel genes, new markers, and genetic diversity. BMC Genom. 2011, 12, 389. [Google Scholar] [CrossRef] [PubMed]
- Ohashi-Ito, K.; Bergmann, D.C. Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY. Development 2007, 134, 2959–2968. [Google Scholar] [CrossRef] [PubMed]
- Sorin, C.; Declerck, M.; Christ, A.; Blein, T.; Ma, L.; Lelandais-Brière, C.; Njo, M.F.; Beeckman, T.; Crespi, M.; Hartmann, C. A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol. 2014, 202, 1197–1211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Qin, G.; Chen, Z.; Gu, H.; Qu, L.J. A gain-of-function mutation of transcriptional factor PTL results in curly leaves, dwarfism and male sterility by affecting auxin homeostasis. Plant Mol. Biol. 2008, 66, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, C. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 2003, 130, 6431–6439. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Favero, D.S.; Peng, H.; Neff, M.M. Arabidopsis thaliana AHL family modulates hypocotyl growth redundantly by interacting with each other via the PPC/DUF296 domain. Proc. Natl. Acad. Sci. USA 2013, 110, E4688–E4697. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Zou, Y.; Feng, N. Overexpression of AHL20 negatively regulates defenses in Arabidopsis. J. Integr. Plant Biol. 2010, 52, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.; Kim, Y.-S.S.; Jung, J.-H.H.; Seo, P.J.; Park, C.-M.M. The AT-hook motif-containing protein AHL22 regulates flowering initiation by modifying FLOWERING LOCUS T chromatin in Arabidopsis. J. Biol. Chem. 2012, 287, 15307–15316. [Google Scholar] [CrossRef] [PubMed]
- Grzebelus, D.; Iorizzo, M.; Senalik, D.; Ellison, S.; Cavagnaro, P.; Macko-Podgorni, A.; Heller-Uszynska, K.; Kilian, A.; Nothnagel, T.; Allender, C.; et al. Diversity, genetic mapping, and signatures of domestication in the carrot (Daucus carota L.) genome, as revealed by Diversity Arrays Technology (DArT) markers. Mol. Breed. 2014, 33, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Hellmann, H. Plant Development: Regulation by Protein Degradation. Science 2002, 297, 793–797. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Parry, G.; Estelle, M. The ubiquitin-proteasome pathway and plant development. Plant Cell 2004, 16, 3181–3195. [Google Scholar] [CrossRef] [PubMed]
- Friso, G.; van Wijk, K.J. Update: Post-translational protein modifications in plant metabolism. Plant Physiol. 2015, 169, 1469–1487. [Google Scholar] [CrossRef] [PubMed]
- Iovine, B.; Iannella, M.L.; Bevilacqua, M.A. Damage-specific DNA binding protein 1 (DDB1): A protein with a wide range of functions. Int. J. Biochem. Cell Biol. 2011, 43, 1664–1667. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Feng, S.; Chen, F.; Chen, H.; Wang, J.; McCall, C.; Xiong, Y.; Deng, X.W. Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell 2008, 20, 1437–1455. [Google Scholar] [CrossRef] [PubMed]
- Catala, R.; Ouyang, J.; Abreu, I.A.; Hu, Y.; Seo, H.; Zhang, X.; Chua, N.-H. The Arabidopsis E3 SUMO Ligase SIZ1 Regulates Plant Growth and Drought Responses. Plant Cell 2007, 19, 2952–2966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durbak, A.; Yao, H.; McSteen, P. Hormone signaling in plant development. Curr. Opin. Plant Biol. 2012, 15, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Considine, M.J.; Foyer, C.H. redox regulation of plant development. Antioxid. Redox Signal. 2014, 21, 1305–1326. [Google Scholar] [CrossRef] [PubMed]
- Camara, B.; Bouvier, F. Oxidative remodeling of plastid carotenoids. Arch. Biochem. Biophys. 2004, 430, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Mabuchi, K.; Maki, H.; Itaya, T.; Suzuki, T.; Nomoto, M.; Sakaoka, S.; Morikami, A.; Higashiyama, T.; Tada, Y.; Busch, W.; et al. MYB30 links ROS signaling, root cell elongation, and plant immune responses. Proc. Natl. Acad. Sci. USA 2018, 115, E4710–E4719. [Google Scholar] [CrossRef] [PubMed]
- Criqui, M.C.; Genschik, P. Mitosis in plants: How far we have come at the molecular level? Curr. Opin. Plant Biol. 2002, 5, 487–493. [Google Scholar] [CrossRef]
- Inzé, D.; De Veylder, L. Cell Cycle Regulation in plant development. Annu. Rev. Genet. 2006, 40, 77–105. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-Y.; Kim, J.-Y. Callose synthesis in higher plants. Plant Signal. Behav. 2009, 4, 489–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussey, P.J.; Hawkins, T.J.; Igarashi, H.; Kaloriti, D.; Smertenko, A. The plant cytoskeleton: Recent advances in the study of the plant microtubule-associated proteins MAP-65, MAP-190 and the Xenopus MAP215-like protein, MOR1. Plant Mol. Biol. 2002, 50, 915–924. [Google Scholar] [CrossRef] [PubMed]
- Bramley, H.; Turner, D.W.; Tyerman, S.D.; Turner, N.C. Water Flow in the Roots of Crop Species: The Influence of Root Structure, Aquaporin Activity, and Waterlogging. Adv. Agron. 2007, 96, 133–196. [Google Scholar] [CrossRef]
- Van Engelen, F.A.; Hartog, M.V.; Thomas, T.L.; Taylor, B.; Sturm, A.; van Kammen, A.; de Vries, S.C. The carrot secreted glycoprotein gene EP1 is expressed in the epidermis and has sequence homology to Brassica S-locus glycoproteins. Plant J. 1993, 4, 855–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rygg, G.L. Sugars in the root of the carrot. Plant Physiol. 1945, 20, 47–50. [Google Scholar] [CrossRef] [PubMed]
- Bufler, G. Accumulation and degradation of starch in carrot roots. Sci. Hortic. (Amsterdam) 2013, 150. [Google Scholar] [CrossRef]
- Rolland, F.; Baena-Gonzalez, E.; Sheen, J. Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 2006, 57, 675–709. [Google Scholar] [CrossRef] [PubMed]
- Bowman, M. Gene Expression and Genetic Analysis of Carotenoid Pigment Accumulation in Carrot (Daucus carota L.). Ph.D. Thesis, University of Wisconsin-Madison, Madison, WI, USA, 2012. [Google Scholar]
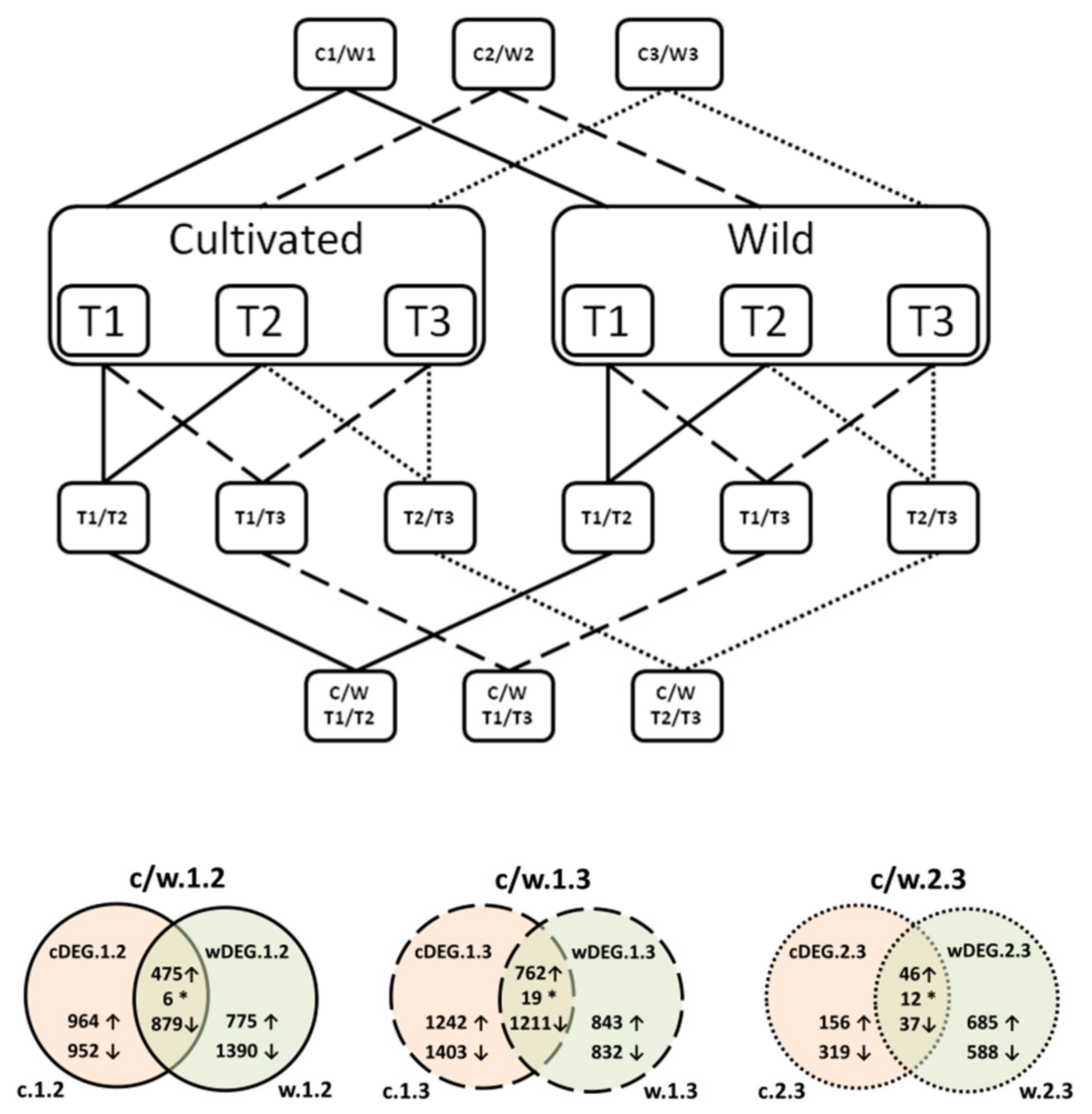
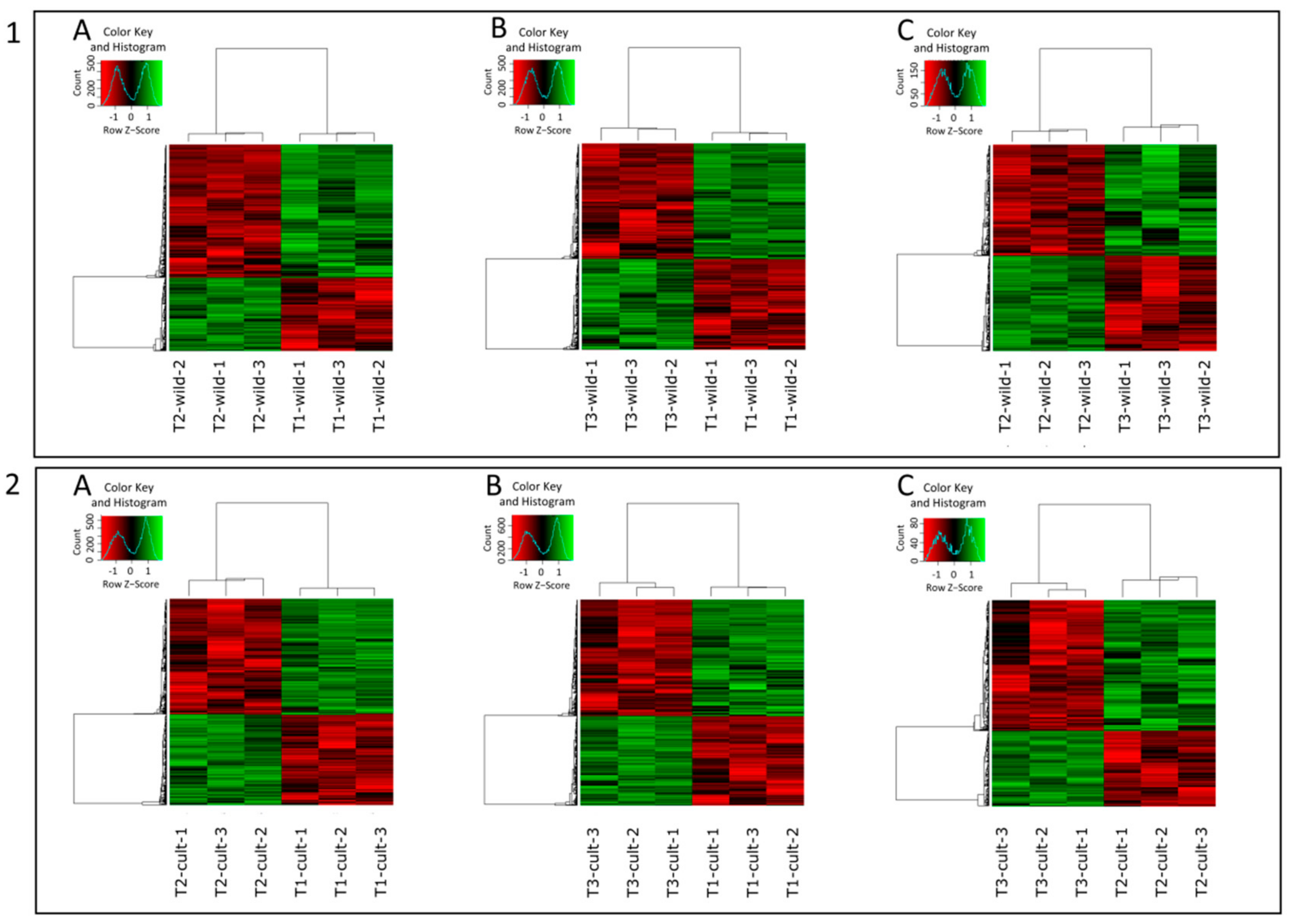
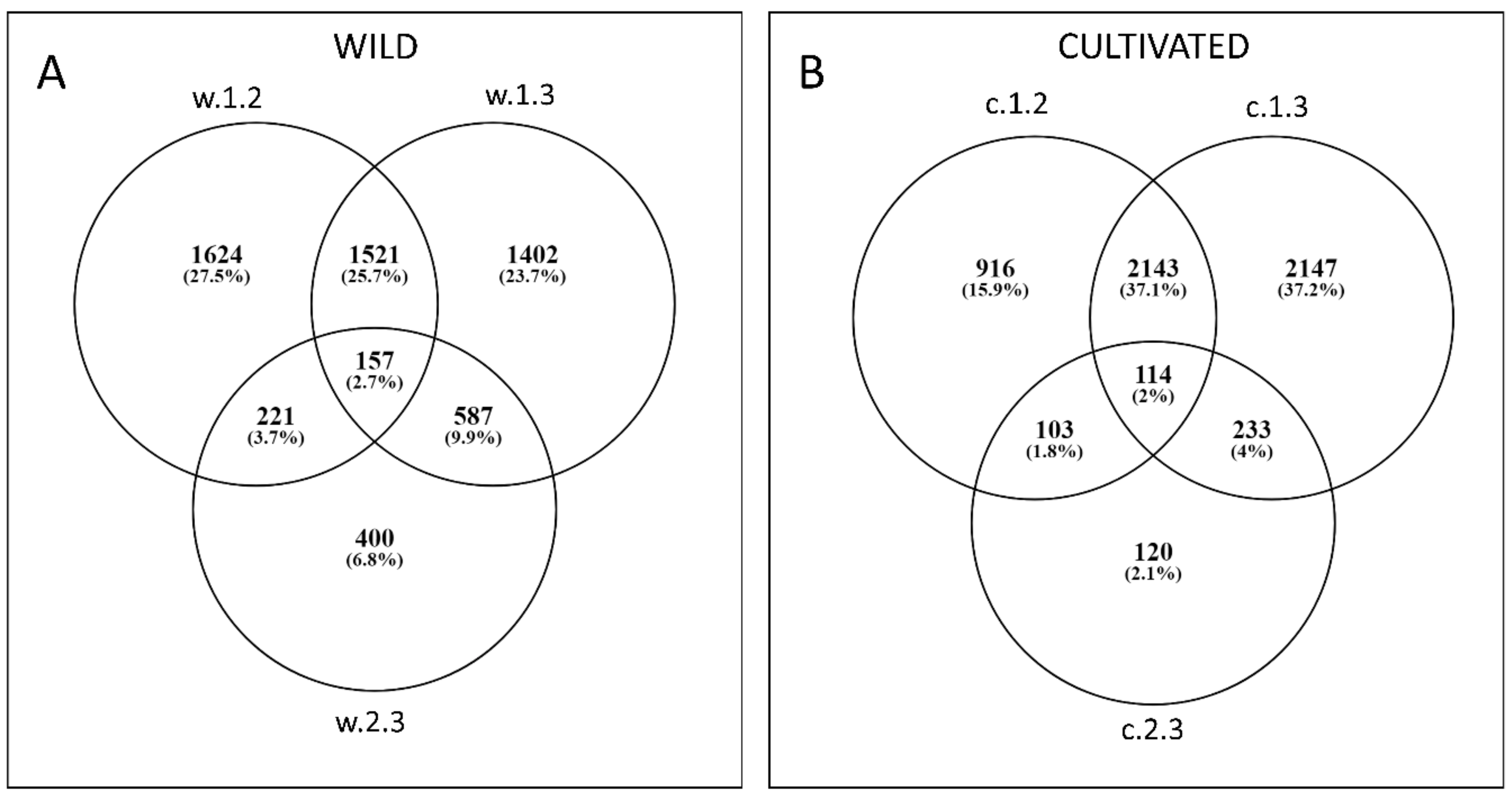
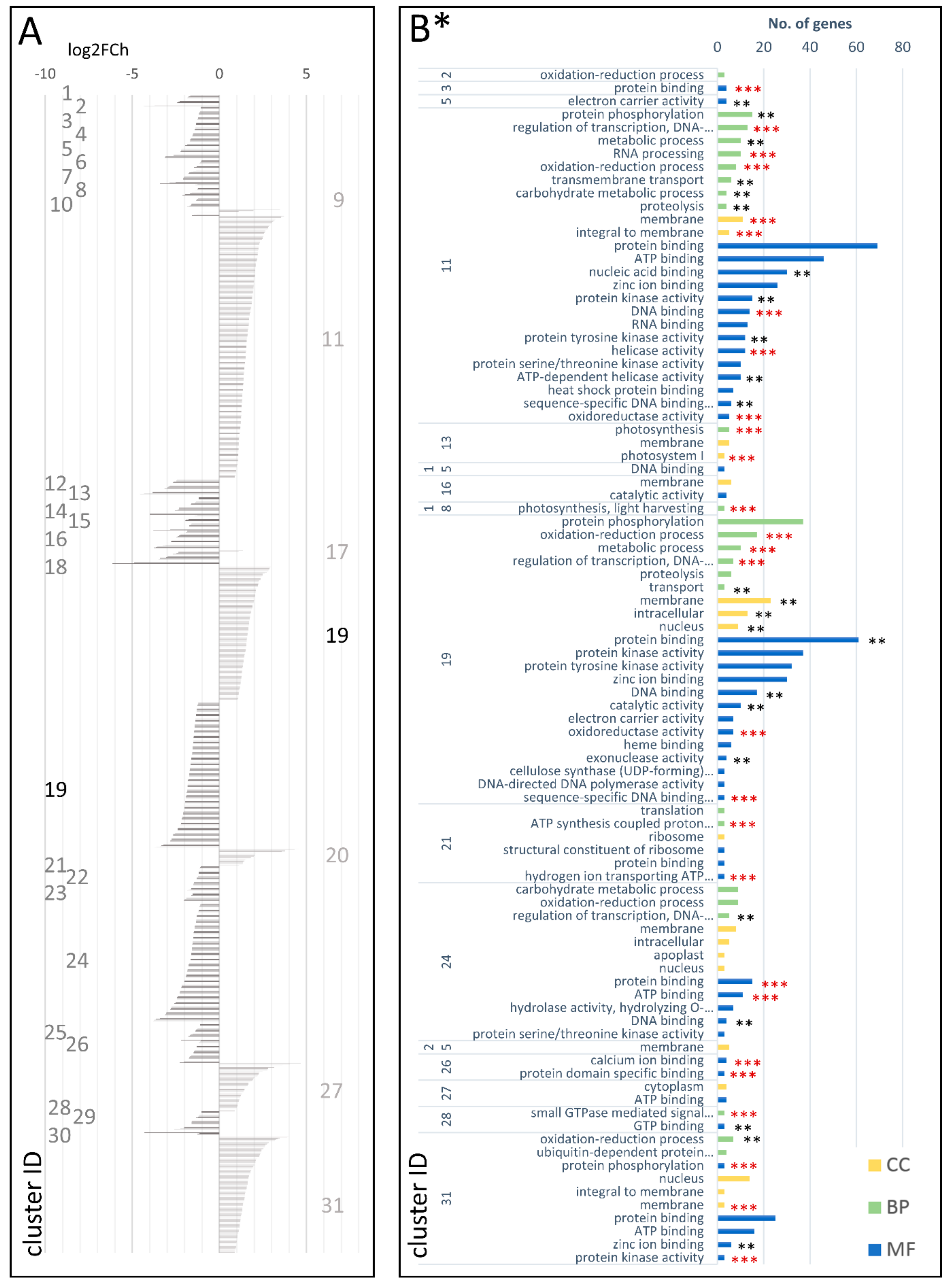
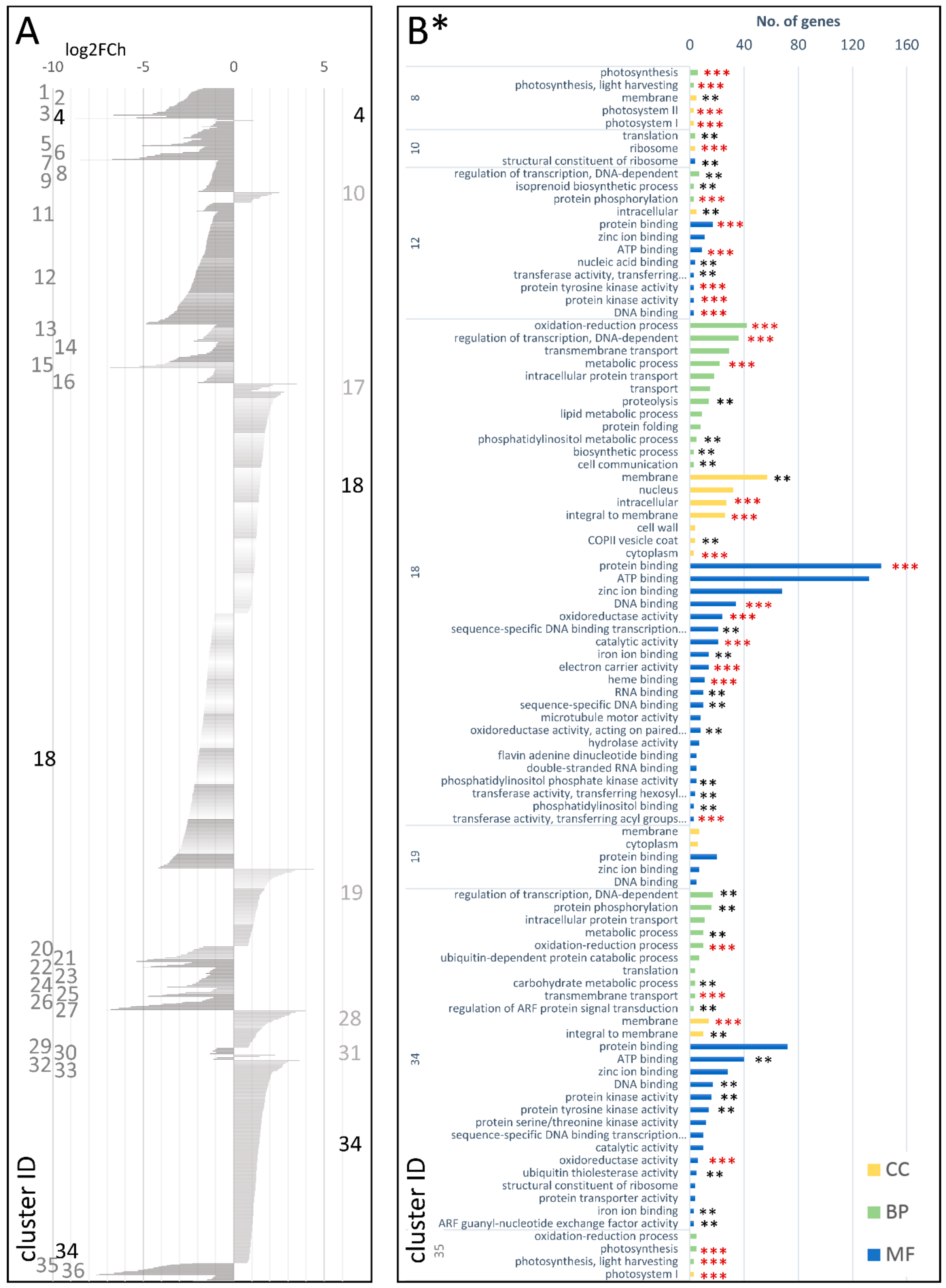
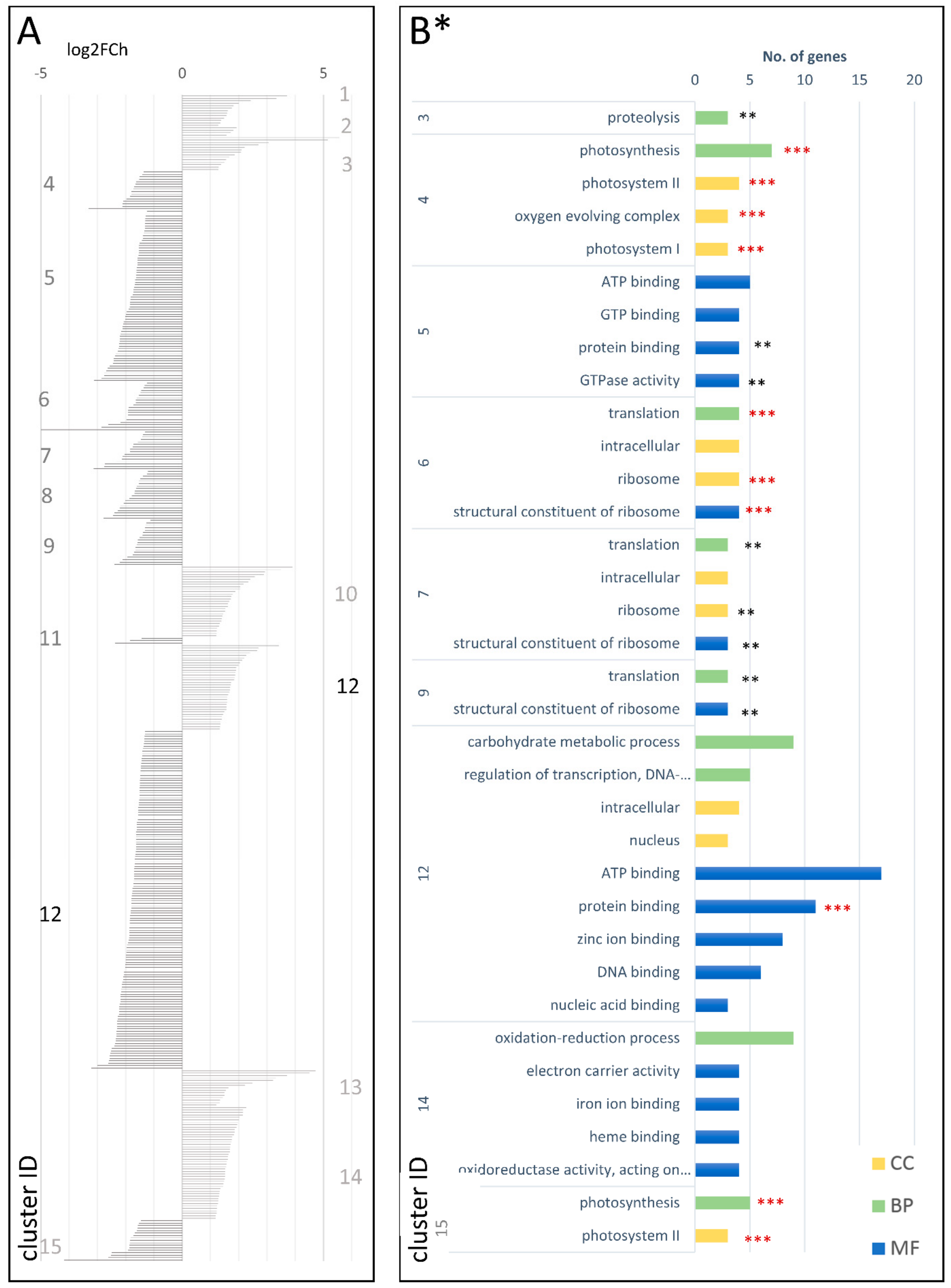

| Type of Comparison | Time Points | Cluster Number | Codes Used in the Text | |||
|---|---|---|---|---|---|---|
| Code | Description | Code | Description | Code | Description | |
| cDEG. | ‘c’ refers to DEGs observed in the cultivated roots but not in the wild roots | 1.2. | Digits refer to the timepoints (T1 vs. T2, T1 vs. T3 or T2 vs. T3) used for comparison | XX | Consecutive numbering resulting from GO enrichment analysis; ‘XX’ stands for the one or two digits representing the clusters | cDEG.1.2.XX |
| 1.3. | cDEG.1.3.XX | |||||
| 2.3. | cDEG.2.3.XX | |||||
| wDEG. | ‘w’ refers to DEGs observed in the wild roots but not in the cultivated roots | 1.2. | wDEG.1.2.XX | |||
| 1.3. | wDEG.1.3.XX | |||||
| 2.3. | wDEG.2.3.XX | |||||
| wcDEG. | ‘wc’ refers to DEGs observed for wild vs. cultivated comparison | 1 | Digit refers to the timepoint (T1, T2, or T3) for which the wild and cultivated transcriptomes were compared | wcDEG.1.XX | ||
| 2 | wcDEG.2.XX | |||||
| 3 | wcDEG.3.XX | |||||
| Comparison | Cluster ID | Gene ID | Functional Annotation | log2FoldChange |
|---|---|---|---|---|
| cDEG.1.2 | 11 | LOC108218660 | transcription factor GTE4-like | 2.218323 |
| cDEG.1.2 | 11 | LOC108218615 | transcription factor TCP8-like | 2.115051 |
| cDEG.1.2 | 11 | LOC108227595 | transcription factor GTE8-like | 2.110012 |
| cDEG.1.2 | 11 | LOC108204589 | trihelix transcription factor GT-2-like | 1.567139 |
| cDEG.1.2 | 11 | LOC108208638 | trihelix transcription factor ASIL1-like | 1.502276 |
| cDEG.1.2 | 11 | LOC108205163 | transcription factor EGL1-like | 1.268485 |
| cDEG.1.2 | 19 | LOC108196018 | transcription factor GTE7-like | 1.534413 |
| cDEG.1.2 | 19 | LOC108215981 | transcription factor IIIB 90 kDa subunit-like | 1.434734 |
| cDEG.1.2 | 19 | LOC108219662 | GATA transcription factor 11-like | 1.210731 |
| cDEG.1.2 | 19 | LOC108205675 | transcription factor TCP8-like | 1.074681 |
| cDEG.1.2 | 20 | LOC108196925 | transcription factor MYB1R1-like | 1.705882 |
| cDEG.1.2 | 27 | LOC108219074 | trihelix transcription factor PTL | 2.193835 |
| cDEG.1.2 | 27 | LOC108209692 | ethylene-responsive transcription factor ERF118-like | 2.100109 |
| cDEG.1.2 | 27 | LOC108209999 | transcription factor MYB1R1-like | 1.675776 |
| cDEG.1.2 | 27 | LOC108200607 | GATA transcription factor 16 | 1.372083 |
| cDEG.1.2 | 27 | LOC108219053 | ethylene-responsive transcription factor RAP2-13 | 1.068196 |
| cDEG.1.2 | 31 | LOC108223963 | bZIP transcription factor 60-like | 1.437431 |
| cDEG.1.2 | 31 | LOC108219044 | transcription factor TCP20 | 1.274651 |
| cDEG.1.2 | 31 | LOC108218833 | bZIP transcription factor 17-like | 1.194341 |
| cDEG.1.3 | 18 | LOC108204549 | general transcription factor 3C polypeptide 3 | 2.767626 |
| cDEG.1.3 | 18 | LOC108208932 | heat stress transcription factor C-1-like | 2.14659 |
| cDEG.1.3 | 18 | LOC108204669 | helicase-like transcription factor CHR28 | 1.576643 |
| cDEG.1.3 | 18 | LOC108219601 | WRKY transcription factor 1-like | 1.371966 |
| cDEG.1.3 | 18 | LOC108205270 | GATA transcription factor 26-like | 1.267808 |
| cDEG.1.3 | 18 | LOC108227355 | transcription factor bHLH130-like | 1.150305 |
| cDEG.1.3 | 19 | LOC108218615 | transcription factor TCP8-like | 3.326214 |
| cDEG.1.3 | 19 | LOC108218436 | nuclear transcription factor Y subunit A-10 | 2.583937 |
| cDEG.1.3 | 19 | LOC108205657 | WRKY transcription factor 21 | 2.093602 |
| cDEG.1.3 | 19 | LOC108213173 | ethylene-responsive transcription factor RAP2-1-like | 1.936345 |
| cDEG.1.3 | 19 | LOC108210769 | WRKY transcription factor 69 | 1.581155 |
| cDEG.1.3 | 19 | LOC108215331 | WRKY transcription factor 21 | 1.340796 |
| cDEG.1.3 | 19 | LOC108205942 | transcription factor MYB1R1-like | 1.16443 |
| cDEG.1.3 | 19 | LOC108213035 | transcription factor bHLH68 | 0.964932 |
| cDEG.1.3 | 19 | LOC108203417 | transcription factor GTE2-like | 0.924269 |
| cDEG.1.3 | 19 | LOC108219684 | trihelix transcription factor ASIL2 | 0.814925 |
| cDEG.1.3 | 28 | LOC108200607 | GATA transcription factor 16 | 1.452332 |
| cDEG.1.3 | 28 | LOC108196925 | transcription factor MYB1R1-like | 1.179838 |
| cDEG.1.3 | 34 | LOC108219539 | transcription factor TGA1-like | 2.492909 |
| cDEG.1.3 | 34 | LOC108200279 | WRKY transcription factor 57 | 1.972727 |
| cDEG.1.3 | 34 | LOC108227595 | transcription factor GTE8-like | 1.901753 |
| cDEG.1.3 | 34 | LOC108223963 | bZIP transcription factor 60-like | 1.775269 |
| cDEG.1.3 | 34 | LOC108206522 | ethylene-responsive transcription factor 4-like | 1.713784 |
| cDEG.1.3 | 34 | LOC108214474 | transcription factor LHW-like | 1.695994 |
| cDEG.1.3 | 34 | LOC108197784 | WRKY transcription factor 28 | 1.669975 |
| cDEG.1.3 | 34 | LOC108208638 | trihelix transcription factor ASIL1-like | 1.472099 |
| cDEG.1.3 | 34 | LOC108218767 | WRKY transcription factor 3 | 1.434899 |
| cDEG.1.3 | 34 | LOC108194726 | trihelix transcription factor ASR3 | 1.425662 |
| cDEG.1.3 | 34 | LOC108227612 | transcription factor 25 | 1.400666 |
| cDEG.1.3 | 34 | LOC108194205 | ethylene-responsive transcription factor ERF008-like | 1.361216 |
| cDEG.1.3 | 34 | LOC108197411 | transcription factor bHLH113-like | 1.250711 |
| cDEG.1.3 | 34 | LOC108205979 | trihelix transcription factor GT-1 | 1.052866 |
| cDEG.2.3 | 3 | LOC108220123 | ethylene-responsive transcription factor 2-like | 1.291574 |
| cDEG.2.3 | 5 | LOC108211036 | transcription factor PCL1-like | −1.98882 |
| cDEG.2.3 | 5 | LOC108224748 | nuclear transcription factor Y subunit C-1-like | −1.72038 |
| cDEG.2.3 | 5 | LOC108218926 | NAC transcription factor 29-like | −1.58027 |
| cDEG.2.3 | 12 | LOC108220417 | transcription factor MYB48-like | −2.28333 |
| cDEG.2.3 | 12 | LOC108214397 | heat stress transcription factor A-3-like | −1.49692 |
| cDEG.2.3 | 14 | LOC108211325 | ethylene-responsive transcription factor ERF010-like | 2.266422 |
| cDEG.2.3 | 14 | LOC108197506 | transcription factor bHLH147 | 1.218184 |
| cDEG.2.3 | 14 | LOC108192438 | nuclear transcription factor Y subunit A-1-like | 1.217534 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Machaj, G.; Bostan, H.; Macko-Podgórni, A.; Iorizzo, M.; Grzebelus, D. Comparative Transcriptomics of Root Development in Wild and Cultivated Carrots. Genes 2018, 9, 431. https://doi.org/10.3390/genes9090431
Machaj G, Bostan H, Macko-Podgórni A, Iorizzo M, Grzebelus D. Comparative Transcriptomics of Root Development in Wild and Cultivated Carrots. Genes. 2018; 9(9):431. https://doi.org/10.3390/genes9090431
Chicago/Turabian StyleMachaj, Gabriela, Hamed Bostan, Alicja Macko-Podgórni, Massimo Iorizzo, and Dariusz Grzebelus. 2018. "Comparative Transcriptomics of Root Development in Wild and Cultivated Carrots" Genes 9, no. 9: 431. https://doi.org/10.3390/genes9090431
APA StyleMachaj, G., Bostan, H., Macko-Podgórni, A., Iorizzo, M., & Grzebelus, D. (2018). Comparative Transcriptomics of Root Development in Wild and Cultivated Carrots. Genes, 9(9), 431. https://doi.org/10.3390/genes9090431






