Olfactory Communication via Microbiota: What Is Known in Birds?
Abstract
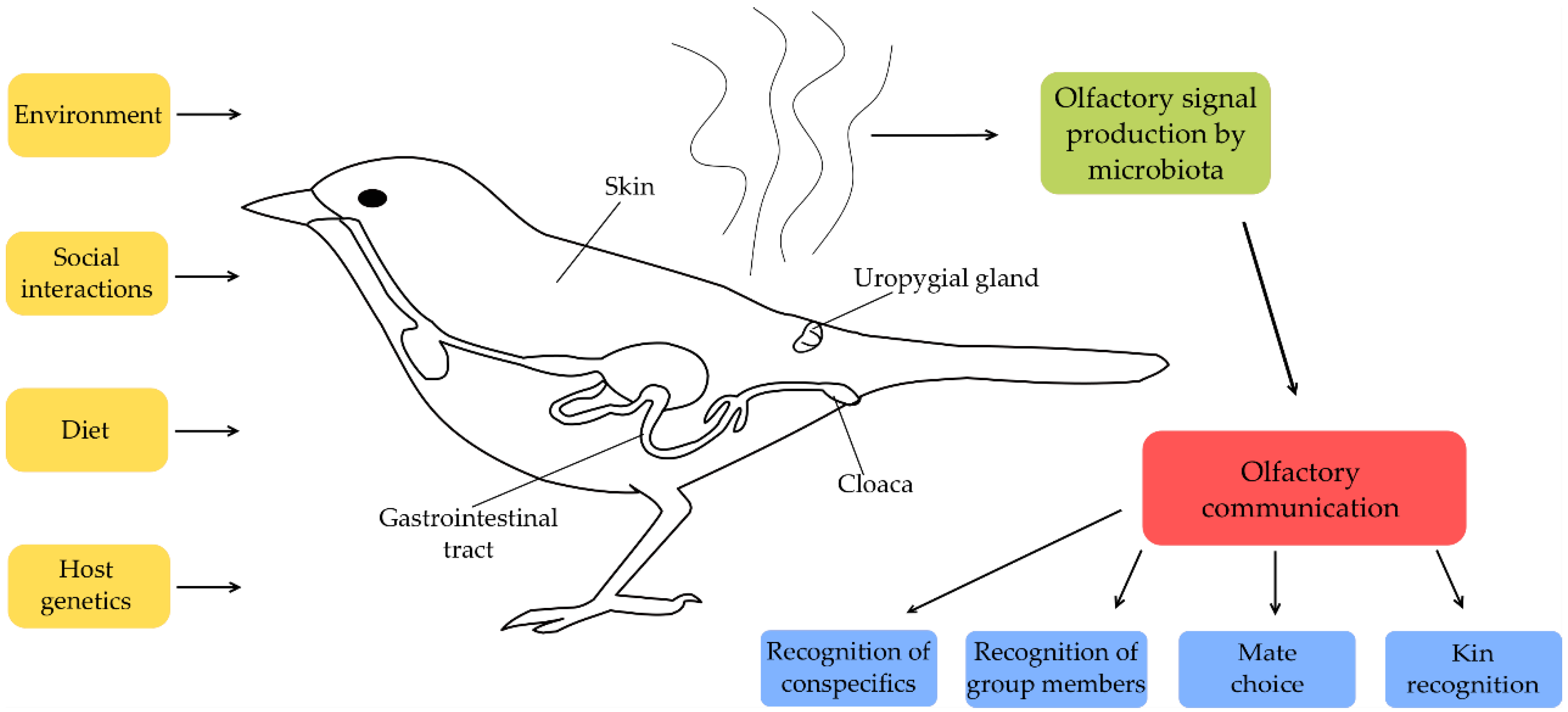
1. Introduction
2. Potential Body Regions of Microbe Induced Odour Production
3. Sources of Microbial Diversity in Birds
3.1. Host-Specific Factors
3.2. Environmental Factors Influencing Microbiota
3.3. Social Interactions Influencing Microbiota
4. Functional Aspects of Microbe Induced Odours in Social Communication
5. Evolutionary Implications and Future Prospects
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Fredrich, E.; Barzantny, H.; Brune, I.; Tauch, A. Daily battle against body odor: Towards the activity of the axillary microbiota. Trends Microbiol. 2013, 21, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Archie, E.A.; Theis, K.R. Animal behaviour meets microbial ecology. Anim. Behav. 2011, 82, 425–436. [Google Scholar] [CrossRef]
- Ezenwa, V.O.; Gerardo, N.M.; Inouye, D.W.; Medina, M.; Xavier, J.B. Microbiology. Animal behavior and the microbiome. Science 2012, 338, 198–199. [Google Scholar] [CrossRef] [PubMed]
- Ezenwa, V.O.; Williams, A.E. Microbes and animal olfactory communication: Where do we go from here? Bioessays 2014, 36, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.J.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of mammals and their gut microbes. Science 2008, 320, 1647–1651. [Google Scholar] [CrossRef] [PubMed]
- Org, E.; Parks, B.W.; Joo, J.W.J.; Emert, B.; Schwartzman, W.; Kang, E.Y.; Mehrabian, M.; Pan, C.; Knight, R.; Gunsalus, R.; et al. Genetic and environmental control of host-gut microbiota interactions. Genome Res. 2015, 25, 1558–1569. [Google Scholar] [CrossRef] [PubMed]
- Buffie, C.G.; Pamer, E.G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 2013, 13, 790–801. [Google Scholar] [CrossRef] [PubMed]
- Archie, E.A.; Tung, J. Social behavior and the microbiome. Curr. Opin. Behav. Sci. 2015, 6, 28–34. [Google Scholar] [CrossRef]
- Bravo, J.A.; Forsythe, P.; Chew, M.V.; Escaravage, E.; Savignac, H.M.; Dinan, T.G.; Bienenstock, J.; Cryan, J.F. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. USA 2011, 108, 16050–16055. [Google Scholar] [CrossRef] [PubMed]
- Gorman, M.; Nedwell, D.B.; Smith, R.M. An analysis of the contents of the anal scent pockets of Herpestes auropunctatus (Carnivora: Viverridae). J. Zool. 1974, 172, 389–399. [Google Scholar] [CrossRef]
- Gorman, M.L. A mechanism for individual recognition by odour in Herpestes auropunctatus (Carnivora: Viverridae). Anim. Behav. 1976, 24, 141–145. [Google Scholar] [CrossRef]
- Bordenstein, S.R.; O’Hara, F.P.; Werren, J.H. Wolbachia-induced incompatibility precedes other hybrid incompatibilities in Nasonia. Nature 2001, 409, 707–710. [Google Scholar] [CrossRef] [PubMed]
- Arbuthnott, D.; Levin, T.C.; Promislow, D.E.L. The impacts of Wolbachia and the microbiome on mate choice in Drosophila melanogaster. J. Evol. Biol. 2016, 29, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Sharon, G.; Segal, D.; Ringo, J.M.; Hefetz, A.; Zilber-Rosenberg, I.; Rosenberg, E. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2010, 107, 20051–20056. [Google Scholar] [CrossRef] [PubMed]
- Lizé, A.; McKay, R.; Lewis, Z. Gut microbiota and kin recognition. Trends Ecol. Evol. 2013, 28, 325–326. [Google Scholar] [CrossRef] [PubMed]
- Theis, K.R.; Venkataraman, A.; Dycus, J.A.; Koonter, K.D.; Schmitt-Matzen, E.N.; Wagner, A.P.; Holekamp, K.E.; Schmidt, T.M. Symbiotic bacteria appear to mediate hyena social odors. Proc. Natl. Acad. Sci. USA 2013, 110, 19832–19837. [Google Scholar] [CrossRef] [PubMed]
- Leclaire, S.; Nielsen, J.F.; Drea, C.M. Bacterial communities in meerkat anal scent secretions vary with host sex, age, and group membership. Behav. Ecol. 2014, 25, 996–1004. [Google Scholar] [CrossRef]
- Albone, E.S.; Gosden, P.E.; Ware, G.C. Bacteria as a Source of Chemical Signals in Mammals. In Chemical Signals in Vertebrates; Müller-Schwarze, D., Mozell, M.M., Eds.; Springer: Boston, MA, USA, 2012; pp. 35–43. [Google Scholar]
- Li, Q.; Korzan, W.J.; Ferrero, D.M.; Chang, R.B.; Roy, D.S.; Buchi, M.; Lemon, J.K.; Kaur, A.W.; Stowers, L.; Fendt, M.; et al. Synchronous evolution of an odor biosynthesis pathway and behavioral response. Curr. Biol. 2013, 23, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.B.; Herbert, J.; Roser, B.; Arnott, L.; Tucker, D.K.; Brown, R.E. Rearing rats in a germ-free environment eliminates their odors of individuality. J. Chem. Ecol. 1990, 16, 1667–1682. [Google Scholar] [CrossRef] [PubMed]
- Albone, E.S.; Perry, G.C. Anal sac secretion of the red fox, Vulpes vulpes; volatile fatty acids and diamines: Implications for a fermentation hypothesis of chemical recognition. J. Chem. Ecol. 1976, 2, 101–111. [Google Scholar] [CrossRef]
- Leclaire, S.; Jacob, S.; Greene, L.K.; Dubay, G.R.; Drea, C.M. Social odours covary with bacterial community in the anal secretions of wild meerkats. Sci. Rep. 2017, 7, 3240. [Google Scholar] [CrossRef] [PubMed]
- Sin, Y.W.; Buesching, C.D.; Burke, T.; Macdonald, D.W. Molecular characterization of the microbial communities in the subcaudal gland secretion of the European badger (Meles meles). FEMS Microbiol. Ecol. 2012, 81, 648–659. [Google Scholar] [CrossRef] [PubMed]
- Theis, K.R.; Schmidt, T.M.; Holekamp, K.E. Evidence for a bacterial mechanism for group-specific social odors among hyenas. Sci. Rep. 2012, 2, 615. [Google Scholar] [CrossRef] [PubMed]
- Voigt, C.C.; Caspers, B.A.; Speck, S. Bats, bacteria, and bat smell: Sex-specific diversity of microbes in a sexually selected scent organ. J. Mammal. 2005, 745–749, 745–749. [Google Scholar] [CrossRef]
- Zechman, J.M.; Martin, I.G.; Wellington, J.L.; Beauchamp, G.K. Perineal scent gland of wild and domestic cavies: Bacterial activity and urine as sources of biologically significant odors. Physiol. Behav. 1984, 32, 269–274. [Google Scholar] [CrossRef]
- Caro, S.P.; Balthazart, J.; Bonadonna, F. The perfume of reproduction in birds: Chemosignaling in avian social life. Horm. Behav. 2015, 68, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Hagelin, J.C.; Jones, I.L. Bird odors and other chemical substances: A defense mechanism or overlooked mode of intraspecific communication? Auk 2007, 124, 741–761. [Google Scholar] [CrossRef]
- Jacob, J.; Ziswiler, V. The uropygial gland. Avian Biol. 1982, 6, 199–324. [Google Scholar]
- Martín-Vivaldi, M.; Peña, A.; Peralta-Sánchez, J.M.; Sánchez, L.; Ananou, S.; Ruiz-Rodríguez, M.; Soler, J.J. Antimicrobial chemicals in hoopoe preen secretions are produced by symbiotic bacteria. Proc. Biol. Sci. 2010, 277, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, D.J.; Theis, K.R. Bacterial communities associated with junco preen glands: Preliminary ramifications for chemical signaling. In Chemical Signals in Vertebrates 13: Bacterial Communities Associated with Junco Preen Glands: Preliminary Ramifications for Chemical Signaling; Schulte, B.A., Goodwin, T.E., Ferkin, M.H., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 105–117. [Google Scholar]
- Law-Brown, J. Chemical defence in the red-billed wood hoopoe: Phoeniculus purpureus. Master’s Thesis, University of Cape Town, Cape Town, South Africa, 2001. [Google Scholar]
- Whittaker, D.J.; Gerlach, N.M.; Slowinski, S.P.; Corcoran, K.P.; Winters, A.D.; Soini, H.A.; Novotny, M.V.; Ketterson, E.D.; Theis, K.R. Social environment has a primary influence on the microbial and odor profiles of a chemically signaling songbird. Front. Ecol. Evol. 2016, 4, 245. [Google Scholar] [CrossRef]
- Dille, J.W.; Rogers, C.M.; Schneegurt, M.A. Isolation and characterization of bacteria from the feathers of wild Dark-eyed Juncos (Junco hyemalis). Auk 2016, 133, 155–167. [Google Scholar] [CrossRef]
- Møller, A.P.; Czirjak, G.Á.; Heeb, P. Feather micro-organisms and uropygial antimicrobial defences in a colonial passerine bird. Funct. Ecol. 2009, 23, 1097–1102. [Google Scholar] [CrossRef]
- Verea, C.; Vitelli-Flores, J.; Dorta, B.; Isturiz, T.; Solórzano, A.; Rodríguez-Lemoine, V.; Bosque, C. Feather-degrading bacteria from the plumage of neotropical spectacled thrushes (Turdus nudigenis). Auk 2014, 131, 100–109. [Google Scholar] [CrossRef]
- Jacob, S.; Sallé, L.; Zinger, L.; Chaine, A.S.; Ducamp, C.; Boutault, L.; Russell, A.F.; Heeb, P. Chemical regulation of body feather microbiota in a wild bird. Mol. Ecol. 2018, 27, 1727–1738. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Rueda, G. Preen oil and bird fitness: A critical review of the evidence. Biol. Rev. Camb. Philos. Soc. 2017, 92, 2131–2143. [Google Scholar] [CrossRef] [PubMed]
- Verhulst, N.O.; Qiu, Y.T.; Beijleveld, H.; Maliepaard, C.; Knights, D.; Schulz, S.; Berg-Lyons, D.; Lauber, C.L.; Verduijn, W.; Haasnoot, G.W.; et al. Composition of human skin microbiota affects attractiveness to malaria mosquitoes. PLoS ONE 2011, 6, e28991. [Google Scholar] [CrossRef] [PubMed]
- Engel, K.; Sauer, J.; Jünemann, S.; Winkler, A.; Wibberg, D.; Kalinowski, J.; Tauch, A.; Caspers, B.A. Individual- and species-specific skin microbiomes in three different estrildid finch species revealed by 16S amplicon sequencing. Microb. Ecol. 2018, 76, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Menon, G.K.; Menon, J. Avian epidermal lipids: Functional considerations and relationship to feathering. Integr. Comp. Biol. 2000, 40, 540–552. [Google Scholar] [CrossRef]
- Purton, M.D. Skin surface topography in the domestic fowl and Japanese quail. Br. Vet. J. 1986, 142, 446–452. [Google Scholar] [CrossRef]
- Caspers, B.A.; Gagliardo, A.; Krause, E.T. Impact of kin odour on reproduction in zebra finches. Behav. Ecol. Sociobiol. 2015, 69, 1827–1833. [Google Scholar] [CrossRef]
- van Veelen, H.P.J.; Falcao Salles, J.; Tieleman, B.I. Multi-level comparisons of cloacal, skin, feather and nest-associated microbiota suggest considerable influence of horizontal acquisition on the microbiota assembly of sympatric woodlarks and skylarks. Microbiome 2017, 5, 156. [Google Scholar] [CrossRef] [PubMed]
- Pearce, D.S.; Hoover, B.A.; Jennings, S.; Nevitt, G.A.; Docherty, K.M. Morphological and genetic factors shape the microbiome of a seabird species (Oceanodroma leucorhoa) more than environmental and social factors. Microbiome 2017, 5, 146. [Google Scholar] [CrossRef] [PubMed]
- Dillon, R.J.; Vennard, C.T.; Charnley, A.K. Exploitation of gut bacteria in the locust. Nature 2000, 403, 851. [Google Scholar] [CrossRef] [PubMed]
- Dillon, R.J.; Vennard, C.T.; Charnley, A.K. A Note: Gut bacteria produce components of a locust cohesion pheromone. J. Appl. Microbiol. 2002, 92, 759–763. [Google Scholar] [CrossRef] [PubMed]
- Dillon, R.J.; Dillon, V.M. The gut bacteria of insects: Nonpathogenic interactions. Annu. Rev. Entomol. 2004, 49, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Wada-Katsumata, A.; Zurek, L.; Nalyanya, G.; Roelofs, W.L.; Zhang, A.; Schal, C. Gut bacteria mediate aggregation in the German cockroach. Proc. Natl. Acad. Sci. USA 2015, 112, 15678–15683. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.K.; Choct, M.; Wu, S.; Swick, R.A. Nutritional effects on odour emissions in broiler production. Worlds Poult. Sci. J. 2017, 73, 257–280. [Google Scholar] [CrossRef]
- Lauzon, C.R.; Sjogren, R.E.; Wright, S.E.; Prokopy, R.J. Attraction of Rhagoletis pomonella (Diptera: Tephritidae) flies to odor of bacteria: Apparent confinement to specialized members of Enterobacteriaceae. Environ. Entomol. 1998, 27, 853–857. [Google Scholar] [CrossRef]
- Sears, H.F. Nesting behavior of the gull-billed tern. Bird-Band. 1978, 49, 1–16. [Google Scholar] [CrossRef]
- Caspers, B.A.; Krause, E.T. Odour-based natal nest recognition in the zebra finch (Taeniopygia guttata), a colony-breeding songbird. Biol. Lett. 2011, 7, 184–186. [Google Scholar] [CrossRef] [PubMed]
- Kruuk, H. Scent marking by otters (Lutra lutra): Signaling the use of resources. Behav. Ecol. 1992, 3, 133–140. [Google Scholar] [CrossRef]
- Marneweck, C.; Jürgens, A.; Shrader, A.M. Dung odours signal sex, age, territorial and oestrous state in white rhinos. Proc. R. Soc. B 2017, 284, 20162376. [Google Scholar] [CrossRef] [PubMed]
- Brooks, A.W.; Kohl, K.D.; Brucker, R.M.; van Opstal, E.J.; Bordenstein, S.R. Phylosymbiosis: Relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 2016, 14, e2000225. [Google Scholar] [CrossRef] [PubMed]
- Brucker, R.M.; Bordenstein, S.R. Speciation by symbiosis. Trends Ecol. Evol. 2012, 27, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Brucker, R.M.; Bordenstein, S.R. The hologenomic basis of speciation: Gut bacteria cause hybrid lethality in the genus Nasonia. Science 2013, 341, 667–669. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Rodríguez, M.; Lucas, F.S.; Heeb, P.; Soler, J.J. Differences in intestinal microbiota between avian brood parasites and their hosts. Biol. J. Linn. Soc. 2009, 96, 406–414. [Google Scholar] [CrossRef]
- Hird, S.M.; Sánchez, C.; Carstens, B.C.; Brumfield, R.T. Comparative gut microbiota of 59 neotropical bird species. Front. Microbiol. 2015, 6, 1403. [Google Scholar] [CrossRef] [PubMed]
- Kropáčková, L.; Těšický, M.; Albrecht, T.; Kubovčiak, J.; Čížková, D.; Tomášek, O.; Martin, J.-F.; Bobek, L.; Králová, T.; Procházka, P.; et al. Codiversification of gastrointestinal microbiota and phylogeny in passerines is not explained by ecological divergence. Mol. Ecol. 2017, 26, 5292–5304. [Google Scholar] [CrossRef] [PubMed]
- Waite, D.W.; Taylor, M.W. Characterizing the avian gut microbiota: Membership, driving influences, and potential function. Front. Microbiol. 2014, 5, 223. [Google Scholar] [CrossRef] [PubMed]
- García-Amado, M.A.; Shin, H.; Sanz, V.; Lentino, M.; Martínez, L.M.; Contreras, M.; Michelangeli, F.; Domínguez-Bello, M.G. Comparison of gizzard and intestinal microbiota of wild neotropical birds. PLoS ONE 2018, 13, e0194857. [Google Scholar] [CrossRef] [PubMed]
- Dewar, M.L.; Arnould, J.P.Y.; Dann, P.; Trathan, P.; Groscolas, R.; Smith, S. Interspecific variations in the gastrointestinal microbiota in penguins. Microbiologyopen 2013, 2, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Grond, K.; Ryu, H.; Baker, A.J.; Santo Domingo, J.W.; Buehler, D.M. Gastro-intestinal microbiota of two migratory shorebird species during spring migration staging in Delaware Bay, USA. J. Ornithol. 2014, 155, 969–977. [Google Scholar] [CrossRef]
- Krause, E.T.; Brummel, C.; Kohlwey, S.; Baier, M.C.; Müller, C.; Bonadonna, F.; Caspers, B.A. Differences in olfactory species recognition in the females of two Australian songbird species. Behav. Ecol. Sociobiol. 2014, 68, 1819–1827. [Google Scholar] [CrossRef]
- Sweeney, R.J.; Lovette, I.J.; Harvey, E.L. Evolutionary variation in feather waxes of passerine birds. Auk 2004, 121, 435–445. [Google Scholar] [CrossRef]
- Jiménez, E.; Marín, M.L.; Martín, R.; Odriozola, J.M.; Olivares, M.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Is meconium from healthy newborns actually sterile? Res. Microbiol. 2008, 159, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Hamady, M.; Knight, R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009, 19, 1141–1152. [Google Scholar] [CrossRef] [PubMed]
- Godoy-Vitorino, F.; Goldfarb, K.C.; Brodie, E.L.; Garcia-Amado, M.A.; Michelangeli, F.; Domínguez-Bello, M.G. Developmental microbial ecology of the crop of the folivorous hoatzin. ISME J. 2010, 4, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Van der Wielen, P.W.J.J.; Keuzenkamp, D.A.; Lipman, L.J.A.; Knapen, F.; Biesterveld, S. Spatial and temporal variation of the intestinal bacterial community in commercially raised broiler chickens during growth. Microb. Ecol. 2002, 44, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Idris, U.; Harmon, B.; Hofacre, C.; Maurer, J.J.; Lee, M.D. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 2003, 69, 6816–6824. [Google Scholar] [CrossRef] [PubMed]
- van Dongen, W.F.D.; White, J.; Brandl, H.B.; Moodley, Y.; Merkling, T.; Leclaire, S.; Blanchard, P.; Danchin, É.; Hatch, S.A.; Wagner, R.H. Age-related differences in the cloacal microbiota of a wild bird species. BMC Ecol. 2013, 13, 11. [Google Scholar] [CrossRef] [PubMed]
- Mueller, S.; Saunier, K.; Hanisch, C.; Norin, E.; Alm, L.; Midtvedt, T.; Cresci, A.; Silvi, S.; Orpianesi, C.; Verdenelli, M.C.; et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: A cross-sectional study. Appl. Environ. Microbiol. 2006, 72, 1027–1033. [Google Scholar] [CrossRef] [PubMed]
- Markle, J.G.M.; Frank, D.N.; Mortin-Toth, S.; Robertson, C.E.; Feazel, L.M.; Rolle-Kampczyk, U.; von Bergen, M.; McCoy, K.D.; Macpherson, A.J.; Danska, J.S. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013, 339, 1084–1088. [Google Scholar] [CrossRef] [PubMed]
- Haro, C.; Rangel-Zúñiga, O.A.; Alcalá-Díaz, J.F.; Gómez-Delgado, F.; Pérez-Martínez, P.; Delgado-Lista, J.; Quintana-Navarro, G.M.; Landa, B.B.; Navas-Cortés, J.A.; Tena-Sempere, M.; et al. Intestinal microbiota is influenced by gender and body mass index. PLoS ONE 2016, 11, e0154090. [Google Scholar] [CrossRef] [PubMed]
- Van Nas, A.; Guhathakurta, D.; Wang, S.S.; Yehya, N.; Horvath, S.; Zhang, B.; Ingram-Drake, L.; Chaudhuri, G.; Schadt, E.E.; Drake, T.A.; et al. Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinology 2009, 150, 1235–1249. [Google Scholar] [CrossRef] [PubMed]
- Yurkovetskiy, L.; Burrows, M.; Khan, A.A.; Graham, L.; Volchkov, P.; Becker, L.; Antonopoulos, D.; Umesaki, Y.; Chervonsky, A.V. Gender bias in autoimmunity is influenced by microbiota. Immunity 2013, 39, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Hird, S.M.; Carstens, B.C.; Cardiff, S.W.; Dittmann, D.L.; Brumfield, R.T. Sampling locality is more detectable than taxonomy or ecology in the gut microbiota of the brood-parasitic Brown-headed Cowbird (Molothrus ater). PeerJ 2014, 2, e321. [Google Scholar] [CrossRef] [PubMed]
- Lumpkins, B.S.; Batal, A.B.; Lee, M. The effect of gender on the bacterial community in the gastrointestinal tract of broilers. Poult. Sci. 2008, 87, 964–967. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-C.; Kil, D.Y.; Sul, W.J. Cecal microbiome divergence of broiler chickens by sex and body weight. J. Microbiol. 2017, 55, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Kreisinger, J.; Čížková, D.; Kropáčková, L.; Albrecht, T. Cloacal microbiome structure in a long-distance migratory bird assessed using deep 16sRNA pyrosequencing. PLoS ONE 2015, 10, e0137401. [Google Scholar] [CrossRef] [PubMed]
- Martín-Vivaldi, M.; Ruiz-Rodríguez, M.; José Soler, J.; Manuel Peralta-Sánchez, J.; Méndez, M.; Valdivia, E.; Manuel Martín-Platero, A.; Martínez-Bueno, M. Seasonal, sexual and developmental differences in hoopoe Upupa epops preen gland morphology and secretions: Evidence for a role of bacteria. J. Avian Biol. 2009, 40, 191–205. [Google Scholar] [CrossRef]
- Rajilić-Stojanović, M.; Smidt, H.; de Vos, W.M. Diversity of the human gastrointestinal tract microbiota revisited. Environ. Microbiol. 2007, 9, 2125–2136. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Zoetendal, E.G.; Akkermans, A.D.L.; Akkermans-van Vliet, W.M.; de Visser, J.A.G.M.; de Vos, W.M. The host genotype affects the bacterial community in the human gastronintestinal tract. Microb. Ecol. Health Dis. 2001, 13, 129–134. [Google Scholar] [CrossRef]
- Banks, J.C.; Cary, S.C.; Hogg, I.D. The phylogeography of Adelie penguin faecal flora. Environ. Microbiol. 2009, 11, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, G.; Siegel, P.; He, C.; Wang, H.; Zhao, W.; Zhai, Z.; Tian, F.; Zhao, J.; Zhang, H.; et al. Quantitative genetic background of the host influences gut microbiomes in chickens. Sci. Rep. 2013, 3, 1163. [Google Scholar] [CrossRef] [PubMed]
- Waite, D.W.; Taylor, M.W. Exploring the avian gut microbiota: Current trends and future directions. Front. Microbiol. 2015, 6, 673. [Google Scholar] [CrossRef] [PubMed]
- Kohl, K.D. Diversity and function of the avian gut microbiota. J. Comp. Physiol. B 2012, 182, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.; Heeb, P. Social and sexual behaviours aid transmission of bacteria in birds. Behav. Processes 2007, 74, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Toivanen, P.; Vaahtovuo, J.; Eerola, E. Influence of major histocompatibility complex on bacterial composition of fecal flora. Infect. Immun. 2001, 69, 2372–2377. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Bach, M.; Asquith, M.; Lee, A.Y.; Akileswaran, L.; Stauffer, P.; Davin, S.; Pan, Y.; Cambronne, E.D.; Dorris, M.; et al. HLA-B27 and human β2-microglobulin affect the gut microbiota of transgenic rats. PLoS ONE 2014, 9, e105684. [Google Scholar] [CrossRef] [PubMed]
- Bolnick, D.I.; Snowberg, L.K.; Caporaso, J.G.; Lauber, C.; Knight, R.; Stutz, W.E. Major Histocompatibility Complex class IIb polymorphism influences gut microbiota composition and diversity. Mol. Ecol. 2014, 23, 4831–4845. [Google Scholar] [CrossRef] [PubMed]
- De Palma, G.; Capilla, A.; Nadal, I.; Nova, E.; Pozo, T.; Varea, V.; Polanco, I.; Castillejo, G.; López, A.; Garrote, J.A.; et al. Interplay between human leukocyte antigen genes and the microbial colonization process of the newborn intestine. Curr. Issues Mol. Biol. 2010, 12, 1–10. [Google Scholar] [PubMed]
- Turnbaugh, P.J.; Quince, C.; Faith, J.J.; McHardy, A.C.; Yatsunenko, T.; Niazi, F.; Affourtit, J.; Egholm, M.; Henrissat, B.; Knight, R.; et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc. Natl. Acad. Sci. USA 2010, 107, 7503–7508. [Google Scholar] [CrossRef] [PubMed]
- Friswell, M.K.; Gika, H.; Stratford, I.J.; Theodoridis, G.; Telfer, B.; Wilson, I.D.; McBain, A.J. Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS ONE 2010, 5, e8584. [Google Scholar] [CrossRef] [PubMed]
- Song, S.J.; Lauber, C.; Costello, E.K.; Lozupone, C.A.; Humphrey, G.; Berg-Lyons, D.; Caporaso, J.G.; Knights, D.; Clemente, J.C.; Nakielny, S.; et al. Cohabiting family members share microbiota with one another and with their dogs. eLife Sci. 2013, 2, e00458. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, F.; Nguyen, T.L.A.; Brinkman, B.; Yunta, R.G.; Cauwe, B.; Vandenabeele, P.; Liston, A.; Raes, J. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013, 14, R4. [Google Scholar] [CrossRef] [PubMed]
- Klomp, J.E.; Murphy, M.T.; Smith, S.B.; McKay, J.E.; Ferrera, I.; Reysenbach, A.-L. Cloacal microbial communities of female spotted towhees Pipilo maculatus: Microgeographic variation and individual sources of variability. J. Avian Biol. 2008, 39, 530–538. [Google Scholar] [CrossRef]
- Funkhouser, L.J.; Bordenstein, S.R. Mom knows best: The universality of maternal microbial transmission. PLoS Biol. 2013, 11, e1001631. [Google Scholar] [CrossRef] [PubMed]
- Lucas, F.S.; Heeb, P. Environmental factors shape cloacal bacterial assemblages in great tit Parus major and blue tit P. caeruleus nestlings. J. Avian Biol. 2005, 36, 510–516. [Google Scholar] [CrossRef]
- Ruiz-Rodríguez, M.; Soler, J.J.; Martín-Vivaldi, M.; Martín-Platero, A.M.; Méndez, M.; Peralta-Sánchez, J.M.; Ananou, S.; Valdivia, E.; Martínez-Bueno, M. Environmental factors shape the community of symbionts in the hoopoe uropygial gland more than genetic factors. Appl. Environ. Microbiol. 2014, 80, 6714–6723. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, Á.; Martín-Vivaldi, M.; Ruiz-Rodríguez, M.; Martínez-Bueno, M.; Arco, L.; Rodríguez-Ruano, S.M.; Peralta-Sánchez, J.M.; Soler, J.J. The microbiome of the uropygial secretion in hoopoes is shaped along the nesting phase. Microb. Ecol. 2016, 72, 252–261. [Google Scholar] [CrossRef] [PubMed]
- González-Braojos, S.; Vela, A.I.; Ruiz-De-Castañeda, R.; Briones, V.; Cantarero, A.; Moreno, J. Is nestling growth affected by nest reuse and skin bacteria in pied flycatchers Ficedula Hypoleuca? Acta Ornithol. 2012, 47, 119–127. [Google Scholar] [CrossRef]
- Glünder, G. Influence of diet on the occurrence of some bacteria in the intestinal flora of wild and pet birds. DTW. Dtsch. Tierarztl. Wochensch. 2002, 109, 266–270. [Google Scholar]
- Blanco, G.; Lemus Jesús, A.; Grande, J. Retracted: Faecal bacteria associated with different diets of wintering red kites: Influence of livestock carcass dumps in microflora alteration and pathogen acquisition. J. Appl. Ecol. 2006, 43, 990–998. [Google Scholar] [CrossRef]
- Hammons, S.; Oh, P.L.; Martínez, I.; Clark, K.; Schlegel, V.L.; Sitorius, E.; Scheideler, S.E.; Walter, J. A small variation in diet influences the Lactobacillus strain composition in the crop of broiler chickens. Syst. Appl. Microbiol. 2010, 33, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Roggenbuck, M.; Bærholm Schnell, I.; Blom, N.; Bælum, J.; Bertelsen, M.F.; Sicheritz-Pontén, T.; Pontén, T.S.; Sørensen, S.J.; Gilbert, M.T.P.; Graves, G.R.; et al. The microbiome of New World vultures. Nat. Commun. 2014, 5, 5498. [Google Scholar] [CrossRef] [PubMed]
- Koch, H.; Schmid-Hempel, P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc. Natl. Acad. Sci. USA 2011, 108, 19288–19292. [Google Scholar] [CrossRef] [PubMed]
- Degnan, P.H.; Pusey, A.E.; Lonsdorf, E.V.; Goodall, J.; Wroblewski, E.E.; Wilson, M.L.; Rudicell, R.S.; Hahn, B.H.; Ochman, H. Factors associated with the diversification of the gut microbial communities within chimpanzees from Gombe National Park. Proc. Natl. Acad. Sci. USA 2012, 109, 13034–13039. [Google Scholar] [CrossRef] [PubMed]
- Moeller, A.H.; Foerster, S.; Wilson, M.L.; Pusey, A.E.; Hahn, B.H.; Ochman, H. Social behavior shapes the chimpanzee pan-microbiome. Sci. Adv. 2016, 2, e1500997. [Google Scholar] [CrossRef] [PubMed]
- Tung, J.; Barreiro, L.B.; Burns, M.B.; Grenier, J.-C.; Lynch, J.; Grieneisen, L.E.; Altmann, J.; Alberts, S.C.; Blekhman, R.; Archie, E.A. Social networks predict gut microbiome composition in wild baboons. eLife Sci. 2015, 4, e05224. [Google Scholar] [CrossRef] [PubMed]
- Perofsky, A.C.; Lewis, R.J.; Abondano, L.A.; Di Fiore, A.; Meyers, L.A. Hierarchical social networks shape gut microbial composition in wild Verreaux’s sifaka. Proc. Biol. Sci. 2017, 284. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, M.P. Access to mutualistic endosymbiotic microbes: An underappreciated benefit of group living. Behav. Ecol. Sociobiol. 2008, 62, 479–497. [Google Scholar] [CrossRef]
- Cooper, R.G. Ostrich (Struthio camelus) chick and grower nutrition. Anim. Sci. J. 2004, 75, 487–490. [Google Scholar] [CrossRef]
- White, J.; Mirleau, P.; Danchin, E.; Mulard, H.; Hatch, S.A.; Heeb, P.; Wagner, R.H. Sexually transmitted bacteria affect female cloacal assemblages in a wild bird. Ecol. Lett. 2010, 13, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.; RAMBO, T.B. Cloacal microbes in house sparrows. Condor 2000, 102, 679–684. [Google Scholar] [CrossRef]
- Westneat, D.F.; Birch Rambo, T. Copulation exposes female red-winged blackbirds to bacteria in male semen. J. Avian Biol. 2000, 31, 1–7. [Google Scholar] [CrossRef]
- Penn, D.; Potts, W. How do Major Histocompatibility complex genes influence odor and mating preferences. Adv. Immunol. 1998, 69, 411–436. [Google Scholar] [CrossRef] [PubMed]
- Shirasu, M.; Touhara, K. The scent of disease: Volatile organic compounds of the human body related to disease and disorder. J. Biochem. 2011, 150, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Ehman, K.D.; Scott, M.E. Female mice mate preferentially with non-parasitized males. Parasitology 2002, 125, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Lizé, A.; Khidr, S.K.; Hardy, I.C.W. Two components of kin recognition influence parasitoid aggression in resource competition. Anim. Behav. 2012, 83, 793–799. [Google Scholar] [CrossRef]
- Lewis, Z.; Lizé, A. Insect behaviour and the microbiome. Curr. Opin. Insect Sci. 2015, 9, 86–90. [Google Scholar] [CrossRef]
- Lizé, A.; McKay, R.; Lewis, Z. Kin recognition in Drosophila: The importance of ecology and gut microbiota. ISME J. 2014, 8, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K. Nestmate recognition mediated by intestinal bacteria in a termite, Reticulitermes speratus. Oikos 2001, 92, 20–26. [Google Scholar] [CrossRef]
- Hongoh, Y.; Deevong, P.; Inoue, T.; Moriya, S.; Trakulnaleamsai, S.; Ohkuma, M.; Vongkaluang, C.; Noparatnaraporn, N.; Kudo, T. Intra- and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl. Environ. Microbiol. 2005, 71, 6590–6599. [Google Scholar] [CrossRef] [PubMed]
- Heys, C.; Lizé, A.; Blow, F.; White, L.; Darby, A.; Lewis, Z.J. The effect of gut microbiota elimination in Drosophila melanogaster: A how-to guide for host-microbiota studies. Ecol. Evol. 2018, 8, 4150–4161. [Google Scholar] [CrossRef] [PubMed]
- Dosmann, A.; Bahet, N.; Gordon, D.M. Experimental modulation of external microbiome affects nestmate recognition in harvester ants (Pogonomyrmex barbatus). PeerJ 2016, 4, e1566. [Google Scholar] [CrossRef] [PubMed]
- Burgener, N.; East, M.L.; Hofer, H.; Dehnhard, M. Do spotted hyena scent marks code for clan membership? Chem. Signals Vertebr. 2008, 11, 192–210. [Google Scholar]
- Roper, T.J. Olfaction in birds. Adv. Stud. Behav. 1999, 28, 247–332. [Google Scholar]
- Caro, S.P.; Balthazart, J. Pheromones in birds: Myth or reality? J. Comp. Phys. A 2010, 196, 751–766. [Google Scholar] [CrossRef] [PubMed]
- Krause, E.T.; Bischof, H.-J.; Engel, K.; Golüke, S.; Maraci, Ö.; Mayer, U.; Sauer, J.; Caspers, B.A. Olfaction in the zebra finch (Taeniopygia guttata): What is known and further perspectives. In Advances in the Study of Behavior; Academic Press: Cambridge, MA, USA, 2017; pp. 37–85. [Google Scholar]
- Caspers, B.A.; Hagelin, J.C.; Paul, M.; Bock, S.; Willeke, S.; Krause, E.T. Zebra Finch chicks recognise parental scent, and retain chemosensory knowledge of their genetic mother, even after egg cross-fostering. Sci. Rep. 2017, 7, 12859. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, D.J.; Richmond, K.M.; Miller, A.K.; Kiley, R.; Bergeon Burns, C.; Atwell, J.W.; Ketterson, E.D. Intraspecific preen oil odor preferences in dark-eyed juncos (Junco hyemalis). Behav. Ecol. 2011, 22, 1256–1263. [Google Scholar] [CrossRef]
- Amo, L.; Avilés, J.M.; Parejo, D.; Peña, A.; Rodríguez, J.; Tomás, G. Sex recognition by odour and variation in the uropygial gland secretion in starlings. J. Anim. Ecol. 2012, 81, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Amo, L.; López-Rull, I.; Pagán, I.; Macías Garcia, C. Male quality and conspecific scent preferences in the house finch, Carpodacus mexicanus. Anim. Behav. 2012, 84, 1483–1489. [Google Scholar] [CrossRef]
- Cook, M.I.; Beissinger, S.R.; Toranzos, G.A.; Rodriguez, R.A.; Arendt, W.J. Microbial infection affects egg viability and incubation behavior in a tropical passerine. Behav. Ecol. 2005, 16, 30–36. [Google Scholar] [CrossRef]
- Cook, M.I.; Beissinger, S.R.; Toranzos, G.A.; Rodriguez, R.A.; Arendt, W.J. Trans-shell infection by pathogenic micro-organisms reduces the shelf life of non-incubated bird’s eggs: A constraint on the onset of incubation? Proc. Biol. Sci. 2003, 270, 2233–2240. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-de-Castañeda, R.; Vela, A.I.; Lobato, E.; Briones, V.; Moreno, J. Bacterial loads on eggshells of the pied flycatcher: environmental and maternal factors. Condor 2011, 113, 200–208. [Google Scholar] [CrossRef]
- Martín-Vivaldi, M.; Soler, J.J.; Peralta-Sánchez, J.M.; Arco, L.; Martín-Platero, A.M.; Martínez-Bueno, M.; Ruiz-Rodríguez, M.; Valdivia, E. Special structures of hoopoe eggshells enhance the adhesion of symbiont-carrying uropygial secretion that increase hatching success. J. Anim. Ecol. 2014, 83, 1289–1301. [Google Scholar] [CrossRef] [PubMed]
- Goodenough, A.E.; Stallwood, B.; Dandy, S.; Nicholson, T.E.; Stubbs, H.; Coker, D.G. Like mother like nest: similarity in microbial communities of adult female pied flycatchers and their nests. J. Ornithol. 2017, 158, 233–244. [Google Scholar] [CrossRef]
- Brandl, H.B.; van Dongen, W.F.D.; Darolová, A.; Krištofík, J.; Majtan, J.; Hoi, H. Composition of bacterial assemblages in different components of reed warbler nests and a possible role of egg incubation in pathogen regulation. PLoS ONE 2014, 9, e114861. [Google Scholar] [CrossRef] [PubMed]
| Study Species | References | |
|---|---|---|
| Host Environment | ||
| Nest Environment | Great tit (Parus major), blue tit (Parus caeruleus) | [102] |
| Woodlark (Lullula arborea), skylark (Alauda arvensis) | [44] | |
| Dark-eyed junco (Junco hyemalis) | [33] | |
| Hoopoe (Upupa epops) | [103] | |
| Pied flycatcher (Ficedula hypoleuca) | [105] | |
| Habitat/Geographic Proximity | Adelie penguin (Pygoscelis adeliae) | [87] |
| Towhee (Pipilo maculatus) | [100] | |
| Brown-headed Cowbird (Molothrus ater) | [79] | |
| Comp. analysis on 59 neotropical bird species | [60] | |
| Zebra finch (Taeniopygia guttata) | [91] | |
| Social Environment | Dark-eyed junco (J. hyemalis) | [31] |
| Zebra finch (T. guttata) | [91] | |
| Black-legged kittiwake (Rissa tridactyla) | [117] | |
| Red-winged blackbird (Agelaius phoeniceus) | [119] | |
| House sparrow (Passer domesticus) | [118] | |
| Barn swallow (Hirundo rustica) | [82] | |
| Host-Specific Factors | ||
| Host Taxonomy | King penguin (Aptenodytes patagonicus), gentoo penguin (Pygoscelis papua), macaroni penguin (Eudyptes chrysolophus), little penguin (Eudyptula minor) | [64] |
| Red knot (Calidris canutus), ruddy turnstone (Arenaria interpres) | [65] | |
| Zebra finch (T. guttata), diamond firetail (Stagonopleura guttata), Bengalese finch (Lonchura striata domestica) | [40] | |
| Magpie (Pica pica), great spotted cuckoo (Clamator glandarius) | [59] | |
| Comp. analysis on 59 neotropical bird species | [60] | |
| Comparative study on 51 bird species | [61] | |
| Metaanalyses | [62] | |
| Comparative analyses on 8 neotropical bird species | [63] | |
| Host Genetics and Genetic Similarity | Chicken (Gallus gallus) | [88] |
| Adelie penguin (P. adeliae) | [87] | |
| Age/Reproductive Status | Hoatzin (Opisthocomus hoazin) | [70] |
| Chicken (G. gallus) | [71,72] | |
| Kittiwake (R. tridactyla) | [73] | |
| Hoopoe (U. epops) | [83] | |
| Sex | Brown-headed cowbird (M. ater) | [79] |
| Chicken (G. gallus) | [80,81] | |
| Leach’s storm petrel (Oceanodroma leucorhoa) | [45] | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maraci, Ö.; Engel, K.; Caspers, B.A. Olfactory Communication via Microbiota: What Is Known in Birds? Genes 2018, 9, 387. https://doi.org/10.3390/genes9080387
Maraci Ö, Engel K, Caspers BA. Olfactory Communication via Microbiota: What Is Known in Birds? Genes. 2018; 9(8):387. https://doi.org/10.3390/genes9080387
Chicago/Turabian StyleMaraci, Öncü, Kathrin Engel, and Barbara A. Caspers. 2018. "Olfactory Communication via Microbiota: What Is Known in Birds?" Genes 9, no. 8: 387. https://doi.org/10.3390/genes9080387
APA StyleMaraci, Ö., Engel, K., & Caspers, B. A. (2018). Olfactory Communication via Microbiota: What Is Known in Birds? Genes, 9(8), 387. https://doi.org/10.3390/genes9080387





