Origins Left, Right, and Centre: Increasing the Number of Initiation Sites in the Escherichia coli Chromosome
Abstract
1. Introduction
2. Material and Methods
2.1. Bacterial Strains and General Methods
2.2. Growth Media
2.3. Marker Frequency Analysis by Deep Sequencing
2.4. Locally Estimated Scatterplot Smoothing (LOESS) Regression
2.5. Growth Curves
2.6. Mathematical Modelling
3. Results
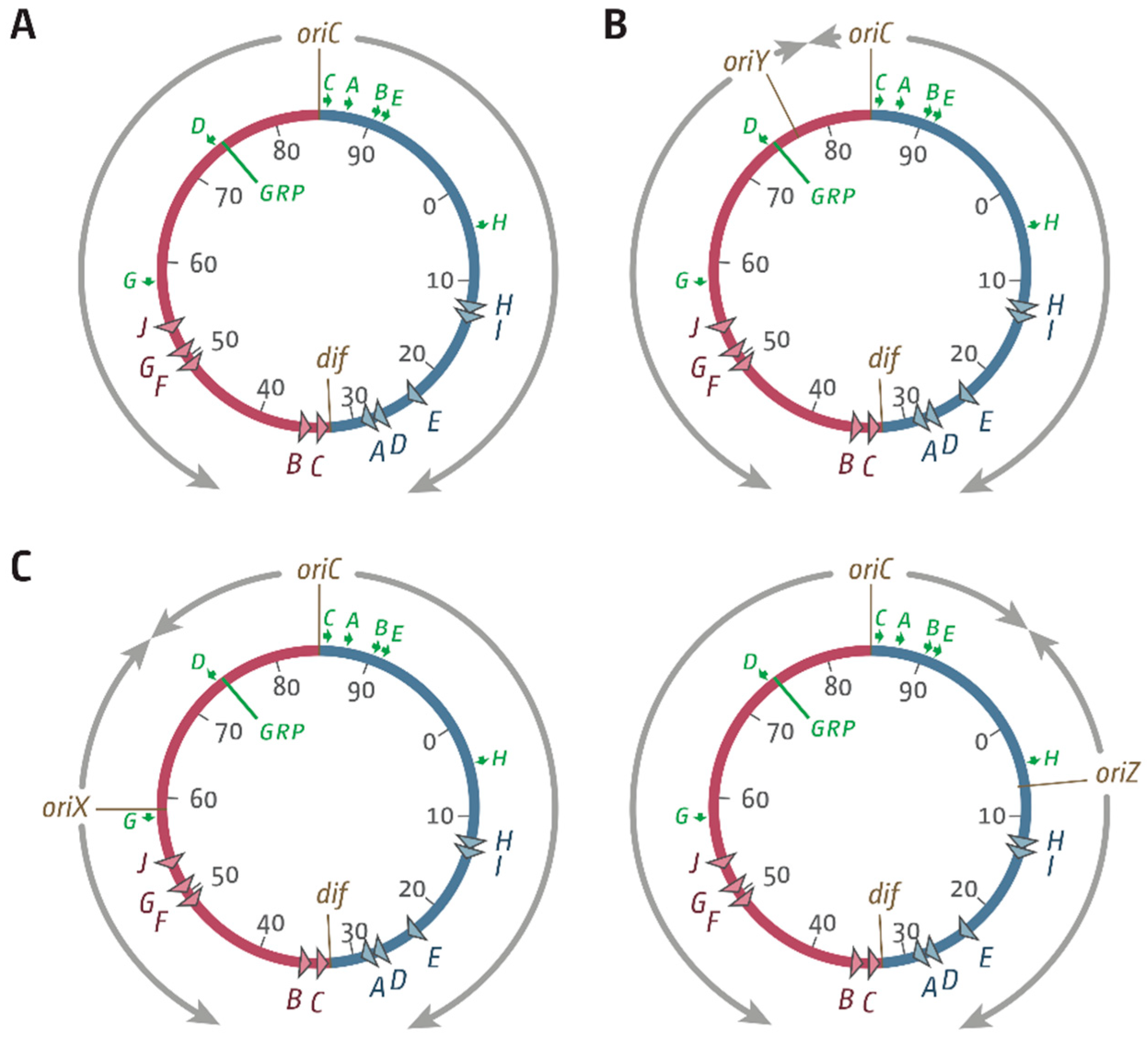
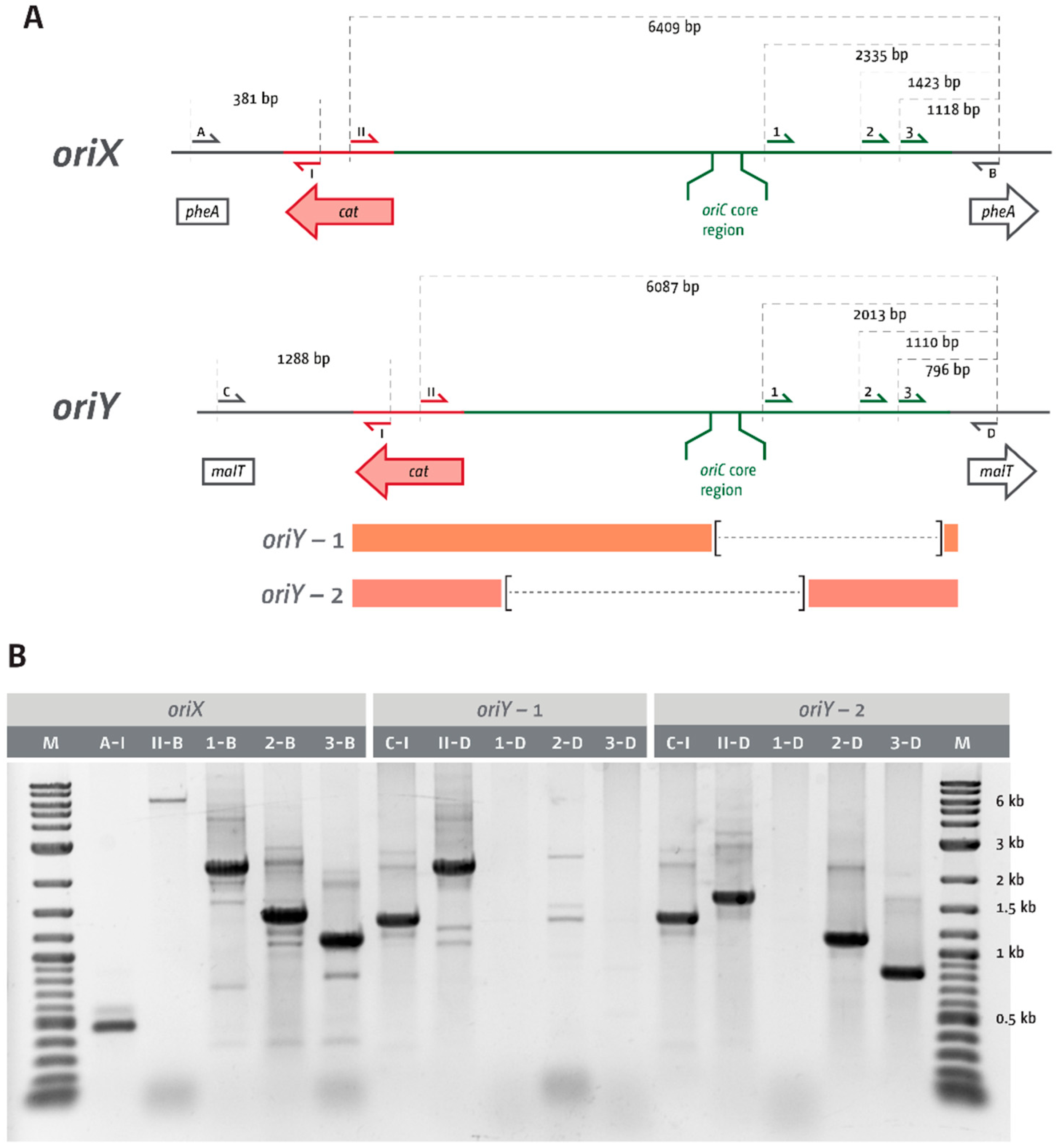
3.1. Ectopic Replication Origins in the Left-Hand Replichore
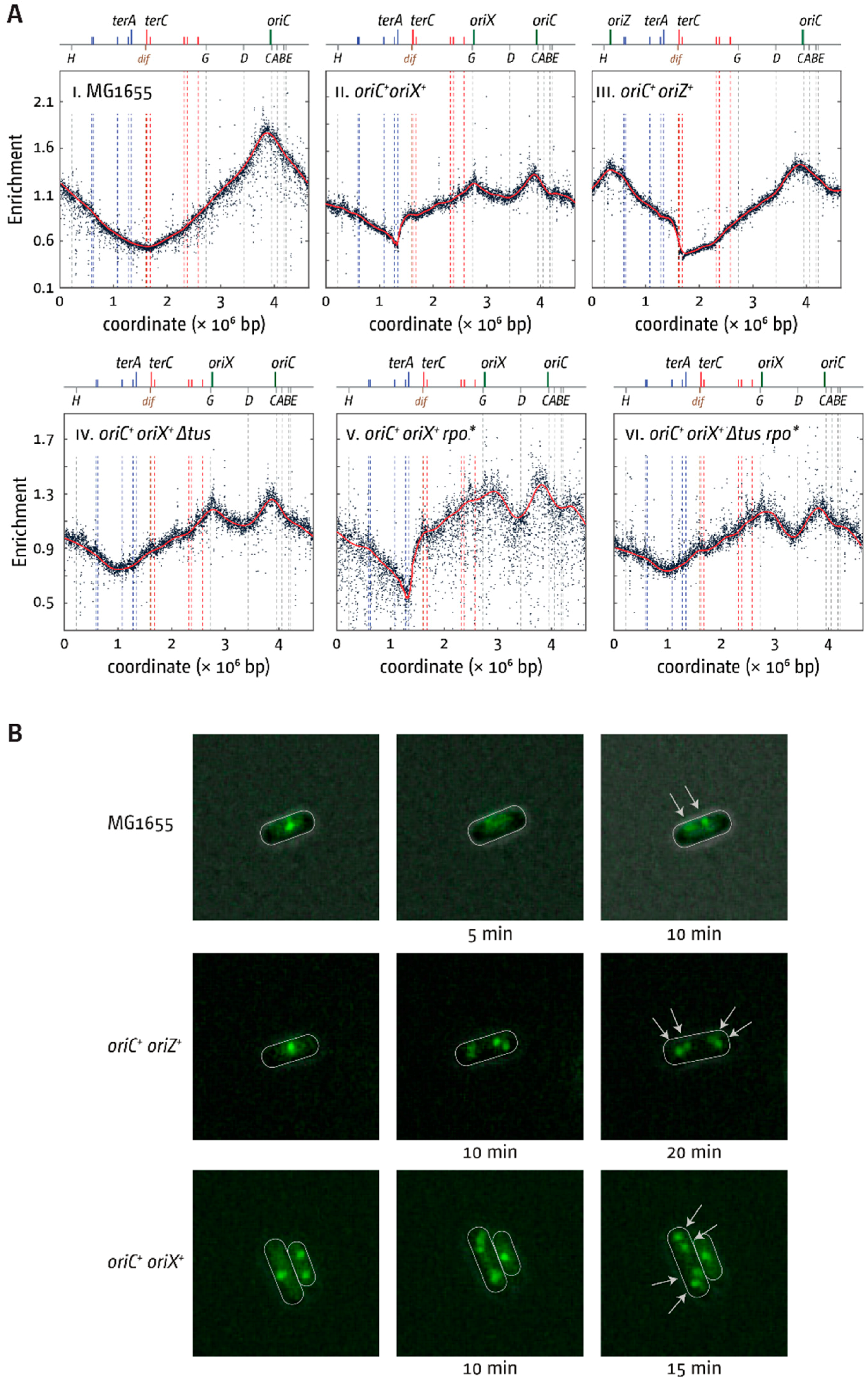
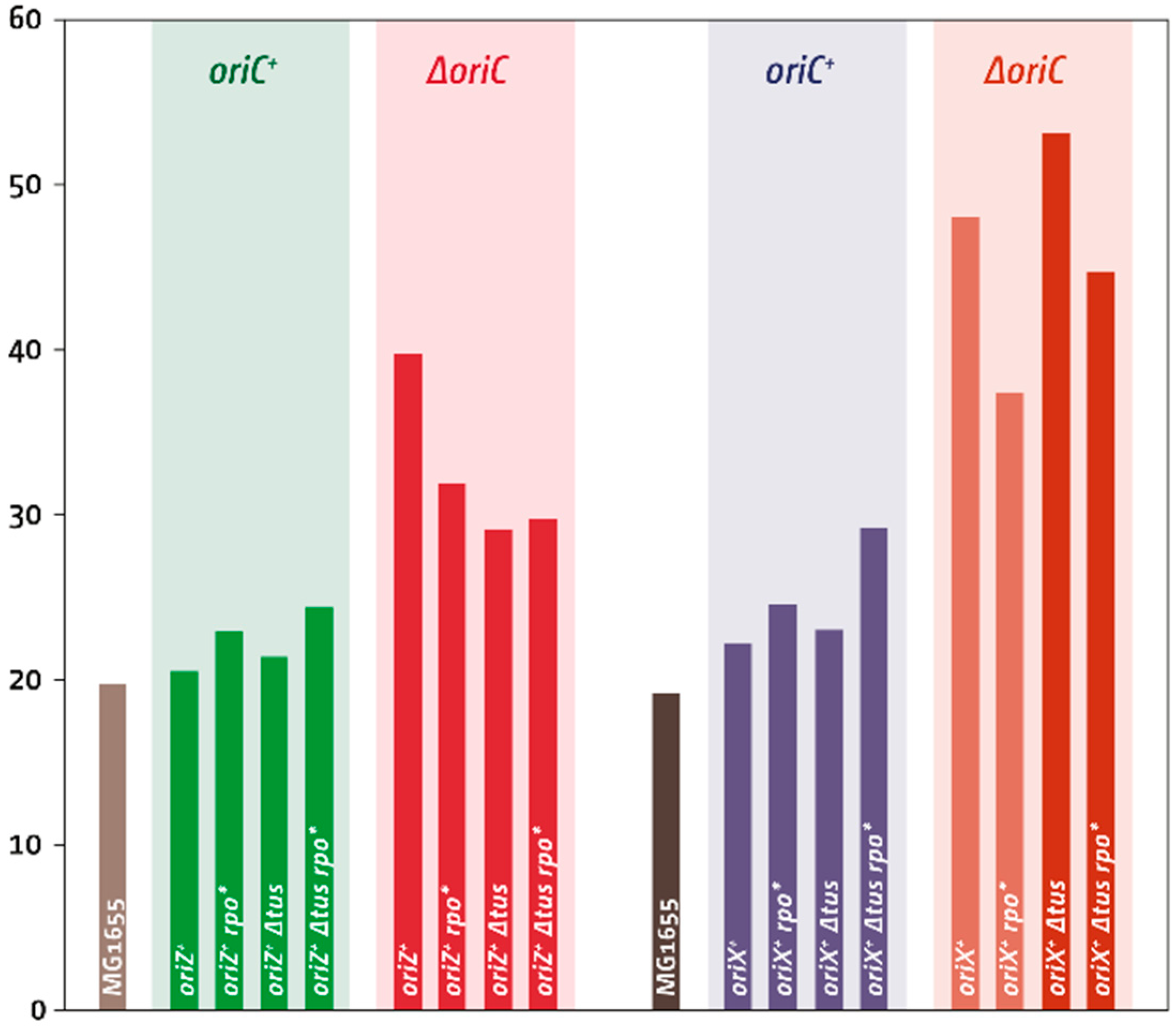
3.2. oriX Is Active in Double-Origin Cells
3.3. Termination and Replication–Transcription Conflicts in Double-Origin Cells
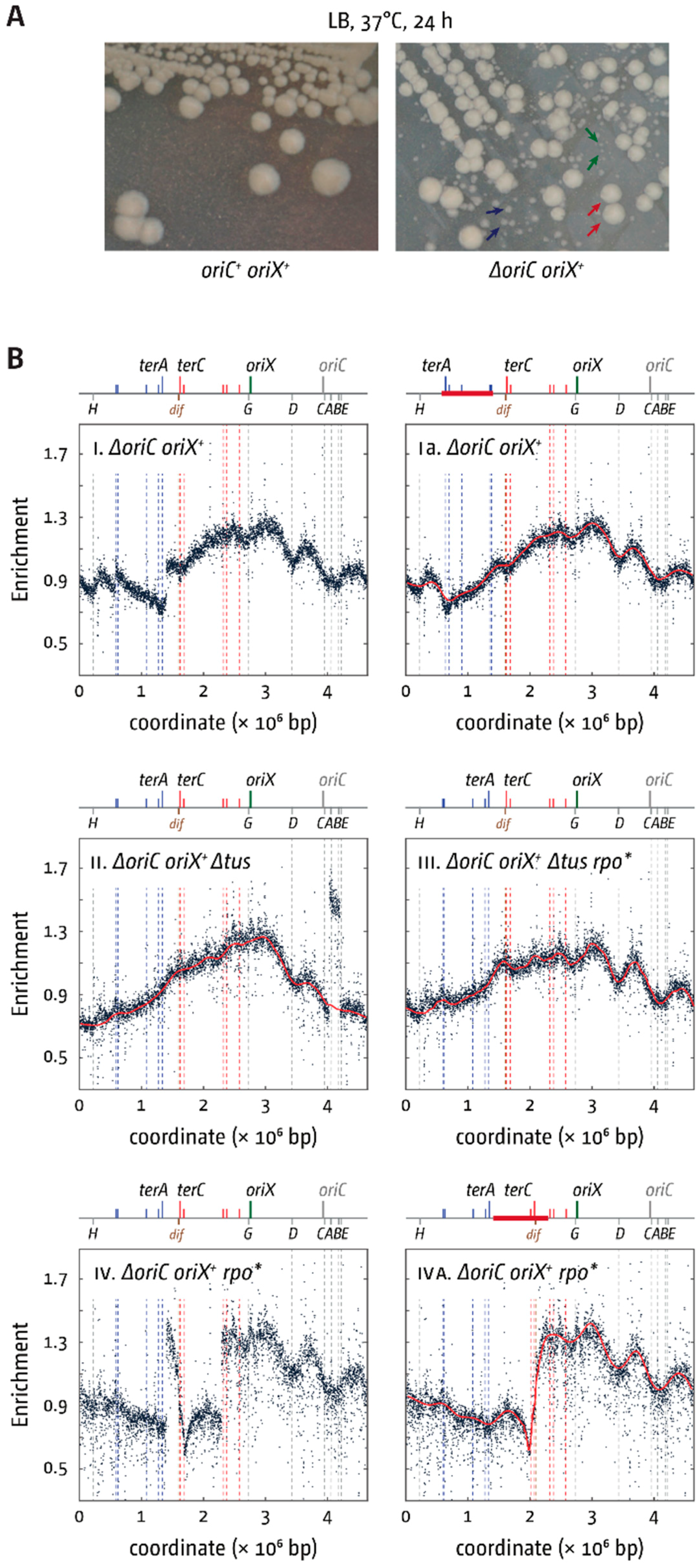
3.4. Deletion of oriC in Double-Origin Mutants Triggers Chromosomal Rearrangements
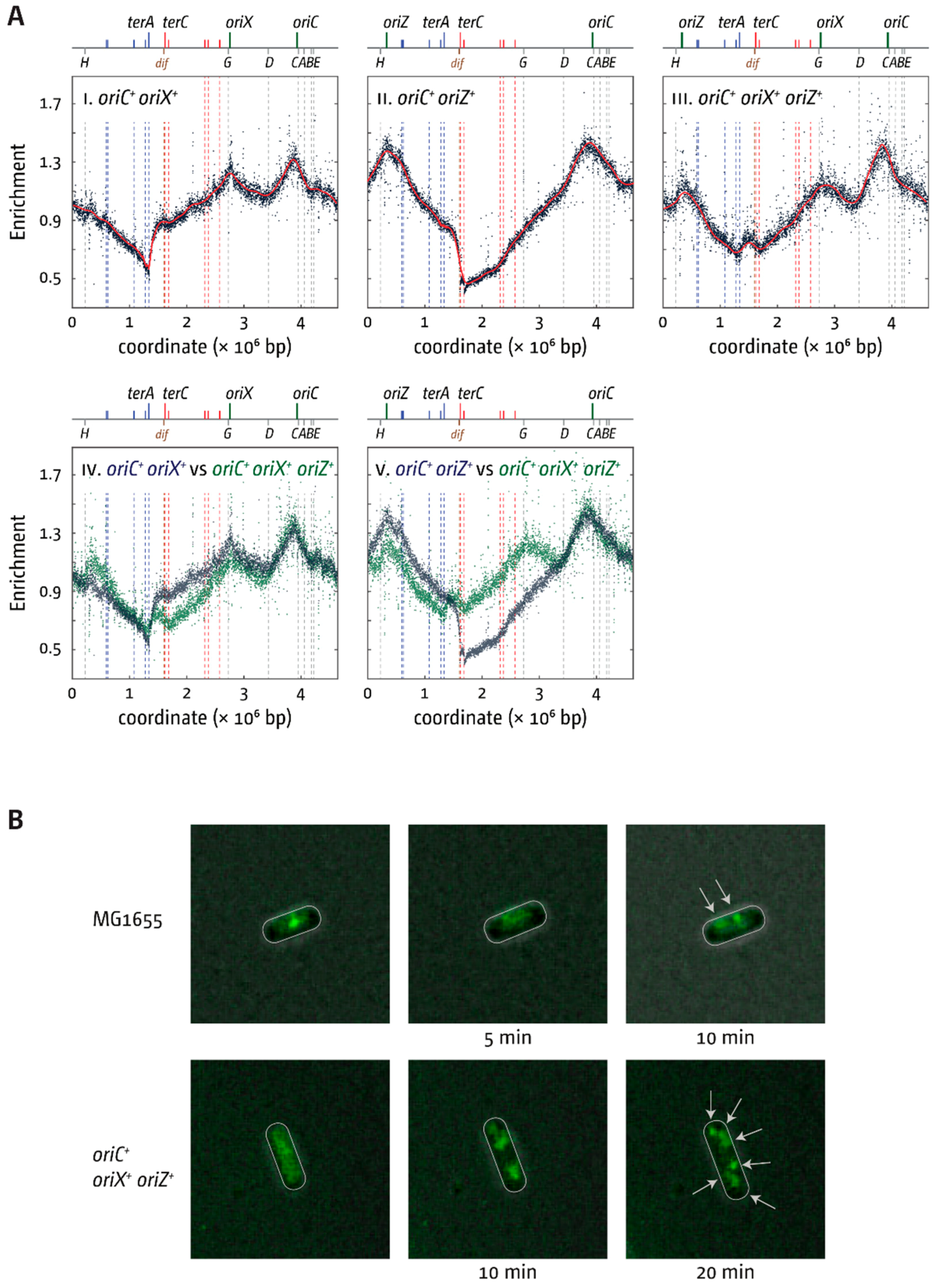
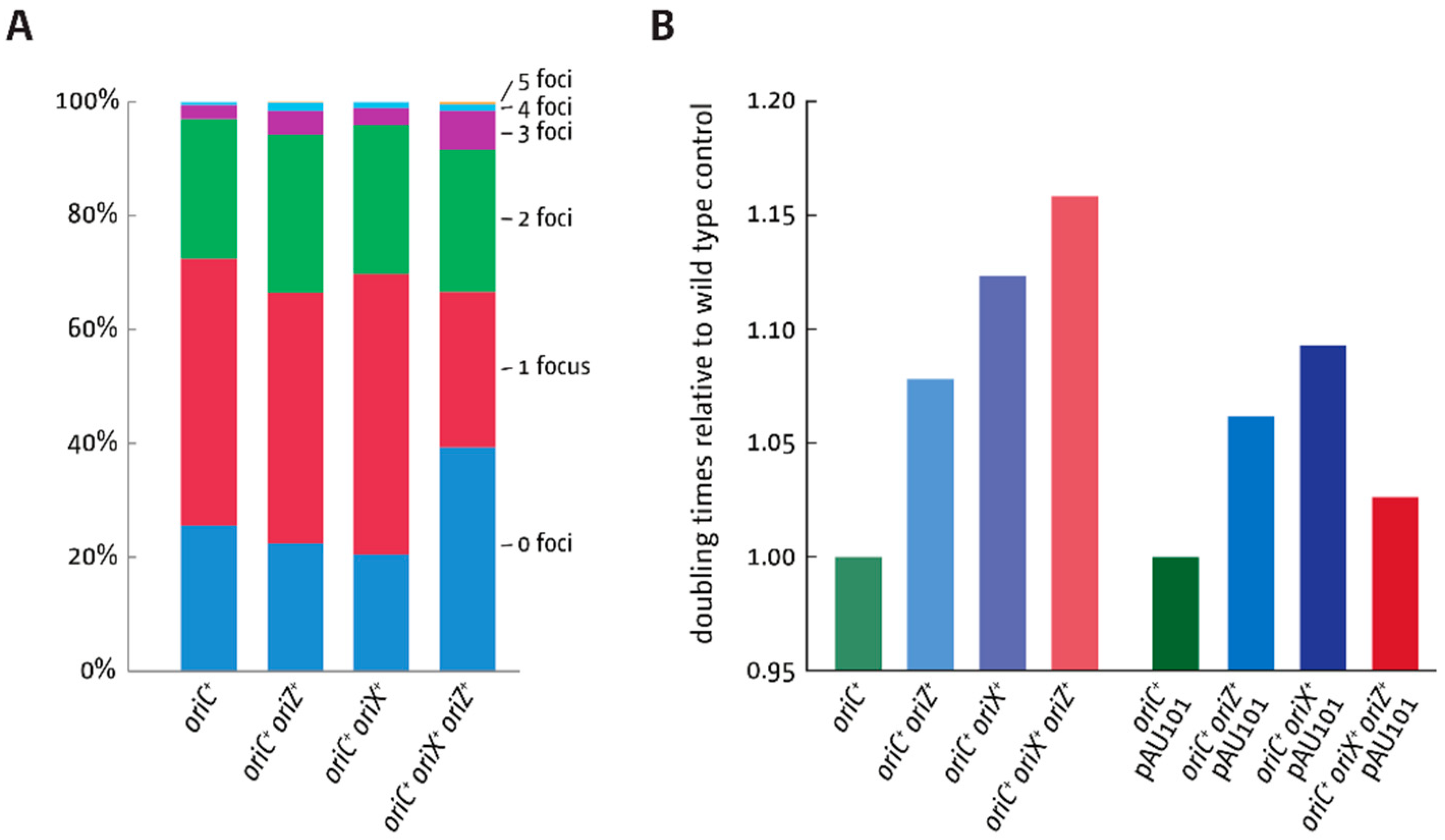
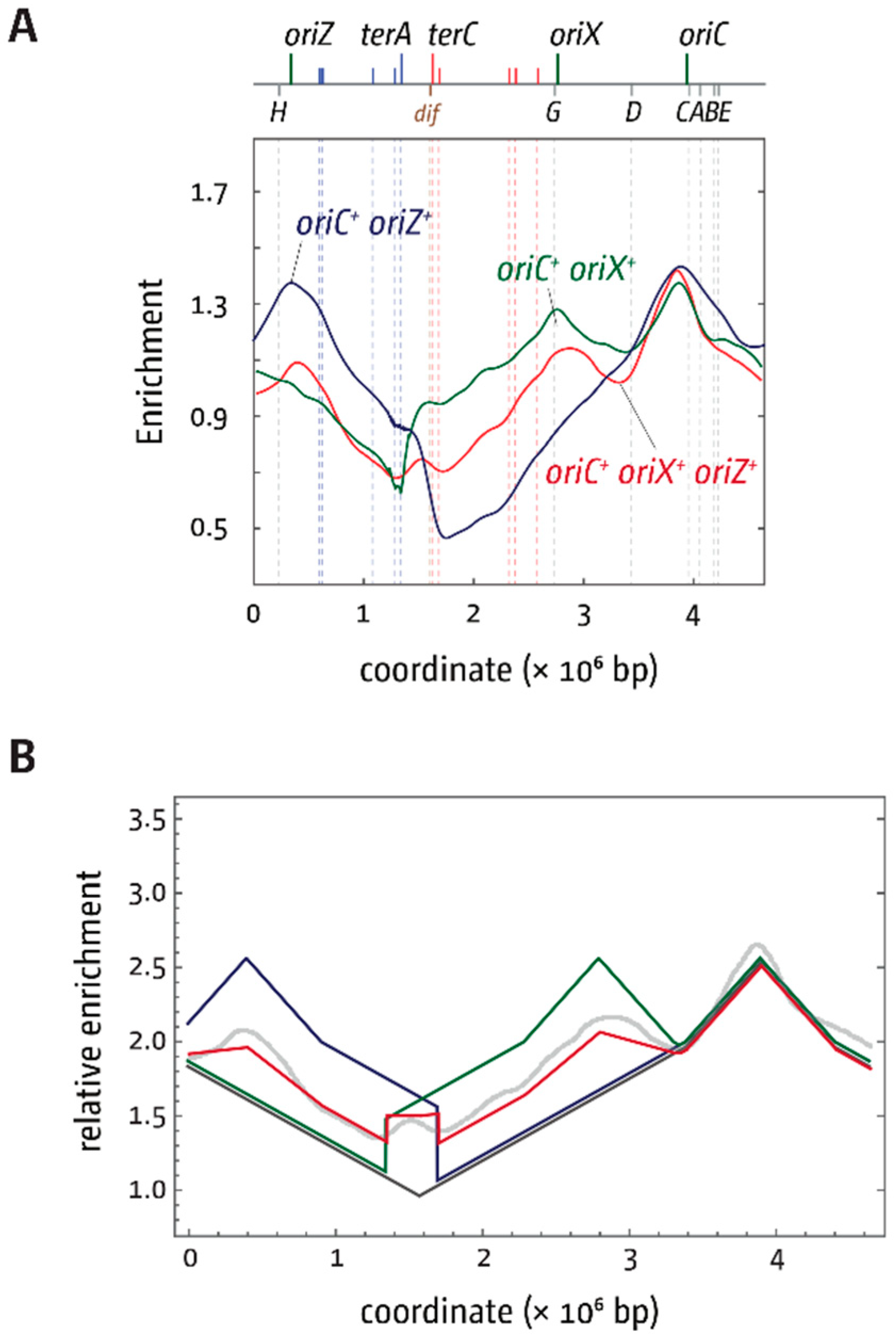
3.5. Replication Initiation in Cells with a Triple-Origin Chromosome
4. Discussion
4.1. Termination and Replication-Transcription Conflicts in Double-Origin Strains
4.2. Large Chromosomal Rearrangements in Double-Origin Cells
4.3. Replication in Cells with Three Functional Replication Origins
5. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Leonard, A.C.; Méchali, M. DNA replication origins. Cold Spring Harb. Perspect. Biol. 2013, 5, a010116. [Google Scholar] [CrossRef] [PubMed]
- Mott, M.L.; Berger, J.M. DNA replication initiation: Mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 2007, 5, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Gao, F. Bacteria may have multiple replication origins. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Zhang, C.-T. Ori-Finder: A web-based system for finding oriC s in unannotated bacterial genomes. BMC Bioinform. 2008, 9, 79. [Google Scholar] [CrossRef] [PubMed]
- Jameson, K.H.; Wilkinson, A.J. Control of initiation of DNA replication in Bacillus subtilis and Escherichia coli. Genes 2017, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Messer, W. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev. 2002, 26, 355–374. [Google Scholar] [PubMed]
- Skarstad, K.; Katayama, T. Regulating DNA replication in bacteria. Cold Spring Harb. Perspect. Biol. 2013, 5, a012922. [Google Scholar] [CrossRef] [PubMed]
- Dimude, J.U.; Midgley-Smith, S.L.; Stein, M.; Rudolph, C.J. Replication termination: Containing fork fusion-mediated pathologies in Escherichia coli. Genes 2016, 7, 40. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Lamothe, R.; Wang, X.; Sherratt, D. Escherichia coli and its chromosome. Trends Microbiol. 2008, 16, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Duggin, I.G.; Wake, R.G.; Bell, S.D.; Hill, T.M. The replication fork trap and termination of chromosome replication. Mol. Microbiol. 2008, 70, 1323–1333. [Google Scholar] [CrossRef] [PubMed]
- Neylon, C.; Kralicek, A.V.; Hill, T.M.; Dixon, N.E. Replication termination in Escherichia coli: Structure and antihelicase activity of the Tus-Ter complex. Microbiol. Mol. Biol. Rev. MMBR 2005, 69, 501–526. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Lamothe, R.; Nicolas, E.; Sherratt, D.J. Chromosome replication and segregation in bacteria. Annu. Rev. Genet. 2012, 46, 121–143. [Google Scholar] [CrossRef] [PubMed]
- Srivatsan, A.; Tehranchi, A.; MacAlpine, D.M.; Wang, J.D. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet. 2010, 6, e1000810. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.D.; Berkmen, M.B.; Grossman, A.D. Genome-wide coorientation of replication and transcription reduces adverse effects on replication in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2007, 104, 5608–5613. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, P.; Savery, N.J.; Dillingham, M.S. The conflict between DNA replication and transcription. Mol. Microbiol. 2012, 85, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Merrikh, H.; Zhang, Y.; Grossman, A.D.; Wang, J.D. Replication-transcription conflicts in bacteria. Nat. Rev. Microbiol. 2012, 10, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Kouzminova, E.A.; Kuzminov, A. Patterns of chromosomal fragmentation due to uracil-DNA incorporation reveal a novel mechanism of replication-dependent double-stranded breaks. Mol. Microbiol. 2008, 68, 202–215. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, D.; Taylor, T.; Smith, S.L.; Dimude, J.U.; Upton, A.L.; Mehrjouy, M.M.; Skovgaard, O.; Sherratt, D.J.; Retkute, R.; Rudolph, C.J. Shaping the landscape of the Escherichia coli chromosome: Replication-transcription encounters in cells with an ectopic replication origin. Nucleic Acids Res. 2015, 43, 7865–7877. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lesterlin, C.; Reyes-Lamothe, R.; Ball, G.; Sherratt, D.J. Replication and segregation of an Escherichia coli chromosome with two replication origins. Proc. Natl. Acad. Sci. USA 2011, 108, E243–E250. [Google Scholar] [CrossRef] [PubMed]
- Thomason, L.C.; Costantino, N.; Court, D.L. E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. 2007, 79. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, B.J. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In Escherichia coli and Salmonella Cellular and Molecular Biology; ASM Press: Washington, DC, USA, 1996. [Google Scholar]
- Rudolph, C.J.; Upton, A.L.; Stockum, A.; Nieduszynski, C.A.; Lloyd, R.G. Avoiding chromosome pathology when replication forks collide. Nature 2013, 500, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Meddows, T.R.; Savory, A.P.; Lloyd, R.G. RecG helicase promotes DNA double-strand break repair. Mol. Microbiol. 2004, 52, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Guy, C.P.; Atkinson, J.; Gupta, M.K.; Mahdi, A.A.; Gwynn, E.J.; Rudolph, C.J.; Moon, P.B.; van Knippenberg, I.C.; Cadman, C.J.; Dillingham, M.S.; et al. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol. Cell 2009, 36, 654–666. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, T.G.; de Boer, P.A.J. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 2003, 48, 1171–1182. [Google Scholar] [CrossRef] [PubMed]
- Luria, S.E.; Burrous, J.W. Hybridization between Escherichia coli and Shigella. J. Bacteriol. 1957, 74, 461–476. [Google Scholar] [PubMed]
- Cleveland, W.S. Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 1979, 74, 829–836. [Google Scholar] [CrossRef]
- Rudolph, C.J.; Dhillon, P.; Moore, T.; Lloyd, R.G. Avoiding and resolving conflicts between DNA replication and transcription. DNA Repair 2007, 6, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Lamothe, R.; Sherratt, D.J.; Leake, M.C. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science 2010, 328, 498–501. [Google Scholar] [CrossRef] [PubMed]
- Moolman, M.C.; Krishnan, S.T.; Kerssemakers, J.W.J.; van den Berg, A.; Tulinski, P.; Depken, M.; Reyes-Lamothe, R.; Sherratt, D.J.; Dekker, N.H. Slow unloading leads to DNA-bound β2-sliding clamp accumulation in live Escherichia coli cells. Nat. Commun. 2014, 5, 5820. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.M.; Tan, K.W.; Sakumura, Y.; Okumura, K.; Maki, H.; Akiyama, M.T. A single-molecule approach to DNA replication in Escherichia coli cells demonstrated that DNA polymerase III is a major determinant of fork speed. Mol. Microbiol. 2013, 90, 584–596. [Google Scholar] [CrossRef] [PubMed]
- Roth, A.; Messer, W. High-affinity binding sites for the initiator protein DnaA on the chromosome of Escherichia coli. Mol. Microbiol. 1998, 28, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Boye, E.; Løbner-Olesen, A.; Skarstad, K. Limiting DNA replication to once and only once. EMBO Rep. 2000, 1, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, R.; Ozaki, T.; Moriya, S.; Ogawa, T. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 1998, 12, 3032–3043. [Google Scholar] [CrossRef] [PubMed]
- Løbner-Olesen, A.; Skarstad, K.; Hansen, F.G.; von Meyenburg, K.; Boye, E. The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell 1989, 57, 881–889. [Google Scholar] [CrossRef]
- Jin, D.J.; Cagliero, C.; Zhou, Y.N. Growth rate regulation in Escherichia coli. FEMS Microbiol. Rev. 2012, 36, 269–287. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Condon, C.; Voulgaris, J.; Zaporojets, D.; Shen, B.; Al-Omar, M.; Squires, C.; Squires, C.L. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J. Bacteriol. 1999, 181, 3803–3809. [Google Scholar] [PubMed]
- Quan, S.; Skovgaard, O.; McLaughlin, R.E.; Buurman, E.T.; Squires, C.L. Markerless Escherichia coli rrn deletion strains for genetic determination of ribosomal binding sites. G3 Genes Genomes Genet. 2015, 5, 2555–2557. [Google Scholar] [CrossRef] [PubMed]
- Babu, V.M.P.; Itsko, M.; Baxter, J.C.; Schaaper, R.M.; Sutton, M.D. Insufficient levels of the nrdAB-encoded ribonucleotide reductase underlie the severe growth defect of the Δhda E. coli strain. Mol. Microbiol. 2017, 104, 377–399. [Google Scholar] [CrossRef] [PubMed]
- Skovgaard, O.; Bak, M.; Løbner-Olesen, A.; Tommerup, N. Genome-wide detection of chromosomal rearrangements, indels, and mutations in circular chromosomes by short read sequencing. Genome Res. 2011, 21, 1388–1393. [Google Scholar] [CrossRef] [PubMed]
- Reams, A.B.; Roth, J.R. Mechanisms of gene duplication and amplification. Cold Spring Harb. Perspect. Biol. 2015, 7, a016592. [Google Scholar] [CrossRef] [PubMed]
- Umenhoffer, K.; Draskovits, G.; Nyerges, Á.; Karcagi, I.; Bogos, B.; Tímár, E.; Csörgő, B.; Herczeg, R.; Nagy, I.; Fehér, T.; et al. Genome-wide abolishment of mobile genetic elements using genome shuffling and CRISPR/Cas-assisted MAGE allows the efficient stabilization of a bacterial chassis. ACS Synth. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Milbredt, S.; Farmani, N.; Sobetzko, P.; Waldminghaus, T. DNA Replication in engineered Escherichia coli genomes with extra replication origins. ACS Synth. Biol. 2016, 5, 1167–1176. [Google Scholar] [CrossRef] [PubMed]
- Dimude, J.U.; Stockum, A.; Midgley-Smith, S.L.; Upton, A.L.; Foster, H.A.; Khan, A.; Saunders, N.J.; Retkute, R.; Rudolph, C.J. The consequences of replicating in the wrong orientation: bacterial chromosome duplication without an active replication origin. mBio 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Maduike, N.Z.; Tehranchi, A.K.; Wang, J.D.; Kreuzer, K.N. Replication of the Escherichia coli chromosome in RNase HI-deficient cells: Multiple initiation regions and fork dynamics. Mol. Microbiol. 2014, 91, 39–56. [Google Scholar] [CrossRef] [PubMed]
- Markovitz, A. A new in vivo termination function for DNA polymerase I of Escherichia coli K12. Mol. Microbiol. 2005, 55, 1867–1882. [Google Scholar] [CrossRef] [PubMed]
- Wendel, B.M.; Courcelle, C.T.; Courcelle, J. Completion of DNA replication in Escherichia coli. Proc. Natl. Acad. Sci. USA 2014, 111, 16454–16459. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, R.G.; Rudolph, C.J. 25 years on and no end in sight: A perspective on the role of RecG protein. Curr. Genet. 2016, 62, 827–840. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, C.J.; Upton, A.L.; Briggs, G.S.; Lloyd, R.G. Is RecG a general guardian of the bacterial genome? DNA Repair 2010, 9, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, C.J.; Mahdi, A.A.; Upton, A.L.; Lloyd, R.G. RecG protein and single-strand DNA exonucleases avoid cell lethality associated with PriA helicase activity in Escherichia coli. Genetics 2010, 186, 473–492. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, C.J.; Upton, A.L.; Lloyd, R.G. Replication fork collisions cause pathological chromosomal amplification in cells lacking RecG DNA translocase. Mol. Microbiol. 2009, 74, 940–955. [Google Scholar] [CrossRef] [PubMed]
- Retkute, R.; Nieduszynski, C.A.; de Moura, A. Mathematical modeling of genome replication. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2012, 86, 031916. [Google Scholar] [CrossRef] [PubMed]
- Sinha, A.K.; Durand, A.; Desfontaines, J.-M.; Iurchenko, I.; Auger, H.; Leach, D.R.F.; Barre, F.-X.; Michel, B. Division-induced DNA double strand breaks in the chromosome terminus region of Escherichia coli lacking RecBCD DNA repair enzyme. PLoS Genet. 2017, 13, e1006895. [Google Scholar] [CrossRef] [PubMed]
- Macaulay, I.C.; Voet, T. Single cell genomics: advances and future perspectives. PLOS Genet. 2014, 10, e1004126. [Google Scholar] [CrossRef] [PubMed]
| Strain Number | Relevant Genotype a | Source |
|---|---|---|
| General P1 donors | ||
| WX297 | AB1157 oriZ-<kan> | [19] |
| RRL190 | AB1157 <kan>-ypet-dnaN | [19] |
| RUC1593 | DY330 pheA::oriX-cat | This study |
| MG1655 derivatives | ||
| MG1655 | F– rph-1 | [22] |
| AS1062 | <kan>-ypet-dnaN | MG1655 × P1.RRL190 to Kmr |
| JD1181 | ΔlacIZYA pheA::oriX-cat | TB28 × P1.RUC1593 to Cmr |
| JD1187 | ΔlacIZYA pheA::oriX-cat ΔoriC::kan | JD1181 × P1.RCe576 to Kmr |
| JD1190 | rpoB*35 ΔlacIZYA pheA::oriX-cat | N5925 × P1.RUC1593 to Cmr |
| JD1197 | rpoB*35 ΔlacIZYA pheA::oriX-cat ΔoriC::kan | JD1190 × P1.RCe576 to Kmr |
| JD1203 | ΔlacIZYA pheA::oriX-cat tus1::dhfr | JD1181 × P1.N6798 to Tmr |
| JD1205 | rpoB*35 ΔlacIZYA pheA::oriX-cat tus1::dhfr | JD1190 × P1.N6798 to Tmr |
| JD1208 | ΔlacIZYA pheA::oriX-cat tus1::dhfr ΔoriC::kan | JD1203 × P1.RCe576 to Kmr |
| JD1209 | rpoB*35 ΔlacIZYA pheA::oriX-cat tus1::dhfr ΔoriC::kan | JD1205 × P1.RCe576 to Kmr |
| JD1332 | ΔlacIZYA pheA::oriX-cat oriZ-<kan> | JD1181 × P1.WX297 to Kmr |
| JD1333 | ΔlacIZYA pheA::oriX-cat oriZ-<kan> | JD1181 × P1.WX297 to Kmr |
| JD1336 | ΔlacIZYA oriZ-<kan> | TB28 × P1.WX297 to Kmr |
| JD1339 | ΔlacIZYA oriZ-<> | JD1336 × pCP20 to Kms Aps |
| JD1341 | ΔlacIZYA oriZ-<> pheA::oriX-cat | JD1339 × P1.RUC1593 to Cmr |
| JD1343 | ΔlacIZYA oriZ-<> pheA::oriX-cat ΔoriC::kan | JD1341 × P1.RCe576 to Kmr |
| JJ1359 | ΔlacIZYA dam1::kan ΔrecG::apra tus1::dhfr | [23] |
| N4560 | ΔrecG265::cat | [24] |
| N5925 | rpoB*35 ΔlacIZYA | [25] |
| N6798 | ΔrecG265::cat tus1::dhfr | N4560 × P1.JJ1359 to Tmr |
| RCe504 | oriZ-<cat> | [18] |
| RCe576 | rpoB*35 oriZ-cat-frt tus1::dhfr ΔoriC::kanb | [18] |
| RCe749 | oriZ-<cat> <kan>-ypet-dnaN | RCe504 × P1.AS1062 to Kmr |
| RCe751 | ΔlacIZYA pheA::oriX-cat <kan>-ypet-dnaN | JD1181 × P1.AS1062 to Kmr |
| RCe753 | ΔlacIZYA oriZ-<> pheA::oriX-cat <kan>-ypet-dnaN | JD1341 × P1.AS1062 to Kmr |
| TB28 | ΔlacIZYA | [26] |
| Strain Background | Doubling Time (min) | SD | r² | Doubling Time oriZ Constructs a |
|---|---|---|---|---|
| MG1655 | 19.3 | ±1.7 | 0.983 | 19.9 |
| oriC+ oriX+ | 22.3 | ±1.2 | 0.981 | 20.6 |
| ΔoriC oriX+ | 48.1 | ±5.6 | 0.969 | 39.8 |
| oriC+ oriX+ Δtus | 23.1 | ±0.7 | 0.985 | 21.5 |
| oriC+ oriX+ rpoB*35 | 24.7 | ±1.5 | 0.986 | 23.1 |
| oriC+ oriX+ Δtus rpoB*35 | 29.3 | ±1.9 | 0.993 | 24.5 |
| ΔoriC oriX+ Δtus | 53.2 | ±9.1 | 0.977 | 29.2 |
| ΔoriC oriX+ rpoB*35 | 37.5 | ±8.4 | 0.980 | 32.0 |
| ΔoriC oriX+ Δtus rpoB*35 | 44.8 | ±9.2 | 0.99 | 29.8 |
| Strain Background | Doubling Time (min) | SD | r² |
|---|---|---|---|
| MG1655 | 19.6 | ±1.0 | 0.999 |
| oriC+ oriZ+ | 21.0 | ±0.8 | 0.997 |
| oriC+ oriX+ | 21.8 | ±0.8 | 0.996 |
| oriC+ oriX+ oriZ+ | 22.7 | ±2.5 | 0.994 |
| ΔoriC oriX+ oriZ+ | 35.3 | ±2.6 | 0.990 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dimude, J.U.; Stein, M.; Andrzejewska, E.E.; Khalifa, M.S.; Gajdosova, A.; Retkute, R.; Skovgaard, O.; Rudolph, C.J. Origins Left, Right, and Centre: Increasing the Number of Initiation Sites in the Escherichia coli Chromosome. Genes 2018, 9, 376. https://doi.org/10.3390/genes9080376
Dimude JU, Stein M, Andrzejewska EE, Khalifa MS, Gajdosova A, Retkute R, Skovgaard O, Rudolph CJ. Origins Left, Right, and Centre: Increasing the Number of Initiation Sites in the Escherichia coli Chromosome. Genes. 2018; 9(8):376. https://doi.org/10.3390/genes9080376
Chicago/Turabian StyleDimude, Juachi U., Monja Stein, Ewa E. Andrzejewska, Mohammad S. Khalifa, Alexandra Gajdosova, Renata Retkute, Ole Skovgaard, and Christian J. Rudolph. 2018. "Origins Left, Right, and Centre: Increasing the Number of Initiation Sites in the Escherichia coli Chromosome" Genes 9, no. 8: 376. https://doi.org/10.3390/genes9080376
APA StyleDimude, J. U., Stein, M., Andrzejewska, E. E., Khalifa, M. S., Gajdosova, A., Retkute, R., Skovgaard, O., & Rudolph, C. J. (2018). Origins Left, Right, and Centre: Increasing the Number of Initiation Sites in the Escherichia coli Chromosome. Genes, 9(8), 376. https://doi.org/10.3390/genes9080376






