Applications of Yeast Synthetic Biology Geared towards the Production of Biopharmaceuticals
Abstract
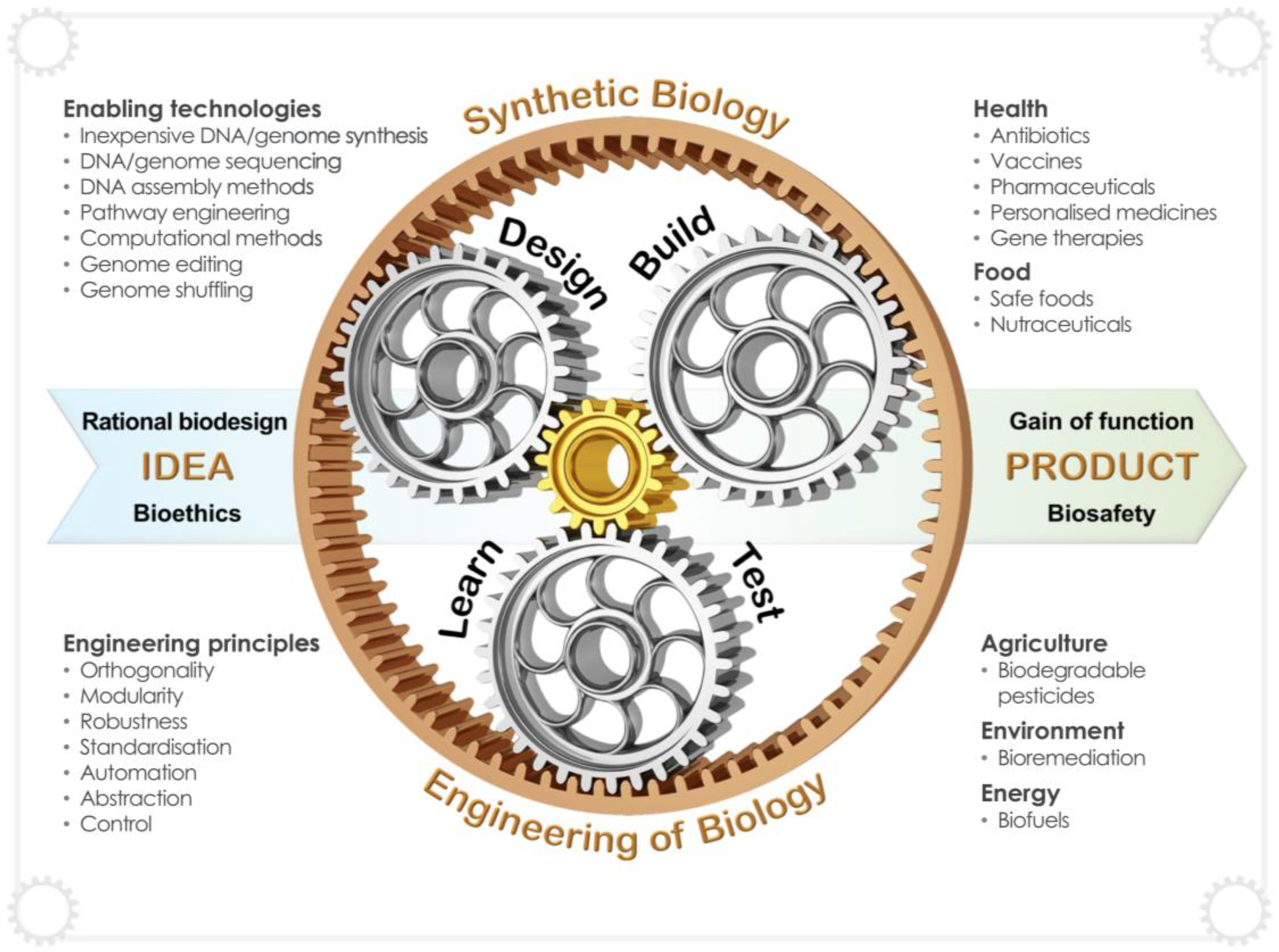
1. Yeast Synthetic Biology Shifts Biopharmaceutical Production Capabilities into High Gear
2. The Importance of Identifying the Optimal Cell Factory for Biopharmaceutical Production
3. Synthetic Biology Tools for Heterologous Production of Biopharmaceuticals
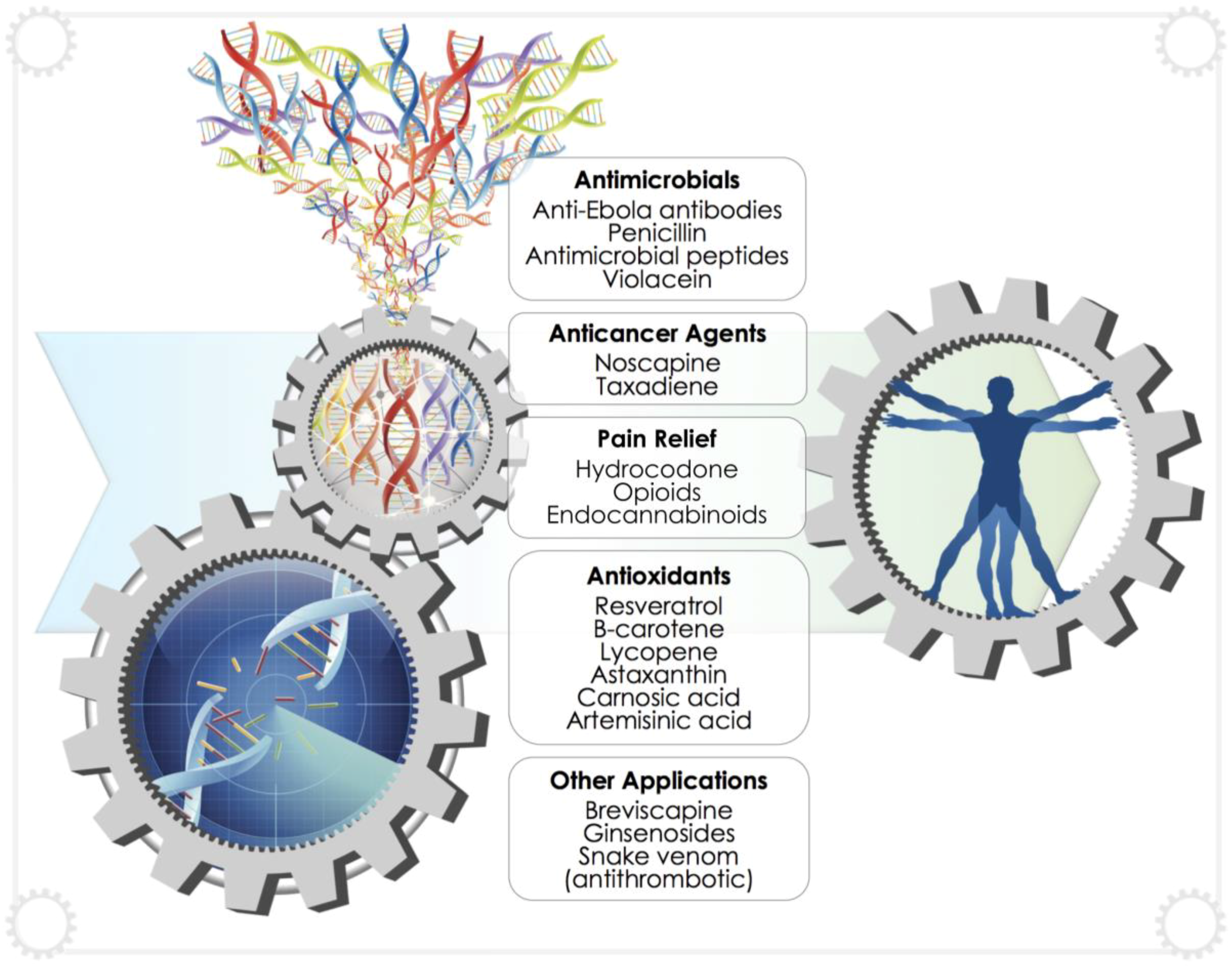
4. Recent Developments in the Production of Biopharmaceuticals
4.1. General Medical Applications
4.2. Anticancer Compounds
4.3. Antivirals, Antibiotics and Antimicrobial Peptides
4.4. Antioxidants
5. Emerging Technologies and Future Trends for the Engineering of Synthetic Yeast Biofactories
5.1. Yeast Synthetic Biology Expands the Biopharmaceutical Repertoire
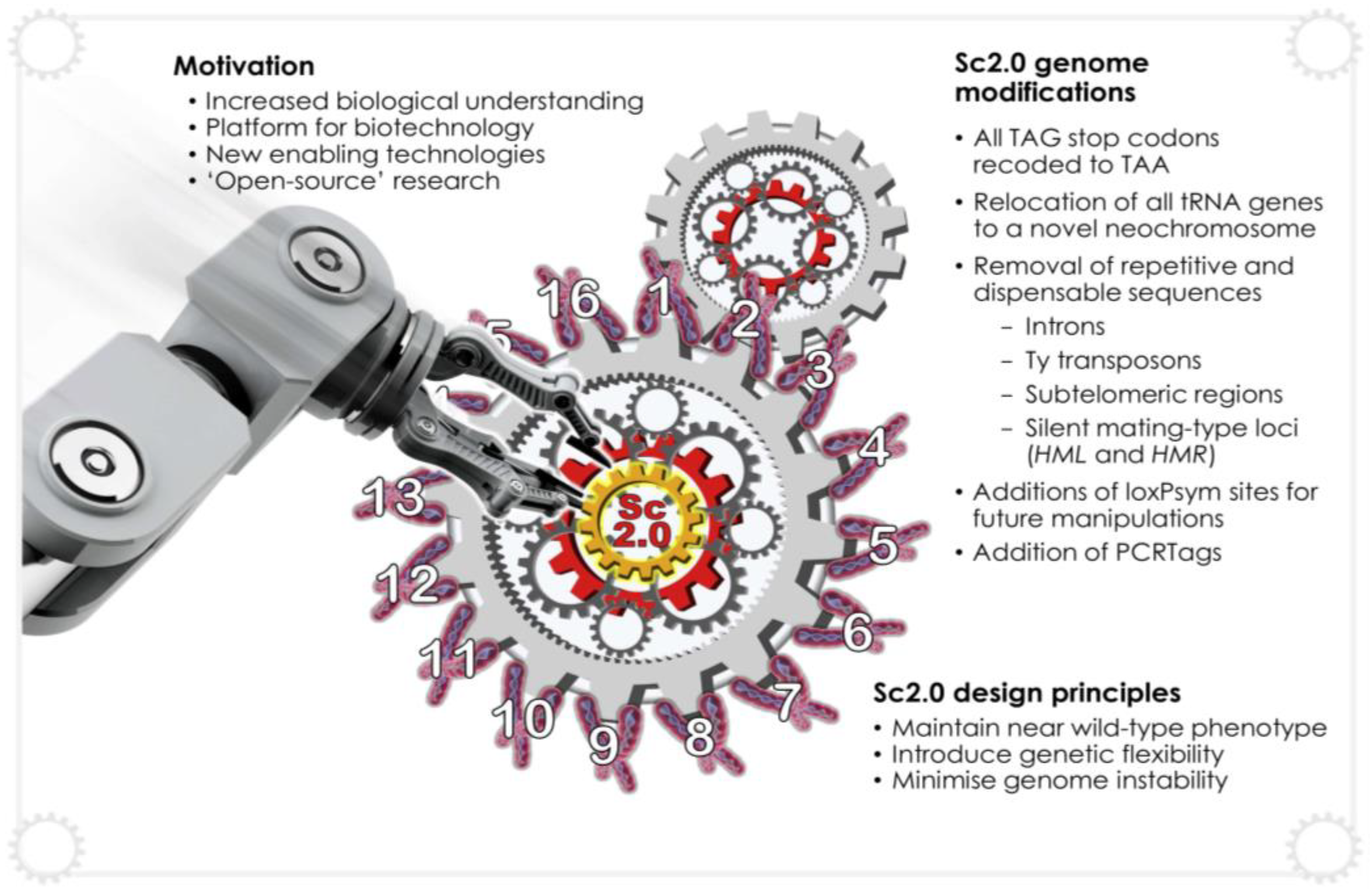
5.2. Synthetic Genomics, Sc2.0 and SCRaMbLE
5.3. Home-Brew Biopharmaceuticals
6. Remaining Hurdles for Biopharmaceutical Production
7. Synthetic Yeast Foundries Awaken Biological Potential
Funding
Acknowledgments
Conflicts of Interest
References
- Solecki, R.S. Shanidar IV, a Neanderthal flower burial in Northern Iraq. Science 1975, 190, 880–881. [Google Scholar] [CrossRef]
- Klockgether-Radke, A.P.F.W. Serturner and the discovery of morphine. 200 years of pain therapy with opioids. Anasthesiol. Intensivmed. Notfallmed. Schmerzther. AINS 2002, 37, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A. On the antibacterial action of cultures of a Penicillium, with special reference to their use in the isolation of B. influenzæ. Br. J. Exp. Pathol. 1929, 10, 226–236. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H.; et al. Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Liu, Z.; Zhao, H.; Ang, E.L. Recent advances in combinatorial biosynthesis for drug discovery. Drug Des. Dev. Ther. 2015, 9, 823–833. [Google Scholar] [CrossRef]
- Cue, B.W.; Zhang, J. Green process chemistry in the pharmaceutical industry. Green Chem. Lett. Rev. 2009, 2, 193–211. [Google Scholar] [CrossRef]
- Luo, Y.; Li, B.Z.; Liu, D.; Zhang, L.; Chen, Y.; Jia, B.; Zeng, B.X.; Zhao, H.; Yuan, Y.J. Engineered biosynthesis of natural products in heterologous hosts. Chem. Soc. Rev. 2015, 44, 5265–5290. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, S.; Thodey, K.; Trenchard, I.; Cravens, A.; Smolke, C.D. Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc. Natl. Acad. Sci. USA 2018. [Google Scholar] [CrossRef] [PubMed]
- Paddon, C.J.; Westfall, P.J.; Pitera, D.J.; Benjamin, K.; Fisher, K.; McPhee, D.; Leavell, M.D.; Tai, A.; Main, A.; Eng, D.; et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013, 496, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Liu, Y.; Guo, J.; Huang, L.; Zhang, X. Yeast synthetic biology for high-value metabolites. FEMS Yeast Res. 2015, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, Y.; Smolke, C.D. Strategies for microbial synthesis of high-value phytochemicals. Nat. Chem. 2018, 10, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L.; Vaishnav, P. Production of recombinant proteins by microbes and higher organisms. Biotechnol. Adv. 2009, 27, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Walker, G.M. Yeast Physiology and Biotechnology; John Wiley & Sons: Chichester, UK; New York, NY, USA, 1998. [Google Scholar]
- Kurtzman, C.P. Biotechnological strains of Komagataella (Pichia) pastoris are Komagataella phaffii as determined from multigene sequence analysis. J. Ind. Microbiol. Biotechnol. 2009, 36, 1435–1438. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Yoo, S.J.; Kang, H.A. Yeast synthetic biology for the production of recombinant therapeutic proteins. FEMS Yeast Res. 2015, 15, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Awan, A.R.; Shaw, W.M.; Ellis, T. Biosynthesis of therapeutic natural products using synthetic biology. Adv. Drug Deliv. Rev. 2016, 105, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.K.; Keasling, J.D. Recent applications of synthetic biology tools for yeast metabolic engineering. FEMS Yeast Res. 2015, 15, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cregg, J.M.; Tolstorukov, I.; Kusari, A.; Sunga, J.; Madden, K.; Chappell, T. Chapter 13 Expression in the Yeast Pichia pastoris. In Methods in Enzymology; Burgess, R.R., Deutscher, M.P., Eds.; Academic Press: Cambridge, MA, USA, 2009; Volume 463, pp. 169–189. [Google Scholar]
- Melmer, G. Production of Recombinant Proteins: Novel Microbial and Eukaryotic Expression Systems; Wiley-VCH: Weinheim, Germany, 2005; pp. 361–383. [Google Scholar] [CrossRef]
- Madhavan, A.; Jose, A.A.; Binod, P.; Sindhu, R.; Sukumaran, R.K.; Pandey, A.; Castro, G.E. Synthetic biology and metabolic engineering approaches and its impact on non-conventional yeast and biofuel production. Front. Energy Res. 2017, 5. [Google Scholar] [CrossRef]
- Ratledge, C.; Wynn, J.P. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv. Appl. Microbiol. 2002, 51, 1–51. [Google Scholar] [PubMed]
- Blazeck, J.; Hill, A.; Liu, L.; Knight, R.; Miller, J.; Pan, A.; Otoupal, P.; Alper, H.S. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat. Commun. 2014, 5, 3131. [Google Scholar] [CrossRef] [PubMed]
- De Pourcq, K.; De Schutter, K.; Callewaert, N. Engineering of glycosylation in yeast and other fungi: Current state and perspectives. Appl. Microbiol. Biotechnol. 2010, 87, 1617–1631. [Google Scholar] [CrossRef] [PubMed]
- Walsh, G. Post-translational modifications of protein biopharmaceuticals. Drug Discov. Today 2010, 15, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Bretthauer, R.K.; Castellino, F.J. Glycosylation of Pichia pastoris-derived proteins. Biotechnol. Appl. Biochem. 1999, 30, 193–200. [Google Scholar] [PubMed]
- Krainer, F.W.; Gmeiner, C.; Neutsch, L.; Windwarder, M.; Pletzenauer, R.; Herwig, C.; Altmann, F.; Glieder, A.; Spadiut, O. Knockout of an endogenous mannosyltransferase increases the homogeneity of glycoproteins produced in Pichia pastoris. Sci. Rep. 2013, 3, 3279. [Google Scholar] [CrossRef] [PubMed]
- Vogl, T.; Hartner, F.S.; Glieder, A. New opportunities by synthetic biology for biopharmaceutical production in Pichia pastoris. Curr. Opin. Biotechnol. 2013, 24, 1094–1101. [Google Scholar] [CrossRef] [PubMed]
- Naatsaari, L.; Mistlberger, B.; Ruth, C.; Hajek, T.; Hartner, F.S.; Glieder, A. Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS ONE 2012, 7, e39720. [Google Scholar] [CrossRef] [PubMed]
- Saraya, R.; Krikken, A.M.; Kiel, J.A.; Baerends, R.J.; Veenhuis, M.; van der Klei, I.J. Novel genetic tools for Hansenula polymorpha. FEMS Yeast Res. 2012, 12, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Verbeke, J.; Beopoulos, A.; Nicaud, J.M. Efficient homologous recombination with short length flanking fragments in Ku70 deficient Yarrowia lipolytica strains. Biotechnol. Lett. 2013, 35, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Giese, B.; Koenigstein, S.; Wigger, H.; Schmidt, J.C.; von Gleich, A. Rational engineering principles in synthetic biology: A framework for quantitative analysis and an initial assessment. Biol. Theory 2013, 8, 324–333. [Google Scholar] [CrossRef]
- Kotula, L.; Curtis, P.J. Evaluation of foreign gene codon optimization in yeast: Expression of a mouse IG κ chain. Nat. Biotechnol. 1991, 9, 1386–1389. [Google Scholar] [CrossRef]
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Lanza, A.M.; Curran, K.A.; Rey, L.G.; Alper, H.S. A condition-specific codon optimization approach for improved heterologous gene expression in Saccharomyces cerevisiae. BMC Syst. Biol. 2014, 8, 33. [Google Scholar] [CrossRef] [PubMed]
- Engler, C.; Kandzia, R.; Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 2008, 3, e3647. [Google Scholar] [CrossRef] [PubMed]
- Awan, A.R.; Blount, B.A.; Bell, D.J.; Shaw, W.M.; Ho, J.C.H.; McKiernan, R.M.; Ellis, T. Biosynthesis of the antibiotic nonribosomal peptide penicillin in baker’s yeast. Nat. Commun. 2017, 8, 15202. [Google Scholar] [CrossRef] [PubMed]
- Larroude, M.; Celinska, E.; Back, A.; Thomas, S.; Nicaud, J.M.; Ledesma-Amaro, R. A synthetic biology approach to transform Yarrowia lipolytica into a competitive biotechnological producer of β-carotene. Biotechnol. Bioeng. 2018, 115, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Stovicek, V.; Holkenbrink, C.; Borodina, I. CRISPR/Cas system for yeast genome engineering: Advances and applications. FEMS Yeast Res. 2017, 17, fox030. [Google Scholar] [CrossRef] [PubMed]
- Si, T.; Chao, R.; Min, Y.; Wu, Y.; Ren, W.; Zhao, H. Automated multiplex genome-scale engineering in yeast. Nat. Commun. 2017, 8, 15187. [Google Scholar] [CrossRef] [PubMed]
- Verwaal, R.; Buiting-Wiessenhaan, N.; Dalhuijsen, S.; Roubos, J.A. CRISPR/Cpf1 enables fast and simple genome editing of Saccharomyces cerevisiae. Yeast 2018, 35, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Lau, Y.H.; Giessen, T.W.; Altenburg, W.J.; Silver, P.A. Prokaryotic nanocompartments form synthetic organelles in a eukaryote. Nat. Commun. 2018, 9, 1311. [Google Scholar] [CrossRef] [PubMed]
- Mellitzer, A.; Ruth, C.; Gustafsson, C.; Welch, M.; Birner-Grunberger, R.; Weis, R.; Purkarthofer, T.; Glieder, A. Synergistic modular promoter and gene optimization to push cellulase secretion by Pichia pastoris beyond existing benchmarks. J. Biotechnol. 2014, 191, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Portela, R.M.; Vogl, T. Synthetic core promoters as universal parts for fine-tuning expression in different yeast species. ACS Synth. Biol. 2017, 6, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Trassaert, M.; Vandermies, M.; Carly, F.; Denies, O.; Thomas, S.; Fickers, P.; Nicaud, J.M. New inducible promoter for gene expression and synthetic biology in Yarrowia lipolytica. Microb. Cell Fact. 2017, 16, 141. [Google Scholar] [CrossRef] [PubMed]
- Weninger, A.; Fischer, J.E.; Raschmanova, H.; Kniely, C.; Vogl, T.; Glieder, A. Expanding the CRISPR/Cas9 toolkit for Pichia pastoris with efficient donor integration and alternative resistance markers. J. Cell. Biochem. 2018, 119, 3183–3198. [Google Scholar] [CrossRef] [PubMed]
- Nambu-Nishida, Y.; Nishida, K.; Hasunuma, T.; Kondo, A. Development of a comprehensive set of tools for genome engineering in a cold- and thermo-tolerant Kluyveromyces marxianus yeast strain. Sci. Rep. 2017, 7, 8993. [Google Scholar] [CrossRef] [PubMed]
- Galanie, S.; Thodey, K.; Trenchard, I.J.; Filsinger Interrante, M.; Smolke, C.D. Complete biosynthesis of opioids in yeast. Science 2015, 349, 1095–1100. [Google Scholar] [CrossRef] [PubMed]
- Meadows, A.L.; Hawkins, K.M.; Tsegaye, Y.; Antipov, E.; Kim, Y.; Raetz, L.; Dahl, R.H.; Tai, A.; Mahatdejkul-Meadows, T.; Xu, L.; et al. Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nature 2016, 537, 694–697. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wu, J.; Jia, H.; Chen, W.; Shao, C.; Zhao, L.; Ma, J.; Li, R.; Zhong, Y.; Fang, F.; et al. Balancing the expression and production of a heterodimeric protein: Recombinant agkisacutacin as a novel antithrombotic drug candidate. Sci. Rep. 2015, 5, 11730. [Google Scholar] [CrossRef] [PubMed]
- Berthold, N.; Hoffmann, R. Cellular uptake of Apidaecin 1b and related analogs in Gram-negative bacteria reveals novel antibacterial mechanism for proline-rich antimicrobial peptides. Protein Pept. Lett. 2014, 21, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.J.; Chang, J.J.; Lin, H.Y.; Thia, C.; Kao, Y.Y.; Huang, C.C.; Li, W.H. Metabolic engineering a yeast to produce astaxanthin. Bioresour. Technol. 2017, 245, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cheng, J.; Zhang, G.; Ding, W.; Duan, L.; Yang, J.; Kui, L.; Cheng, X.; Ruan, J.; Fan, W.; et al. Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat. Commun. 2018, 9, 448. [Google Scholar] [CrossRef] [PubMed]
- Ignea, C.; Athanasakoglou, A.; Andreadelli, A.; Apostolaki, M.; Iakovides, M.; Stephanou, E.G.; Makris, A.M.; Kampranis, S.C. Overcoming the plasticity of plant specialized metabolism for selective diterpene production in yeast. Sci. Rep. 2017, 7, 8855. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xiao, W.; Wang, Y.; Liu, H.; Li, X.; Yuan, Y. Lycopene overproduction in Saccharomyces cerevisiae through combining pathway engineering with host engineering. Microb. Cell Fact. 2016, 15, 113. [Google Scholar] [CrossRef] [PubMed]
- Purcell, O.; Opdensteinen, P.; Chen, W.; Lowenhaupt, K.; Brown, A.; Hermann, M.; Cao, J.; Tenhaef, N.; Kallweit, E.; Kastilan, R.; et al. Production of functional anti-Ebola antibodies in Pichia pastoris. ACS Synth. Biol. 2017, 6, 2183–2190. [Google Scholar] [CrossRef] [PubMed]
- Blount, B.A.; Gowers, G.O.F.; Ho, J.C.H.; Ledesma-Amaro, R.; Jovicevic, D.; McKiernan, R.M.; Xie, Z.X.; Li, B.Z.; Yuan, Y.J.; Ellis, T. Rapid host strain improvement by in vivo rearrangement of a synthetic yeast chromosome. Nat. Commun. 2018, 9, 1932. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Schneider, K.; Kristensen, M.; Borodina, I.; Nielsen, J. Engineering yeast for high-level production of stilbenoid antioxidants. Sci. Rep. 2016, 6, 36827. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.; Clastre, M.; Courdavault, V.; O’Connor, S.E. De novo production of the plant-derived alkaloid strictosidine in yeast. Proc. Natl. Acad. Sci. USA 2015, 112, 3205–3210. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.Z.; Yan, H.F.; Li, L.F.; Zhai, F.; Shang, L.Q.; Yin, Z.; Yuan, Y.J. Biosynthesis of Taxadiene in Saccharomyces cerevisiae: Selection of geranylgeranyl diphosphate synthase directed by a computer-aided docking strategy. PLoS ONE 2014, 9, e109348. [Google Scholar] [CrossRef] [PubMed]
- Zirpel, B.; Degenhardt, F.; Zammarelli, C.; Wibberg, D.; Kalinowski, J.; Stehle, F.; Kayser, O. Optimization of Delta(9)-tetrahydrocannabinolic acid synthase production in Komagataella phaffii via post-translational bottleneck identification. J. Biotechnol. 2018, 272, 40–47. [Google Scholar] [CrossRef] [PubMed]
- McKeague, M.; Wang, Y.H.; Cravens, A.; Win, M.N.; Smolke, C.D. Engineering a microbial platform for de novo biosynthesis of diverse methylxanthines. Metab. Eng. 2016, 38, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Easson, M.L.A.E.; Froese, J.; Simionescu, R.; Hudlicky, T.; De Luca, V. Completion of the seven-step pathway from tabersonine to the anticancer drug precursor vindoline and its assembly in yeast. Proc. Natl. Acad. Sci. USA 2015, 112, 6224–6229. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Luo, Z.; Wang, Y.; Pham, N.T.; Tuck, L.; Pérez-Pi, I.; Liu, L.; Shen, Y.; French, C.; Auer, M.; et al. Rapid pathway prototyping and engineering using in vitro and in vivo synthetic genome SCRaMbLE-in methods. Nat. Commun. 2018, 9, 1936. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.H.; Chen, K.P.; Chen, S.X.; Liu, K.Z.; Fan, T.L.; Chen, Y.C. Breviscapine, a traditional Chinese medicine, alleviates myocardial ischaemia reperfusion injury in diabetic rats. Acta Cardiol. 2008, 63, 757–762. [Google Scholar] [CrossRef] [PubMed]
- Li, X.L.; Li, Y.Q.; Yan, W.M.; Li, H.Y.; Xu, H.; Zheng, X.X.; Guo, D.W.; Tang, L.K. A study of the cardioprotective effect of breviscapine during hypoxia of cardiomyocytes. Planta Med. 2004, 70, 1039–1044. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Chen, G.; He, H.; Liu, C.; Xiong, X.; Li, J.; Wang, J. Therapeutic effects of breviscapine in cardiovascular diseases: A Review. Front. Pharmacol. 2017, 8, 289. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yang, S.C.; Zhang, G.H.; Su, B. Research on situation and countermeasure of Erigeron breviscapus plant production. China J. Chin. Mater. Med. 2013, 38, 2227–2230. [Google Scholar]
- Leung, K.W.; Wong, A.S. Pharmacology of ginsenosides: A literature review. Chin. Med. 2010, 5, 20. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Wang, B.; Liu, Y.; Shi, M.; Wang, D.; Zhang, X.; Liu, T.; Huang, L.; Zhang, X. Producing aglycons of ginsenosides in bakers’ yeast. Sci. Rep. 2014, 4, 3698. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Ji, H.; Cheng, X.; Li, B.X.; Ng, T.B. Antithrombotic and thrombolytic activities of Agkisacutacin, a snake venom proteinase, in experimental models. Gen. Pharmacol. 2000, 35, 179–187. [Google Scholar] [CrossRef]
- Whiting, P.F.; Wolff, R.F.; Deshpande, S.; Di Nisio, M.; Duffy, S.; Hernandez, A.V.; Keurentjes, J.C.; Lang, S.; Misso, K.; Ryder, S.; et al. Cannabinoids for medical use: A systematic review and meta-analysis. JAMA 2015, 313, 2456–2473. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.; Hansen, E.H.; Kayser, O.; Carlsen, S.; Stehle, F. Designing microorganisms for heterologous biosynthesis of cannabinoids. FEMS Yeast Res. 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Zirpel, B.; Stehle, F.; Kayser, O. Production of Delta9-tetrahydrocannabinolic acid from cannabigerolic acid by whole cells of Pichia (Komagataella) pastoris expressing Δ9-tetrahydrocannabinolic acid synthase from Cannabis sativa L. Biotechnol. Lett. 2015, 37, 1869–1875. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Naghavi, M.; Allen, C.; Barber, R.M.; Bhutta, Z.A.; Carter, A.; Casey, D.C.; Charlson, F.J.; Chen, A.Z.; Coates, M.M.; et al. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1459–1544. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. The discovery of anticancer drugs from natural sources. In Natural Products: Drug Discovery and Therapeutic Medicine; Zhang, L., Demain, A.L., Eds.; Humana Press: Totowa, NJ, USA, 2005; pp. 129–168. [Google Scholar]
- Camidge, R. The Story of Taxol: Nature and politics in the pursuit of an anti-cancer drug. BMJ Br. Med. J. 2001, 323, 115. [Google Scholar] [CrossRef]
- Malik, S.; Cusidó, R.M.; Mirjalili, M.H.; Moyano, E.; Palazón, J.; Bonfill, M. Production of the anticancer drug taxol in Taxus baccata suspension cultures: A review. Process Biochem. 2011, 46, 23–34. [Google Scholar] [CrossRef]
- Exposito, O.; Bonfill, M.; Moyano, E.; Onrubia, M.; Mirjalili, M.H.; Cusido, R.M.; Palazon, J. Biotechnological production of taxol and related taxoids: Current state and prospects. Anti-Cancer Agents Med. Chem. 2009, 9, 109–121. [Google Scholar] [CrossRef]
- Engels, B.; Dahm, P.; Jennewein, S. Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards Taxol (Paclitaxel) production. Metab. Eng. 2008, 10, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Ajikumar, P.K.; Xiao, W.H.; Tyo, K.E.; Wang, Y.; Simeon, F.; Leonard, E.; Mucha, O.; Phon, T.H.; Pfeifer, B.; Stephanopoulos, G. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 2010, 330, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Ye, K.; Grossniklaus, H.E.; Archer, D.R.; Joshi, H.C.; Kapp, J.A. Noscapine inhibits tumor growth with little toxicity to normal tissues or inhibition of immune responses. Cancer Immunol. Immunother. 2000, 49, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Gupta, K.; Yao, J.; Ye, K.; Panda, D.; Giannakakou, P.; Joshi, H.C. Paclitaxel-resistant human ovarian cancer cells undergo c-Jun NH2-terminal kinase-mediated apoptosis in response to noscapine. J. Biol. Chem. 2002, 277, 39777–39785. [Google Scholar] [CrossRef] [PubMed]
- Doddapaneni, R.; Patel, K.; Chowdhury, N.; Singh, M. Reversal of drug-resistance by noscapine chemo-sensitization in docetaxel resistant triple negative breast cancer. Sci. Rep. 2017, 7, 15824. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Smolke, C.D. Engineering biosynthesis of the anticancer alkaloid noscapine in yeast. Nat. Commun. 2016, 7, 12137. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Davis, K. The potential of plants as a system for the development and production of human biologics. F1000Research 2016, 5, 912. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Li, Y. Recombinant production of antimicrobial peptides in Escherichia coli: A review. Protein Expr. Purif. 2011, 80, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; de la Fuente-Nunez, C.; Ou, R.W.; Torres, M.T.; Pande, S.G.; Sinskey, A.J.; Lu, T.K. Yeast-Based synthetic biology platform for antimicrobial peptide production. ACS Synth. Biol. 2018, 7, 896–902. [Google Scholar] [CrossRef] [PubMed]
- Baur, J.A.; Pearson, K.J.; Price, N.L.; Jamieson, H.A.; Lerin, C.; Kalra, A.; Prabhu, V.V.; Allard, J.S.; Lopez-Lluch, G.; Lewis, K.; et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature 2006, 444, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Catalgol, B.; Batirel, S.; Taga, Y.; Ozer, N.K. Resveratrol: French paradox revisited. Front. Pharmacol. 2012, 3, 141. [Google Scholar] [CrossRef] [PubMed]
- Smoliga, J.M.; Baur, J.A.; Hausenblas, H.A. Resveratrol and health—A comprehensive review of human clinical trials. Mol. Nutr. Food Res. 2011, 55, 1129–1141. [Google Scholar] [CrossRef] [PubMed]
- Fiedor, J.; Burda, K. Potential role of carotenoids as antioxidants in human health and disease. Nutrients 2014, 6, 466–488. [Google Scholar] [CrossRef] [PubMed]
- Grune, T.; Lietz, G.; Palou, A.; Ross, A.C.; Stahl, W.; Tang, G.; Thurnham, D.; Yin, S.-A.; Biesalski, H.K. β-Carotene is an important vitamin A source for humans. J. Nutr. 2010, 140, 2268S–2285S. [Google Scholar] [CrossRef] [PubMed]
- Sies, H.; Prof, W. Lycopene: Antioxidant and biological effects and its bioavailability in the human. Proc. Soc. Exp. Biol. Med. 1998, 218, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.J.; Ho, C.Y.; Ho, F.J.; Tsai, T.Y.; Ke, H.M.; Wang, C.H.; Chen, H.L.; Shih, M.C.; Huang, C.C.; Li, W.H. PGASO: A synthetic biology tool for engineering a cellulolytic yeast. Biotechnol. Biofuels 2012, 5, 53. [Google Scholar] [CrossRef] [PubMed]
- Birtić, S.; Dussort, P.; Pierre, F.-X.; Bily, A.C.; Roller, M. Carnosic acid. Phytochemistry 2015, 115, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Scheler, U.; Brandt, W.; Porzel, A.; Rothe, K.; Manzano, D.; Bozic, D.; Papaefthimiou, D.; Balcke, G.U.; Henning, A.; Lohse, S.; et al. Elucidation of the biosynthesis of carnosic acid and its reconstitution in yeast. Nat. Commun. 2016, 7, 12942. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.D.; Wang, R.; Yang, J.; Lien, E.J. Preclinical and clinical examinations of Salvia miltiorrhiza and its tanshinones in ischemic conditions. Chin. Med. 2006, 1, 3. [Google Scholar] [CrossRef] [PubMed]
- Fukui, H.; Koshimizu, K.; Egawa, H. A new Diterpene with antimicrobial activity from Chamaecyparis pisifera Endle. Agric. Biol. Chem. 1978, 42, 1419–1423. [Google Scholar] [CrossRef]
- Breitling, R.; Takano, E. Synthetic biology advances for pharmaceutical production. Curr. Opin. Biotechnol. 2015, 35, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.; Heal, J.R.; Hamilton, W.D.O.; Boussemghoune, T.; Tange, T.Ø.; Delegrange, F.; Jaeschke, G.; Hatsch, A.; Heim, J. Yeast synthetic biology platform generates novel chemical structures as scaffolds for drug discovery. ACS Synth. Biol. 2014, 3, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Casini, A.; Chang, F.-Y.; Eluere, R.; King, A.M.; Young, E.M.; Dudley, Q.M.; Karim, A.; Pratt, K.; Bristol, C.; Forget, A.; et al. A pressure test to make 10 molecules in 90 days: External evaluation of methods to engineer biology. J. Am. Chem. Soc. 2018, 140, 4302–4316. [Google Scholar] [CrossRef] [PubMed]
- Harvey, C.J.B.; Tang, M.; Schlecht, U.; Horecka, J.; Fischer, C.R.; Lin, H.C.; Li, J.; Naughton, B.; Cherry, J.; Miranda, M.; et al. HEx: A heterologous expression platform for the discovery of fungal natural products. Sci. Adv. 2018, 4, eaar5459. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.S.; Buss, A.D. Natural products—The future scaffolds for novel antibiotics? Biochem. Pharmacol. 2006, 71, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Yuzawa, S.; Backman, T.W.H.; Keasling, J.D.; Katz, L. Synthetic biology of polyketide synthases. J. Ind. Microbiol. Biotechnol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Li, Y.; Fang, L.; Pfeifer, B.A. Tailoring pathway modularity in the biosynthesis of erythromycin analogs heterologously engineered in E. coli. Sci. Adv. 2015, 1, e1500077. [Google Scholar] [CrossRef] [PubMed]
- Yim, G.; Wang, W.; Thaker, M.N.; Tan, S.; Wright, G.D. How to make a glycopeptide: A synthetic biology approach to expand antibiotic chemical diversity. ACS Infect. Dis. 2016, 2, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Pretorius, I.S.; Boeke, J.D. Yeast 2.0—Connecting the dots in the construction of the world’s first functional synthetic eukaryotic genome. FEMS Yeast Res. 2018, 18, foy032. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Wang, Y.; Chen, T.; Gao, F.; Gong, J.; Abramczyk, D.; Walker, R.; Zhao, H.; Chen, S.; Liu, W.; et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome. Science 2017, 355. [Google Scholar] [CrossRef] [PubMed]
- Annaluru, N.; Muller, H.; Mitchell, L.A.; Ramalingam, S.; Stracquadanio, G.; Richardson, S.M.; Dymond, J.S.; Kuang, Z.; Scheifele, L.Z.; Cooper, E.M.; et al. Total synthesis of a functional designer eukaryotic chromosome. Science 2014, 344, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.X.; Li, B.Z.; Mitchell, L.A.; Wu, Y.; Qi, X.; Jin, Z.; Jia, B.; Wang, X.; Zeng, B.X.; Liu, H.M.; et al. “Perfect” designer chromosome V and behavior of a ring derivative. Science 2017, 355. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, L.A.; Wang, A.; Stracquadanio, G.; Kuang, Z.; Wang, X.; Yang, K.; Richardson, S.; Martin, J.A.; Zhao, Y.; Walker, R.; et al. Synthesis, debugging, and effects of synthetic chromosome consolidation: SynVI and beyond. Science 2017, 355. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, B.Z.; Zhao, M.; Mitchell, L.A.; Xie, Z.X.; Lin, Q.H.; Wang, X.; Xiao, W.H.; Wang, Y.; Zhou, X.; et al. Bug mapping and fitness testing of chemically synthesized chromosome X. Science 2017, 355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhao, G.; Luo, Z.; Lin, Y.; Wang, L.; Guo, Y.; Wang, A.; Jiang, S.; Jiang, Q.; Gong, J.; et al. Engineering the ribosomal DNA in a megabase synthetic chromosome. Science 2017, 355. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Stracquadanio, G.; Wang, Y.; Yang, K.; Mitchell, L.A.; Xue, Y.; Cai, Y.; Chen, T.; Dymond, J.S.; Kang, K.; et al. SCRaMbLE generates designed combinatorial stochastic diversity in synthetic chromosomes. Genome Res. 2016, 26, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.J.; Wu, Y.; Yang, K.; Li, Y.; Xu, H.; Zhang, H.; Li, B.-Z.; Li, X.; Xiao, W.-H.; Zhou, X.; et al. Heterozygous diploid and interspecies SCRaMbLEing. Nat. Commun. 2018, 9, 1934. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhu, R.-Y.; Mitchell, L.A.; Ma, L.; Liu, R.; Zhao, M.; Jia, B.; Xu, H.; Li, Y.-X.; Yang, Z.-M.; et al. In vitro DNA SCRaMbLE. Nat. Commun. 2018, 9, 1935. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Wang, L.; Wang, Y.; Zhang, W.; Guo, Y.; Shen, Y.; Jiang, L.; Wu, Q.; Zhang, C.; Cai, Y.; et al. Identifying and characterizing SCRaMbLEd synthetic yeast using ReSCuES. Nat. Commun. 2018, 9, 1930. [Google Scholar] [CrossRef] [PubMed]
- Hochrein, L.; Mitchell, L.A.; Schulz, K.; Messerschmidt, K.; Mueller-Roeber, B. L-SCRaMbLE as a tool for light-controlled CRE-mediated recombination in yeast. Nat. Commun. 2018, 9, 1931. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Wu, Y.; Li, B.-Z.; Mitchell, L.A.; Liu, H.; Pan, S.; Wang, J.; Zhang, H.-R.; Jia, N.; Li, B.; et al. Precise control of SCRaMbLE in synthetic haploid and diploid yeast. Nat. Commun. 2018, 9, 1933. [Google Scholar] [CrossRef] [PubMed]
- Gray, A.; Manasse, H.R., Jr. Shortages of medicines: A complex global challenge. Bull. World Health Organ. 2012, 90, 158. [Google Scholar] [CrossRef] [PubMed]
- Perez-Pinera, P.; Han, N.; Cleto, S.; Cao, J.; Purcell, O.; Shah, K.A.; Lee, K.; Ram, R.; Lu, T.K. Synthetic biology and microbioreactor platforms for programmable production of biologics at the point-of-care. Nat. Commun. 2016, 7, 12211. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Perez-Pinera, P.; Lowenhaupt, K.; Wu, M.R.; Purcell, O.; de la Fuente-Nunez, C.; Lu, T.K. Versatile and on-demand biologics co-production in yeast. Nat. Commun. 2018, 9, 77. [Google Scholar] [CrossRef] [PubMed]
- Oye, K.A.; Lawson, J.C.; Bubela, T. Drugs: Regulate ‘home-brew’ opiates. Nature 2015, 521, 281–283. [Google Scholar] [CrossRef] [PubMed]
- Endy, D.; Galanie, S.; Smolke, C.D. Complete absence of thebaine biosynthesis under home-brew fermentation conditions. bioRxiv 2015. [Google Scholar] [CrossRef]
- Denby, C.M.; Li, R.A.; Vu, V.T.; Costello, Z.; Lin, W.; Chan, L.J.G.; Williams, J.; Donaldson, B.; Bamforth, C.W.; Petzold, C.J.; et al. Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer. Nat. Commun. 2018, 9, 965. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Lloyd, N.D.; Pretorius, I.S.; Borneman, A.R. Heterologous production of raspberry ketone in the wine yeast Saccharomyces cerevisiae via pathway engineering and synthetic enzyme fusion. Microb. Cell Fact. 2016, 15, 49. [Google Scholar] [CrossRef] [PubMed]
- Varman, A.M.; Xiao, Y.; Leonard, E.; Tang, Y.J. Statistics-based model for prediction of chemical biosynthesis yield from Saccharomyces cerevisiae. Microb. Cell Fact. 2011, 10, 45. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Du, J.; Eriksen, D.T.; Zhao, H. Combinatorial design of a highly efficient xylose-utilizing pathway in Saccharomyces cerevisiae for the production of cellulosic biofuels. Appl. Environ. Microbiol. 2013, 79, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Chao, R.; Mishra, S.; Si, T.; Zhao, H. Engineering biological systems using automated biofoundries. Metab. Eng. 2017, 42, 98–108. [Google Scholar] [CrossRef] [PubMed]




| Compound | Application | Compound Class | Chassis Organism | Titre | Natural Source |
|---|---|---|---|---|---|
| Agkisacutacin (Snake venom) | Antithrombotic | Protein | Pichia pastoris | 100 mg/L [49] | Agkistrodon acutus (Pit viper) |
| Apidaecin Ia | Antimicrobial | Antimicrobial peptide | P. pastoris | >700 mg/L [50] | Apis (Honeybee) |
| Artemisinic acid | Artemisinin (anti-malaria) precursor | Sesquiterpene | S. cerevisiae | 25 g/L [9] | Artemisia annua (Sweet wormwood) |
| Astaxanthin | Antioxidant | Carotenoid | Kluyveromyces marxianus | 1 mg/g DCW [51] | Various, including krill and shrimp |
| Breviscapine (Scutellarin and apigenin-7-O-glucuronide) | Chinese medicine. Cardiovascular and cerebrovascular disease. | Flavanoid | S. cerevisiae | 105 and 185 mg/L [52] | Erigeron breviscapus |
| Carnosic acid | Antioxidant | Diterpene | S. cerevisiae | 18 mg/L [53] | Rosmarinus officinalis (Rosemary) and Salvia officinalis (Sage) |
| β-Carotene | Antioxidant | Carotenoid | Yarrowia lipolytica | 6.5 g/L (90 mg/g) [37] | Various, including carrots |
| Hydrocodone | Pain relief (opioid) | Benzylisoquinoline alkaloids (BIA) | S. cerevisiae | <1 μg/L [47] | N/A (Semi-synthetic from Codeine) |
| Lycopene | Antioxidant, anti-cancer | Carotenoid | S. cerevisiae | 55.56 mg/g DCW [54] | Solanum lycopersicum (Tomato) |
| Anti-Ebola monoclonal antibodies | Antiviral | Monoclonal antibody | P. pastoris | 1 to 10 mg/L [55] | N/A |
| Noscapine | Anticancer | Benzylisoquinoline alkaloids (BIA) | S. cerevisiae | 2.2 mg/L [8] | Papaver somniferum (Poppy plant) |
| Penicillin | Antibiotic | Beta-lactam nonribosomal peptide | S. cerevisiae | 14.9 ng/mL [56] | Penicillium fungi |
| Pisiferic acid | Antimicrobial agent | Diterpene | S. cerevisiae | 2.65 mg/L [53] | Chamaecyparis Pisifera (Sawara cypress) |
| Resveratrol | Several; antioxidant | Stilbenoid | S. cerevisiae | 800 mg/L [57] | Polygonum cupidatum (Japanese knotweed) |
| Salviol | Established bioactivity, awaiting further evaluation | Diterpene | S. cerevisiae | 15 mg/L [53] | Salvia miltiorrhiza (Chinese sage) |
| Strictosidine | Intermediate | Monoterpene indole alkaloid | S. cerevisiae | 0.8 mg/L [58] | N/A (chemical synthesis) |
| Taxadiene | Anticancer Taxol precursor | Diterpenoid | S. cerevisiae | 72.8 mg/L [59] | Taxus brevifolia (Pacific yew) |
| Δ9-tetrahydrocannabinolic acid | Tetrahydrocannabinol precursor | Cannabinoid | P. pastoris | 3.05 g/L [60] | Cannabis sativa (Cannabis) |
| Thebaine | Opioid precursor | Benzylisoquinoline alkaloids (BIA) | S. cerevisiae | <1 μg/L [47] | Papaver somniferum (Poppy straw) |
| Theophylline | Anti-asthma medication | Methylxanthine | S. cerevisiae | 61 μg/L [61] | Camellia sinensis (Tea) and Theobroma cacao (Cocoa) |
| Vindoline | Anticancer (vinblastine and vincristine) precursor | Monterpenoid indole alkaloid | S. cerevisiae | 2.7 mg/L [62] | Catharanthus roseus (Madagascar periwinkle) |
| Violacein | Antibiotic | Bis-indole pigment | S. cerevisiae | 16.8 mg/L [63] | Chromobacterium violaceum |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walker, R.S.K.; Pretorius, I.S. Applications of Yeast Synthetic Biology Geared towards the Production of Biopharmaceuticals. Genes 2018, 9, 340. https://doi.org/10.3390/genes9070340
Walker RSK, Pretorius IS. Applications of Yeast Synthetic Biology Geared towards the Production of Biopharmaceuticals. Genes. 2018; 9(7):340. https://doi.org/10.3390/genes9070340
Chicago/Turabian StyleWalker, Roy S. K., and Isak S. Pretorius. 2018. "Applications of Yeast Synthetic Biology Geared towards the Production of Biopharmaceuticals" Genes 9, no. 7: 340. https://doi.org/10.3390/genes9070340
APA StyleWalker, R. S. K., & Pretorius, I. S. (2018). Applications of Yeast Synthetic Biology Geared towards the Production of Biopharmaceuticals. Genes, 9(7), 340. https://doi.org/10.3390/genes9070340





