Above the Epitranscriptome: RNA Modifications and Stem Cell Identity
Abstract
1. Epigenetics and Epitranscriptomics
2. RNA Modifications
2.1. State of the Art
2.2. Coding RNA Modifications: The Messenger RNA
2.2.1. Canonical Modifications in mRNA
2.2.2. Non-Canonical mRNA Modifications
2.2.3. Mitochondrial mRNA
2.3. Noncoding-RNA Modifications
2.3.1. Transfer RNA
2.3.2. Ribosomal RNA
2.3.3. Long Noncoding-RNA
2.3.4. microRNAs
2.3.5. circRNAs
3. Bioinformatics for Epitranscriptomics
3.1. Databases for RNA Modifications
3.2. Bioinformatic Tools for Predicting RNA Modifications
4. Epitranscriptomics and Stem Cells
4.1. Stem Cells
4.2. RNA Modifications and Stem Cells
4.2.1. N6-methyl-adenosine mRNA Modification and Stem Cells
4.2.2. 5-methylcytosine and 5-hydroxymethylcytosine Modifications and Stem Cells
4.2.3. Pseudouridine Modification and Stem Cells
5. Concluding Remarks
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 3′-UTR | Three prime untranslated region |
| ALKB | Alpha-ketoglutarate-dependent dioxygenase AlkB |
| ALKBH1 | Nucleic acid dioxygenase |
| ALKBH5 | RNA demethylase ALKBH5 |
| ALYREF | THO complex subunit 4 |
| BCL2 | B-cell lymphoma 2 |
| CDS | CoDing Sequence |
| DKC1 | H/ACA ribonucleoprotein complex subunit 4 |
| DNA | Deoxyribonucleic acid |
| DNMT2 | tRNA (cytosine(38)-C(5))-methyltransferase |
| EGFR | Epidermal growth factor receptor |
| ELP3 | Elongator complex protein 3 |
| FOXM1 | Forkhead box protein M1 |
| HEN1 | Small RNA 2′-O-methyltransferase |
| HNRNPA2B1 | Heterogeneous nuclear ribonucleoproteins A2/B1 |
| hTRMT9 | Human Probable tRNA methyltransferase 9B |
| IME4 | N6-adenosine-methyltransferase |
| MED2 | Mediator of RNA polymerase II transcription subunit 2 |
| MED4 | Mediator of RNA polymerase II transcription subunit 4 |
| METTL2 | Methyltransferase-like protein 2 |
| METTL3 | Methyltransferase-like protein 3 |
| METTL6 | Methyltransferase-like protein 6 |
| METTL14 | Methyltransferase-like protein 14 |
| MYB | Myb proto-oncogene protein |
| MYC | Myc proto-oncogene protein |
| MyoD | Myogenic differentiation 1 |
| NOTCH1 | Neurogenic locus notch homolog protein 1 |
| NSUN2 | NOP2/Sun domain family, member 2 |
| NSUN4 | NOL1/NOP2/Sun domain family member 4 |
| p16 | Cyclin-dependent kinase inhibitor 2A, multiple tumor suppressor 1 |
| PTEN | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase |
| PUS | Pseudouridine synthase A |
| RBM15 | Putative RNA-binding protein 15 |
| RluE | Ribosomal large subunit Pseudouridine synthase E |
| ROS | Reactive oxygen specie |
| RPUSD3 | Mitochondrial mRNA Pseudouridine synthase RPUSD3 |
| RPUSD4 | Mitochondrial RNA Pseudouridine synthase RPUSD4 |
| SMAD2 | Mothers against decapentaplegic homolog 2 |
| SMAD3 | Mothers against decapentaplegic homolog 2 |
| SOD1 | Superoxide dismutase [Cu-Zn] |
| SPI1 | Transcription factor PU.1 |
| SRSF2 | Serine/arginine-rich splicing factor 2 |
| TAZ | Tafazzin |
| TET1 | methylcytosine dioxygenase TET1 |
| TET3 | methylcytosine dioxygenase TET3 |
| TGFβ | Transforming growth factor beta |
| TRM4 | Multisite-specific tRNA:(cytosine-C(5))-methyltransferase |
| TrmL | tRNA(cytidine(34)-2′-O)-methyltransferase |
| TRMT10C | tRNA methyltransferase 10 homolog C |
| TRMT61A | tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit |
| TRMT61B | tRNA (adenine(58)-N(1))-methyltransferase, mitochondrial |
| TRUB2 | Mitochondrial mRNA Pseudouridine synthase TRUB2 |
| U1 | U1 spliceosomal RNA |
| U2 | U2 spliceosomal RNA |
| U3 | U3 spliceosomal RNA |
| U4 | U4 spliceosomal RNA |
| U5 | U5 spliceosomal RNA |
| U6 | U6 spliceosomal RNA |
| U8 | U8 spliceosomal RNA |
| UHRF2 | E3 ubiquitin-protein ligase UHRF2 |
| WTAP | Wilms tumor 1 associated protein |
| YTDCH1 | YTH domain-containing protein 1 |
| YTHDF1 | YTH domain-containing family protein 1 |
| YTHDF2 | YTH domain-containing family protein 2 |
| YTHDF3 | YTH domain-containing family protein 2 |
| ZNF217 | Zinc finger protein 217 |
References
- Martino, S.; Morena, F.; Barola, C.; Bicchi, I.; Emiliani, C. Proteomics and epigenetic mechanisms in stem cells. Curr. Proteom. 2014, 11, 193–209. [Google Scholar] [CrossRef]
- Shakya, K.; O’Connell, M.J.; Ruskin, H.J. The landscape for epigenetic/epigenomic biomedical resources. Epigenetics 2012, 7, 982–986. [Google Scholar] [CrossRef] [PubMed]
- Sen, P.; Shah, P.P.; Nativio, R.; Berger, S.L. Epigenetic mechanisms of longevity and aging. Cell 2016, 166, 822–839. [Google Scholar] [CrossRef] [PubMed]
- Jaenisch, R.; Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; He, C. Nucleic acid modifications with epigenetic significance. Curr. Opin. Chem. Biol. 2012, 16, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, Z. Linking DNA replication to heterochromatin silencing and epigenetic inheritance. Acta Biochim. Biophys. Sin. 2012, 44, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Francis, N.J. Mechanisms of epigenetic inheritance: Copying of polycomb repressed chromatin. Cell Cycle 2009, 8, 3521–3526. [Google Scholar] [CrossRef] [PubMed]
- Roloff, T.C.; Nuber, U.A. Chromatin, epigenetics and stem cells. Eur. J. Cell Biol. 2005, 84, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Krishnakumar, R.; Blelloch, R.H. Epigenetics of cellular reprogramming. Curr. Opin. Genet. Dev. 2013, 23, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Ng, R.K.; Gurdon, J.B. Epigenetic inheritance of cell differentiation status. Cell Cycle 2008, 7, 1173–1177. [Google Scholar] [CrossRef] [PubMed]
- Probst, A.V.; Dunleavy, E.; Almouzni, G. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 2009, 10, 192–206. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Beard, C.; Jaenisch, R. Role for DNA methylation in genomic imprinting. Nature 1993, 366, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Groth, A.; Rocha, W.; Verreault, A.; Almouzni, G. Chromatin challenges during DNA replication and repair. Cell 2007, 128, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, S.; Van Emburgh, B.O.; Robertson, K.D. DNA methylation in development and human disease. Mutat. Res. 2008, 647, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Hamm, C.A.; Costa, F.F. Epigenomes as therapeutic targets. Pharmacol. Ther. 2015, 151, 72–86. [Google Scholar] [CrossRef] [PubMed]
- Saletore, Y.; Chen-Kiang, S.; Mason, C.E. Novel RNA regulatory mechanisms revealed in the epitranscriptome. RNA Biol. 2013, 10, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Bicchi, I.; Morena, F.; Montesano, S.; Polidoro, M.; Martino, S. MicroRNAs and molecular mechanisms of neurodegeneration. Genes 2013, 4, 244–263. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S. Cracking the epitranscriptome. RNA 2016, 22, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fu, Y.; Klungland, A.; Yang, Y.-G.; He, C. Sprouts of RNA epigenetics: The discovery of mammalian RNA demethylases. RNA Biol. 2013, 10, 915–918. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Jia, G.; Pang, X.; Wang, R.N.; Wang, X.; Li, C.J.; Smemo, S.; Dai, Q.; Bailey, K.A.; Nobrega, M.A.; et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 2013, 4, 1798. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.J. From birth to death: The complex lives of eukaryotic mRNAs. Science 2005, 309, 1514–1518. [Google Scholar] [CrossRef] [PubMed]
- Jaffrey, S.R. An expanding universe of mRNA modifications. Nat. Struct. Mol. Biol. 2014, 21, 945–946. [Google Scholar] [CrossRef] [PubMed]
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012, 40, 5023–5033. [Google Scholar] [CrossRef] [PubMed]
- Khoddami, V.; Cairns, B.R. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 2013, 31, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Yu, Y.-T. RNA pseudouridylation: New insights into an old modification. Trends Biochem. Sci. 2013, 38, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Hoang, C.; Ferré-D’Amaré, A.R. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: Nucleotide flipping by an RNA-modifying enzyme. Cell 2001, 107, 929–939. [Google Scholar] [CrossRef]
- Xiang, Y.; Laurent, B.; Hsu, C.-H.; Nachtergaele, S.; Lu, Z.; Sheng, W.; Xu, C.; Chen, H.; Ouyang, J.; Wang, S.; et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 2017, 543, 573–576. [Google Scholar] [CrossRef] [PubMed]
- Incarnato, D.; Oliviero, S. The RNA epistructurome: uncovering rna function by studying structure and post-transcriptional modifications. Trends Biotechnol. 2017, 35, 318–333. [Google Scholar] [CrossRef] [PubMed]
- Gatsiou, A.; Vlachogiannis, N.; Lunella, F.F.; Sachse, M.; Stellos, K. Adenosine-to-Inosine RNA editing in health and disease. Antioxid. Redox Signal. 2017. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, W.V.; Bell, T.A.; Schaening, C. Messenger RNA modifications: Form, distribution, and function. Science 2016, 352, 1408–1412. [Google Scholar] [CrossRef] [PubMed]
- Ben-Haim, M.S.; Moshitch-Moshkovitz, S.; Rechavi, G. FTO: Linking m6A demethylation to adipogenesis. Cell Res. 2015, 25, 3–4. [Google Scholar] [CrossRef] [PubMed]
- Licht, K.; Jantsch, M.F. Rapid and dynamic transcriptome regulation by RNA editing and RNA modifications. J. Cell Biol. 2016, 213, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Machnicka, M.A.; Milanowska, K.; Osman Oglou, O.; Purta, E.; Kurkowska, M.; Olchowik, A.; Januszewski, W.; Kalinowski, S.; Dunin-Horkawicz, S.; Rother, K.M.; et al. MODOMICS: A database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013, 41, D262–D267. [Google Scholar] [CrossRef] [PubMed]
- Perry, R.P.; Kelley, D.E.; Friderici, K.; Rottman, F. The methylated constituents of L cell messenger RNA: Evidence for an unusual cluster at the 5′ terminus. Cell 1975, 4, 387–394. [Google Scholar] [CrossRef]
- Batista, P.J. The RNA Modification N6-methyladenosine and its implications in human disease. Genom. Proteom. Bioinform. 2017, 15, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Helm, M.; Motorin, Y. Detecting RNA modifications in the epitranscriptome: Predict and validate. Nat. Rev. Genet. 2017, 18, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Pan, T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem. Sci. 2013, 38, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Dubin, D.T.; Taylor, R.H. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975, 2, 1653–1668. [Google Scholar] [CrossRef] [PubMed]
- Delatte, B.; Wang, F.; Ngoc, L.V.; Collignon, E.; Bonvin, E.; Deplus, R.; Calonne, E.; Hassabi, B.; Putmans, P.; Awe, S.; et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 2016, 351, 282–285. [Google Scholar] [CrossRef] [PubMed]
- Rosa-Mercado, N.A.; Withers, J.B.; Steitz, J.A. Settling the m6A debate: Methylation of mature mRNA is not dynamic but accelerates turnover. Genes Dev. 2017, 31, 957–958. [Google Scholar] [CrossRef] [PubMed]
- Roignant, J.-Y.; Soller, M. m6A in mRNA: An ancient mechanism for fine-tuning gene expression. Trends Genet. 2017, 33, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Nachtergaele, S.; Moshitch-Moshkovitz, S.; Peer, E.; Kol, N.; Ben-Haim, M.S.; Dai, Q.; Di Segni, A.; Salmon-Divon, M.; Clark, W.C.; et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016, 530, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xiong, X.; Wang, K.; Wang, L.; Shu, X.; Ma, S.; Yi, C. Transcriptome-wide mapping reveals reversible and dynamic N1-methyladenosine methylome. Nat. Chem. Biol. 2016, 12, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Harcourt, E.M.; Kietrys, A.M.; Kool, E.T. Chemical and structural effects of base modifications in messenger RNA. Nature 2017, 541, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Morse, D.P.; Bass, B.L. Detection of inosine in messenger RNA by inosine-specific cleavage. Biochemistry 1997, 36, 8429–8434. [Google Scholar] [CrossRef] [PubMed]
- Copp, W.; Denisov, A.Y.; Xie, J.; Noronha, A.M.; Liczner, C.; Safaee, N.; Wilds, C.J.; Gehring, K. Influence of nucleotide modifications at the C2′ position on the Hoogsteen base-paired parallel-stranded duplex of poly(A) RNA. Nucleic Acids Res. 2017, 45, 10321–10331. [Google Scholar] [CrossRef] [PubMed]
- Byszewska, M.; Śmietański, M.; Purta, E.; Bujnicki, J.M. RNA methyltransferases involved in 5′ cap biosynthesis. RNA Biol. 2014, 11, 1597–1607. [Google Scholar] [CrossRef] [PubMed]
- Bawankar, P.; Shaw, P.J.; Sardana, R.; Babar, P.H.; Patankar, S. 5′ and 3′ end modifications of spliceosomal RNAs in Plasmodium falciparum. Mol. Biol. Rep. 2010, 37, 2125–2133. [Google Scholar] [CrossRef] [PubMed]
- Crick, F. Split genes and RNA splicing. Science 1979, 204, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Muthukrishnan, S.; Filipowicz, W.; Sierra, J.M.; Both, G.W.; Shatkin, A.J.; Ochoa, S. mRNA methylation and protein synthesis in extracts from embryos of brine shrimp, Artemia salina. J. Biol. Chem. 1975, 250, 9336–9341. [Google Scholar] [PubMed]
- Mao, X.; Schwer, B.; Shuman, S. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol. Cell. Biol. 1995, 15, 4167–4174. [Google Scholar] [CrossRef] [PubMed]
- Wurth, L.; Gribling-Burrer, A.-S.; Verheggen, C.; Leichter, M.; Takeuchi, A.; Baudrey, S.; Martin, F.; Krol, A.; Bertrand, E.; Allmang, C. Hypermethylated-capped selenoprotein mRNAs in mammals. Nucleic Acids Res. 2014, 42, 8663–8677. [Google Scholar] [CrossRef] [PubMed]
- Furuichi, Y.; Shatkin, A.J. Viral and cellular mRNA capping: Past and prospects. Adv. Virus Res. 2000, 55, 135–184. [Google Scholar] [PubMed]
- Zhao, J.; Hyman, L.; Moore, C. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999, 63, 405–445. [Google Scholar] [PubMed]
- Darnell, J.E. Implications of RNA-RNA splicing in evolution of eukaryotic cells. Science 1978, 202, 1257–1260. [Google Scholar] [CrossRef] [PubMed]
- Schellenberg, M.J.; Ritchie, D.B.; MacMillan, A.M. Pre-mRNA splicing: A complex picture in higher definition. Trends Biochem. Sci. 2008, 33, 243–246. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Rio, D.C. Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef] [PubMed]
- Dezi, V.; Ivanov, C.; Haussmann, I.U.; Soller, M. Nucleotide modifications in messenger RNA and their role in development and disease. Biochem. Soc. Trans. 2016, 44, 1385–1393. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Kuzmenko, A.; Atkinson, G.C.; Levitskii, S.; Zenkin, N.; Tenson, T.; Hauryliuk, V.; Kamenski, P. Mitochondrial translation initiation machinery: Conservation and diversification. Biochimie 2014, 100, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Salemi, M.; Giambirtone, M.; Barone, C.; Salluzzo, M.G.; Russo, R.; Giudice, M.L.; Cutuli, S.; Ridolfo, F.; Romano, C. Mitochondrial mRNA expression in fibroblasts of Down syndrome subjects. Hum. Cell 2018, 31, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Perry, R.; Kelley, D. Existence of methylated messenger RNA in mouse L cells. Cell 1974, 1, 37–42. [Google Scholar] [CrossRef]
- Bujnicki, J.M.; Feder, M.; Radlinska, M.; Blumenthal, R.M. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA: m6A methyltransferase. J. Mol. Evol. 2002, 55, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huang, J.; Zou, T.; Yin, P. Human m6A writers: Two subunits, 2 roles. RNA Biol. 2017, 14, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Alarcón, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a mediator of m6A-dependent nuclear RNA processing events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Edupuganti, R.R.; Geiger, S.; Lindeboom, R.G.H.; Shi, H.; Hsu, P.J.; Lu, Z.; Wang, S.-Y.; Baltissen, M.P.A.; Jansen, P.W.T.C.; Rossa, M.; et al. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 2017, 24, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; He, C. RNA epigenetics—Chemical messages for posttranscriptional gene regulation. Curr. Opin. Chem. Biol. 2016, 30, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA modifications in gene expression regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Loos, R.J.F.; Yeo, G.S.H. The bigger picture of FTO: The first GWAS-identified obesity gene. Nat. Rev. Endocrinol. 2014, 10, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.J.T.; Pan, T.; Kalsotra, A. RNA modifications and structures cooperate to guide RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 2017, 18, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T.K.; de Crécy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef] [PubMed]
- Klungland, A.; Dahl, J.A. Dynamic RNA modifications in disease. Curr. Opin. Genet. Dev. 2014, 26, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.-B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Samanta, D.; Lu, H.; Bullen, J.W.; Zhang, H.; Chen, I.; He, X.; Semenza, G.L. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. USA 2016, 113, E2047–E2056. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.; Agarwala, S.D.; Mumbach, M.R.; Jovanovic, M.; Mertins, P.; Shishkin, A.; Tabach, Y.; Mikkelsen, T.S.; Satija, R.; Ruvkun, G.; et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 2013, 155, 1409–1421. [Google Scholar] [CrossRef] [PubMed]
- Dunn, D.B. The occurrence of 1-methyladenine in ribonucleic acid. Biochim. Biophys. Acta 1961, 46, 198–200. [Google Scholar] [CrossRef]
- Dai, X.; Wang, T.; Gonzalez, G.; Wang, Y. Identification of YTH Domain-containing proteins as the readers for N1-Methyladenosine in RNA. Anal. Chem. 2018, 90, 6380–6384. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yang, Y.; Sun, B.-F.; Chen, Y.-S.; Xu, J.-W.; Lai, W.-Y.; Li, A.; Wang, X.; Bhattarai, D.P.; Xiao, W.; et al. 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017, 27, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Kool, E.T. Origins of the large differences in stability of DNA and RNA helices: C-5 methyl and 2′-hydroxyl effects. Biochemistry 1995, 34, 4125–4132. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Fan, X.; Xing, J.; Liu, Z.; Jiang, B.; Dou, Y.; Gorospe, M.; Wang, W. NSun2 delays replicative senescence by repressing p27 (KIP1) translation and elevating CDK1 translation. Aging 2015, 7, 1143–1158. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.; Bernstein, D.A.; Mumbach, M.R.; Jovanovic, M.; Herbst, R.H.; León-Ricardo, B.X.; Engreitz, J.M.; Guttman, M.; Satija, R.; Lander, E.S.; et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014, 159, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Carlile, T.M.; Rojas-Duran, M.F.; Zinshteyn, B.; Shin, H.; Bartoli, K.M.; Gilbert, W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014, 515, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, P.P.; AlSadhan, I.; Merriman, D.K.; Al-Hashimi, H.M.; Herschlag, D. Pseudouridine and N6-methyladenosine modifications weaken PUF protein/RNA interactions. RNA 2017, 23, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Lim, V.I.; Curran, J.F. Analysis of codon: Anticodon interactions within the ribosome provides new insights into codon reading and the genetic code structure. RNA 2001, 7, 942–957. [Google Scholar] [CrossRef] [PubMed]
- Keegan, L.; Khan, A.; Vukic, D.; O’Connell, M. ADAR RNA editing below the backbone. RNA 2017, 23, 1317–1328. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, M.T.; Sloan, K.E. The mitochondrial epitranscriptome: The roles of RNA modifications in mitochondrial translation and human disease. Cell. Mol. Life Sci. 2018, 75, 241–260. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Huang, C.; Zhang, D.; Wang, J.; Li, J.; Jin, H.; Huang, C. Mitochondrial catalase induces cells transformation through nucleolin-dependent Cox-2 mRNA stabilization. Free Radic. Biol. Med. 2017, 113, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Safra, M.; Sas-Chen, A.; Nir, R.; Winkler, R.; Nachshon, A.; Bar-Yaacov, D.; Erlacher, M.; Rossmanith, W.; Stern-Ginossar, N.; Schwartz, S. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 2017, 551, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.K.; Rajasekharan, R. The m6A methyltransferase Ime4 and mitochondrial functions in yeast. Curr. Genet. 2018, 64, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Rüdinger, M.; Funk, H.T.; Rensing, S.A.; Maier, U.G.; Knoop, V. RNA editing: Only eleven sites are present in the Physcomitrella patens mitochondrial transcriptome and a universal nomenclature proposal. Mol. Genet. Genom. 2009, 281, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.D.; Parsons, C.; Walker, L.; Zhang, W.C.; Slack, F.J. Targeting noncoding RNAs in disease. J. Clin. Investig. 2017, 127, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Xiong, M.; Xu, C.; Xiang, P.; Zhong, X. Long noncoding RNAs: An overview. Methods Mol. Biol. 2016, 1402, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Pan, T. Modifications and functional genomics of human transfer RNA. Cell Res. 2018, 28, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, K.; Nostramo, R.T.; Wan, Y.; Hopper, A.K. tRNA dynamics between the nucleus, cytoplasm and mitochondrial surface: Location, location, location. Biochim. Biophys. Acta 2018, 1861, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Sissler, M.; González-Serrano, L.E.; Westhof, E. Recent advances in mitochondrial aminoacyl-tRNA synthetases and disease. Trends Mol. Med. 2017, 23, 693–708. [Google Scholar] [CrossRef] [PubMed]
- Greco, C.M.; Condorelli, G. Epigenetic modifications and noncoding RNAs in cardiac hypertrophy and failure. Nat. Rev. Cardiol. 2015, 12, 488–497. [Google Scholar] [CrossRef] [PubMed]
- Shafik, A.; Schumann, U.; Evers, M.; Sibbritt, T.; Preiss, T. The emerging epitranscriptomics of long noncoding RNAs. Biochim. Biophys. Acta 2016, 1859, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Pepin, G.; Gantier, M.P. microRNA Decay: Refining microRNA regulatory activity. Microrna 2016, 5, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Tuorto, F.; Lyko, F. Genome recoding by tRNA modifications. Open Biol. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Zhao, X.; Wu, Y.-S.; Li, M.-M.; Wang, X.-J.; Yang, Y.-G. N6-methyl-adenosine (m6A) in RNA: An old modification with a novel epigenetic function. Genom. Proteom. Bioinform. 2013, 11, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Indrisiunaite, G.; DeMirci, H.; Ieong, K.-W.; Wang, J.; Petrov, A.; Prabhakar, A.; Rechavi, G.; Dominissini, D.; He, C.; et al. 2′-O-methylation in mRNA disrupts tRNA decoding during translation elongation. Nat. Struct. Mol. Biol. 2018, 25, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Asano, K.; Suzuki, T.; Saito, A.; Wei, F.-Y.; Ikeuchi, Y.; Numata, T.; Tanaka, R.; Yamane, Y.; Yamamoto, T.; Goto, T.; et al. Metabolic and chemical regulation of tRNA modification associated with taurine deficiency and human disease. Nucleic Acids Res. 2018, 46, 1565–1583. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.-J.; Zhou, M.; Fang, Z.-P.; Wang, M.; Zhou, X.-L.; Wang, E.-D. The tRNA recognition mechanism of the minimalist SPOUT methyltransferase, TrmL. Nucleic Acids Res. 2013, 41, 7828–7842. [Google Scholar] [CrossRef] [PubMed]
- Begley, U.; Sosa, M.S.; Avivar-Valderas, A.; Patil, A.; Endres, L.; Estrada, Y.; Chan, C.T.Y.; Su, D.; Dedon, P.C.; Aguirre-Ghiso, J.A.; et al. A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-α. EMBO Mol. Med. 2013, 5, 366–383. [Google Scholar] [CrossRef] [PubMed]
- Lentini, J.M.; Ramos, J.; Fu, D. Monitoring the 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) modification in eukaryotic tRNAs via the γ-toxin endonuclease. RNA 2018, 24, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Oerum, S.; Dégut, C.; Barraud, P.; Tisné, C. m1A Post-transcriptional modification in tRNAs. Biomolecules 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Kawarada, L.; Suzuki, T.; Ohira, T.; Hirata, S.; Miyauchi, K.; Suzuki, T. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 2017, 45, 7401–7415. [Google Scholar] [CrossRef] [PubMed]
- Pang, P.; Deng, X.; Wang, Z.; Xie, W. Structural and biochemical insights into the 2′-O-methylation of pyrimidines 34 in tRNA. FEBS J. 2017, 284, 2251–2263. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.M.; Wang, J.; Zhang, Y.F.; Gao, Y.H. Ovarian cancer proliferation and apoptosis are regulated by human transfer RNA methyltransferase 9-likevia LIN9. Oncol. Lett. 2017, 14, 4461–4466. [Google Scholar] [CrossRef] [PubMed]
- Bourgeois, G.; Létoquart, J.; van Tran, N.; Graille, M. Trm112, a protein activator of methyltransferases modifying actors of the eukaryotic translational apparatus. Biomolecules 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Liu, X.; Sheng, N.; Oo, K.S.; Liang, J.; Chionh, Y.H.; Xu, J.; Ye, F.; Gao, Y.-G.; Dedon, P.C.; et al. Three distinct 3-methylcytidine (m3C) methyltransferases modify tRNA and mRNA in mice and humans. J. Biol. Chem. 2017, 292, 14695–14703. [Google Scholar] [CrossRef] [PubMed]
- Bednářová, A.; Hanna, M.; Durham, I.; VanCleave, T.; England, A.; Chaudhuri, A.; Krishnan, N. Lost in translation: defects in transfer RNA modifications and neurological disorders. Front. Mol. Neurosci. 2017, 10, 135. [Google Scholar] [CrossRef] [PubMed]
- Bento-Abreu, A.; Jager, G.; Swinnen, B.; Rué, L.; Hendrickx, S.; Jones, A.; Staats, K.A.; Taes, I.; Eykens, C.; Nonneman, A.; et al. Elongator subunit 3 (ELP3) modifies ALS through tRNA modification. Hum. Mol. Genet. 2018, 27, 1276–1289. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Nagao, A.; Suzuki, T. Human mitochondrial tRNAs: Biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 2011, 45, 299–329. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, M.T. Genetics: The Importance of Mitochondrial tRNA. Available online: https://www.nature.com/articles/nrc3997 (accessed on 12 June 2018).
- Morscher, R.J.; Ducker, G.S.; Li, S.H.-J.; Mayer, J.A.; Gitai, Z.; Sperl, W.; Rabinowitz, J.D. Mitochondrial translation requires folate-dependent tRNA methylation. Nature 2018, 554, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Trixl, L.; Amort, T.; Wille, A.; Zinni, M.; Ebner, S.; Hechenberger, C.; Eichin, F.; Gabriel, H.; Schoberleitner, I.; Huang, A.; et al. RNA cytosine methyltransferase Nsun3 regulates embryonic stem cell differentiation by promoting mitochondrial activity. Cell. Mol. Life Sci. 2018, 75, 1483–1497. [Google Scholar] [CrossRef] [PubMed]
- Erales, J.; Marchand, V.; Panthu, B.; Gillot, S.; Belin, S.; Ghayad, S.E.; Garcia, M.; Laforêts, F.; Marcel, V.; Baudin-Baillieu, A.; et al. Evidence for rRNA 2′-O-methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proc. Natl. Acad. Sci. USA 2017, 114, 12934–12939. [Google Scholar] [CrossRef] [PubMed]
- Garone, C.; D’Souza, A.R.; Dallabona, C.; Lodi, T.; Rebelo-Guiomar, P.; Rorbach, J.; Donati, M.A.; Procopio, E.; Montomoli, M.; Guerrini, R.; et al. Defective mitochondrial rRNA methyltransferase MRM2 causes MELAS-like clinical syndrome. Hum. Mol. Genet. 2017, 26, 4257–4266. [Google Scholar] [CrossRef] [PubMed]
- Sergiev, P.V.; Aleksashin, N.A.; Chugunova, A.A.; Polikanov, Y.S.; Dontsova, O.A. Structural and evolutionary insights into ribosomal RNA methylation. Nat. Chem. Biol. 2018, 14, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Natchiar, S.K.; Myasnikov, A.G.; Kratzat, H.; Hazemann, I.; Klaholz, B.P. Visualization of chemical modifications in the human 80S ribosome structure. Nature 2017, 551, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Tillault, A.-S.; Schultz, S.K.; Wieden, H.-J.; Kothe, U. Molecular Determinants for 23S rRNA Recognition and modification by the E. coli Pseudouridine Synthase RluE. J. Mol. Biol. 2018, 430, 1284–1294. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, W.; Hirota, K.; Wan, H.; Sumi, N.; Miyata, M.; Araoi, S.; Nomura, N.; Kako, K.; Fukamizu, A. rRNA adenine methylation requires T07A9.8 gene as rram-1 in Caenorhabditis elegans. J. Biochem. 2018, 163, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Suzuki, T.; Yokoyama, W.; Yagi, S.; Matsumura, K.; Nakajima, Y.; Harigae, H.; Fukamizu, A.; Motohashi, H. Nucleomethylin deficiency impairs embryonic erythropoiesis. J. Biochem. 2018, 163, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Bar-Yaacov, D.; Frumkin, I.; Yashiro, Y.; Chujo, T.; Ishigami, Y.; Chemla, Y.; Blumberg, A.; Schlesinger, O.; Bieri, P.; Greber, B.; et al. Correction: Mitochondrial 16S rRNA is methylated by tRNA methyltransferase TRMT61B in all vertebrates. PLoS Biol. 2017, 15, e1002594. [Google Scholar] [CrossRef] [PubMed]
- Antonicka, H.; Choquet, K.; Lin, Z.-Y.; Gingras, A.-C.; Kleinman, C.L.; Shoubridge, E.A. A pseudouridine synthase module is essential for mitochondrial protein synthesis and cell viability. EMBO Rep. 2017, 18, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Chigusa, S.; Moroi, T.; Shoji, Y. State-of-the-art calculation of the decay rate of electroweak vacuum in the standard model. Phys. Rev. Lett. 2017, 119, 211801. [Google Scholar] [CrossRef] [PubMed]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, biology and functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Meyers, B.C. Deep sequencing from hen1 mutants to identify small RNA 3′ modifications. Cold Spring Harb. Symp. Quant. Biol. 2012, 77, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Shoshan, E.; Mobley, A.K.; Braeuer, R.R.; Kamiya, T.; Huang, L.; Vasquez, M.E.; Salameh, A.; Lee, H.J.; Kim, S.J.; Ivan, C.; et al. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat. Cell Biol. 2015, 17, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Molinie, B.; Daneshvar, K.; Pondick, J.V.; Wang, J.; Van Wittenberghe, N.; Xing, Y.; Giallourakis, C.C.; Mullen, A.C. Genome-wide maps of m6A circRNAs Identify widespread and Cell-Type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 2017, 20, 2262–2276. [Google Scholar] [CrossRef] [PubMed]
- Cantara, W.A.; Crain, P.F.; Rozenski, J.; McCloskey, J.A.; Harris, K.A.; Zhang, X.; Vendeix, F.A.P.; Fabris, D.; Agris, P.F. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011, 39, D195–D201. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Flores, M.A.; Meng, J.; Zhang, L.; Zhao, X.; Rao, M.K.; Chen, Y.; Huang, Y. MeT-DB: A database of transcriptome methylation in mammalian cells. Nucleic Acids Res. 2015, 43, D197–D203. [Google Scholar] [CrossRef] [PubMed]
- Xuan, J.-J.; Sun, W.-J.; Lin, P.-H.; Zhou, K.-R.; Liu, S.; Zheng, L.-L.; Qu, L.-H.; Yang, J.-H. RMBase v2.0: Deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res 2018, 46, D327–D334. [Google Scholar] [CrossRef] [PubMed]
- Crain, P.F.; McCloskey, J.A. The RNA modification database. Nucleic Acids Res. 1997, 25, 126–127. [Google Scholar] [CrossRef] [PubMed]
- Aduri, R.; Psciuk, B.T.; Saro, P.; Taniga, H.; Schlegel, H.B.; SantaLucia, J. AMBER force field parameters for the naturally occurring modified nucleosides in RNA. J. Chem. Theory Comput. 2007, 3, 1464–1475. [Google Scholar] [CrossRef] [PubMed]
- McDowell, S.E.; Špačková, N.; Šponer, J.; Walter, N.G. Molecular Dynamics simulations of RNA: An in silico single molecule approach. Biopolymers 2007, 85, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Dudek, M.; Trylska, J. Molecular Dynamics Simulations of l-RNA involving homo- and heterochiral complexes. J. Chem. Theory Comput. 2017, 13, 1244–1253. [Google Scholar] [CrossRef] [PubMed]
- Verona, M.D.; Verdolino, V.; Palazzesi, F.; Corradini, R. Focus on PNA Flexibility and RNA binding using molecular dynamics and metadynamics. Sci. Rep. 2017, 7, 42799. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Salmon-Divon, M.; Amariglio, N.; Rechavi, G. Transcriptome-wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 2013, 8, 176–189. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Cui, X.; Rao, M.K.; Chen, Y.; Huang, Y. Exome-based analysis for RNA epigenome sequencing data. Bioinformatics 2013, 29, 1565–1567. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Liu, T.; Qin, B.; Zhang, Y.; Liu, X.S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012, 7, 1728–1740. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.-J.; Li, J.-H.; Liu, S.; Wu, J.; Zhou, H.; Qu, L.-H.; Yang, J.-H. RMBase: A resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res. 2016, 44, D259–D265. [Google Scholar] [CrossRef] [PubMed]
- Vapnik, V.N. An overview of statistical learning theory. IEEE Trans. Neural Netw. 1999, 10, 988–999. [Google Scholar] [CrossRef] [PubMed]
- Kuksa, P.P.; Leung, Y.Y.; Vandivier, L.E.; Anderson, Z.; Gregory, B.D.; Wang, L.-S. In silico identification of rna modifications from high-throughput sequencing data using HAMR. Methods Mol. Biol. 2017, 1562, 211–229. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Lin, H. Recent Advances in Identification of RNA Modifications. Noncoding RNA 2016, 3. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Feng, P.; Ding, H.; Lin, H. PAI: Predicting adenosine to inosine editing sites by using pseudo nucleotide compositions. Sci. Rep. 2016, 6, 35123. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Feng, P.; Yang, H.; Ding, H.; Lin, H.; Chou, K.-C. iRNA-AI: Identifying the adenosine to inosine editing sites in RNA sequences. Oncotarget 2017, 8, 4208–4217. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Feng, P.; Tang, H.; Ding, H.; Lin, H. RAMPred: Identifying the N1-methyladenosine sites in eukaryotic transcriptomes. Sci. Rep. 2016, 6, 31080. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Feng, P.; Yang, H.; Ding, H.; Lin, H.; Chou, K.-C. iRNA-3typeA: Identifying three types of modification at RNA’s adenosine sites. Mol. Ther. Nucleic Acids 2018, 11, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.; Ding, H.; Yang, H.; Chen, W.; Lin, H.; Chou, K.-C. iRNA-PseColl: Identifying the occurrence sites of different RNA modifications by incorporating collective effects of nucleotides into PseKNC. Mol. Ther. Nucleic Acids 2017, 7, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.-R.; Jiang, S.-Y.; Xu, Z.-C.; Xiao, X.; Chou, K.-C. iRNAm5C-PseDNC: Identifying RNA 5-methylcytosine sites by incorporating physical-chemical properties into pseudo dinucleotide composition. Oncotarget 2017, 8, 41178–41188. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Feng, P.; Ding, H.; Lin, H.; Chou, K.-C. iRNA-Methyl: Identifying N6-methyladenosine sites using pseudo nucleotide composition. Anal. Biochem. 2015, 490, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Tran, H.; Liang, Z.; Lin, H.; Zhang, L. Identification and analysis of the N6-methyladenosine in the Saccharomyces cerevisiae transcriptome. Sci. Rep. 2015, 5, 13859. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Tang, H.; Lin, H. MethyRNA: A web server for identification of N6-methyladenosine sites. J. Biomol. Struct. Dyn. 2017, 35, 683–687. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zeng, P.; Li, Y.-H.; Zhang, Z.; Cui, Q. SRAMP: Prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016, 44, e91. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Xing, P.; Zou, Q. Detecting N6-methyladenosine sites from RNA transcriptomes using ensemble Support Vector Machines. Sci. Rep. 2017, 7, 40242. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-H.; Zhang, G.; Cui, Q. PPUS: A web server to predict PUS-specific pseudouridine sites. Bioinformatics 2015, 31, 3362–3364. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Tang, H.; Ye, J.; Lin, H.; Chou, K.-C. iRNA-PseU: Identifying RNA pseudouridine sites. Mol. Ther. Nucleic Acids 2016, 5, e332. [Google Scholar] [CrossRef] [PubMed]
- Panwar, B.; Raghava, G.P.S. Prediction of uridine modifications in tRNA sequences. BMC Bioinform. 2014, 15, 326. [Google Scholar] [CrossRef] [PubMed]
- Machnicka, M.A.; Dunin-Horkawicz, S.; de Crécy-Lagard, V.; Bujnicki, J.M. tRNAmodpred: A computational method for predicting posttranscriptional modifications in tRNAs. Methods 2016, 107, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.; Boue, S.; Izpisua Belmonte, J.C. Dedifferentiation, transdifferentiation and reprogramming: Three routes to regeneration. Nat. Rev. Mol. Cell Biol. 2011, 12, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Orlacchio, A.; Bernardi, G.; Orlacchio, A.; Martino, S. Stem cells and neurological diseases. Discov. Med. 2010, 9, 546–553. [Google Scholar] [PubMed]
- Orlacchio, A.; Bernardi, G.; Orlacchio, A.; Martino, S. Stem cells: An overview of the current status of therapies for central and peripheral nervous system diseases. Curr. Med. Chem. 2010, 17, 595–608. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, L.; Ayyash, M.; Novershtern, N.; Hanna, J.H. Dynamic stem cell states: Naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 2016, 17, 155–169. [Google Scholar] [CrossRef] [PubMed]
- Morena, F.; Armentano, I.; Montanucci, P.; Argentati, C.; Fortunati, E.; Montesano, S.; Bicchi, I.; Pescara, T.; Pennoni, I.; Mattioli, S.; et al. Design of a nanocomposite substrate inducing adult stem cell assembly and progression toward an Epiblast-like or Primitive Endoderm-like phenotype via mechanotransduction. Biomaterials 2017, 144, 211–229. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.; Smith, A. Naive and primed pluripotent states. Cell Stem Cell 2009, 4, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.J.; Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Doğan, A. Embryonic stem cells in development and regenerative medicine. Adv. Exp. Med. Biol. 2018. [Google Scholar] [CrossRef]
- Clevers, H.; Watt, F.M. Defining adult stem cells by function, not by phenotype. Annu. Rev. Biochem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Meissner, A.; Wernig, M.; Jaenisch, R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat. Biotechnol. 2007, 25, 1177–1181. [Google Scholar] [CrossRef] [PubMed]
- Martin, U. Therapeutic application of pluripotent stem cells: challenges and risks. Front. Med. 2017, 4, 229. [Google Scholar] [CrossRef] [PubMed]
- De Lázaro, I.; Kostarelos, K. Engineering cell fate for tissue regeneration by in vivo transdifferentiation. Stem Cell Rev. Rep. 2016, 12, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Hu, X.; Wang, J. Concise review: optimized strategies for stem cell-based therapy in myocardial repair: Clinical translatability and potential limitation. Stem Cells 2018. [Google Scholar] [CrossRef] [PubMed]
- Satthenapalli, V.R.; Lamberts, R.R.; Katare, R.G. Concise review: challenges in regenerating the diabetic Heart: A comprehensive review. Stem Cells 2017, 35, 2009–2026. [Google Scholar] [CrossRef] [PubMed]
- Ciccocioppo, R.; Cangemi, G.C.; Kruzliak, P.; Corazza, G.R. Concise Review: Cellular Therapies: The potential to regenerate and restore tolerance in immune-mediated intestinal diseases. Stem Cells 2016, 34, 1474–1486. [Google Scholar] [CrossRef] [PubMed]
- Martino, S.; Montesano, S.; di Girolamo, I.; Tiribuzi, R.; Di Gregorio, M.; Orlacchio, A.; Datti, A.; Calabresi, P.; Sarchielli, P.; Orlacchio, A. Expression of cathepsins S and D signals a distinctive biochemical trait in CD34+ hematopoietic stem cells of relapsing-remitting multiple sclerosis patients. Mult. Scler. 2013, 19, 1443–1453. [Google Scholar] [CrossRef] [PubMed]
- Sessa, M.; Lorioli, L.; Fumagalli, F.; Acquati, S.; Redaelli, D.; Baldoli, C.; Canale, S.; Lopez, I.D.; Morena, F.; Calabria, A.; et al. Lentiviral haemopoietic stem-cell gene therapy in early-onset metachromatic leukodystrophy: An ad-hoc analysis of a non-randomised, open-label, phase 1/2 trial. Lancet 2016, 388, 476–487. [Google Scholar] [CrossRef]
- Ricca, A.; Rufo, N.; Ungari, S.; Morena, F.; Martino, S.; Kulik, W.; Alberizzi, V.; Bolino, A.; Bianchi, F.; Del Carro, U.; et al. Combined gene/cell therapies provide long-term and pervasive rescue of multiple pathological symptoms in a murine model of globoid cell leukodystrophy. Hum. Mol. Genet. 2015, 24, 3372–3389. [Google Scholar] [CrossRef] [PubMed]
- Ungari, S.; Montepeloso, A.; Morena, F.; Cocchiarella, F.; Recchia, A.; Martino, S.; Gentner, B.; Naldini, L.; Biffi, A. Design of a regulated lentiviral vector for hematopoietic stem cell gene therapy of globoid cell leukodystrophy. Mol. Ther. Methods Clin. Dev. 2015, 2, 15038. [Google Scholar] [CrossRef] [PubMed]
- Biffi, A.; Montini, E.; Lorioli, L.; Cesani, M.; Fumagalli, F.; Plati, T.; Baldoli, C.; Martino, S.; Calabria, A.; Canale, S.; et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 2013, 341, 1233158. [Google Scholar] [CrossRef] [PubMed]
- Visigalli, I.; Ungari, S.; Martino, S.; Park, H.; Cesani, M.; Gentner, B.; Sergi Sergi, L.; Orlacchio, A.; Naldini, L.; Biffi, A. The galactocerebrosidase enzyme contributes to the maintenance of a functional hematopoietic stem cell niche. Blood 2010, 116, 1857–1866. [Google Scholar] [CrossRef] [PubMed]
- Morena, F.; di Girolamo, I.; Emiliani, C.; Gritti, A.; Biffi, A.; Martino, S. A new analytical bench assay for the determination of arylsulfatase a activity toward galactosyl-3-sulfate ceramide: Implication for metachromatic leukodystrophy diagnosis. Anal. Chem. 2014, 86, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Meneghini, V.; Frati, G.; Sala, D.; De Cicco, S.; Luciani, M.; Cavazzin, C.; Paulis, M.; Mentzen, W.; Morena, F.; Giannelli, S.; et al. Generation of human induced pluripotent stem cell-derived bona fide neural stem cells for ex vivo gene therapy of metachromatic leukodystrophy. Stem Cells Transl. Med. 2017, 6, 352–368. [Google Scholar] [CrossRef] [PubMed]
- Martino, S.; di Girolamo, I.; Cavazzin, C.; Tiribuzi, R.; Galli, R.; Rivaroli, A.; Valsecchi, M.; Sandhoff, K.; Sonnino, S.; Vescovi, A.; et al. Neural precursor cell cultures from GM2 gangliosidosis animal models recapitulate the biochemical and molecular hallmarks of the brain pathology. J. Neurochem. 2009, 109, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Calbi, V.; Fumagalli, F.; Consiglieri, G.; Penati, R.; Acquati, S.; Redaelli, D.; Attanasio, V.; Marcella, F.; Cicalese, M.P.; Migliavacca, M.; et al. Use of Defibrotide to help prevent post-transplant endothelial injury in a genetically predisposed infant with metachromatic leukodystrophy undergoing hematopoietic stem cell gene therapy. Bone Marrow Transplant. 2018. [Google Scholar] [CrossRef] [PubMed]
- Martino, S.; D’Angelo, F.; Armentano, I.; Kenny, J.M.; Orlacchio, A. Stem cell-biomaterial interactions for regenerative medicine. Biotechnol. Adv. 2012, 30, 338–351. [Google Scholar] [CrossRef] [PubMed]
- Morena, F.; Argentati, C.; Calzoni, E.; Cordellini, M.; Emiliani, C.; D’Angelo, F.; Martino, S. Ex-Vivo tissues engineering modeling for reconstructive surgery using human adult adipose stem cells and polymeric Nanostructured Matrix. Nanomaterials 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Martino, S.; D’Angelo, F.; Armentano, I.; Tiribuzi, R.; Pennacchi, M.; Dottori, M.; Mattioli, S.; Caraffa, A.; Cerulli, G.G.; Kenny, J.M.; et al. Hydrogenated amorphous carbon nanopatterned film designs drive human bone marrow mesenchymal stem cell cytoskeleton architecture. Tissue Eng. Part A 2009, 15, 3139–3149. [Google Scholar] [CrossRef] [PubMed]
- Tarpani, L.; Morena, F.; Gambucci, M.; Zampini, G.; Massaro, G.; Argentati, C.; Emiliani, C.; Martino, S.; Latterini, L. The influence of modified silica nanomaterials on adult stem cell culture. Nanomaterials 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Argentati, C.; Morena, F.; Montanucci, P.; Rallini, M.; Basta, G.; Calabrese, N.; Calafiore, R.; Cordellini, M.; Emiliani, C.; Armentano, I.; et al. Surface hydrophilicity of poly (l-Lactide) acid polymer film changes the human adult adipose stem cell architecture. Polymers 2018, 10, 140. [Google Scholar] [CrossRef]
- Islam, F.; Qiao, B.; Smith, R.A.; Gopalan, V.; Lam, A.K.-Y. Cancer stem cell: Fundamental experimental pathological concepts and updates. Exp. Mol. Pathol. 2015, 98, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Sampieri, K.; Fodde, R. Cancer stem cells and metastasis. Semin. Cancer Biol. 2012, 22, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Badve, S.; Nakshatri, H. Breast-cancer stem cells-beyond semantics. Lancet Oncol. 2012, 13, e43–e48. [Google Scholar] [CrossRef]
- Barrero, M.J.; Boué, S.; Izpisúa Belmonte, J.C. Epigenetic mechanisms that regulate cell identity. Cell Stem Cell 2010, 7, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Tee, W.-W.; Reinberg, D. Chromatin features and the epigenetic regulation of pluripotency states in ESCs. Development 2014, 141, 2376–2390. [Google Scholar] [CrossRef] [PubMed]
- Takata, N.; Eiraku, M. Stem cells and genome editing: Approaches to tissue regeneration and regenerative medicine. J. Hum. Genet. 2018, 63, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Paksa, A.; Rajagopal, J. The epigenetic basis of cellular plasticity. Curr. Opin. Cell Biol. 2018, 49, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.P.; Park, J.; Roh, T.-Y. Epigenetic regulation in cell reprogramming revealed by genome-wide analysis. Epigenomics 2011, 3, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Basanta-Sanchez, M.; Temple, S.; Ansari, S.A.; D’Amico, A.; Agris, P.F. Attomole quantification and global profile of RNA modifications: Epitranscriptome of human neural stem cells. Nucleic Acids Res. 2016, 44, e26. [Google Scholar] [CrossRef] [PubMed]
- Hattangadi, S.M.; Wong, P.; Zhang, L.; Flygare, J.; Lodish, H.F. From stem cell to red cell: Regulation of erythropoiesis at multiple levels by multiple proteins, RNAs, and chromatin modifications. Blood 2011, 118, 6258–6268. [Google Scholar] [CrossRef] [PubMed]
- Frye, M.; Blanco, S. Post-transcriptional modifications in development and stem cells. Development 2016, 143, 3871–3881. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Liu, X.; Zheng, X.; Huang, Y.; Chen, X. m6A RNA modification determines cell fate by regulating mRNA Degradation. Cell Reprogr. 2017, 19, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Aguilo, F.; Walsh, M.J. The N6-Methyladenosine RNA modification in pluripotency and reprogramming. Curr. Opin. Genet. Dev. 2017, 46, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.-L.; Wang, X.; An, R.; Cassin, J.; Vissers, C.; Liu, Y.; Liu, Y.; Xu, T.; Wang, X.; Wong, S.Z.H.; et al. Epitranscriptomic m6A Regulation of axon regeneration in the adult mammalian nervous system. Neuron 2018, 97, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Geula, S.; Moshitch-Moshkovitz, S.; Dominissini, D.; Mansour, A.A.; Kol, N.; Salmon-Divon, M.; Hershkovitz, V.; Peer, E.; Mor, N.; Manor, Y.S.; et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 2015, 347, 1002–1006. [Google Scholar] [CrossRef] [PubMed]
- Aguilo, F.; Zhang, F.; Sancho, A.; Fidalgo, M.; Di Cecilia, S.; Vashisht, A.; Lee, D.-F.; Chen, C.-H.; Rengasamy, M.; Andino, B.; et al. Coordination of m6A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell 2015, 17, 689–704. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Yue, M.; Wang, J.; Kumar, S.; Wechsler-Reya, R.J.; Zhang, Z.; Ogawa, Y.; Kellis, M.; Duester, G.; et al. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 2018, 21, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Katsaros, D.; de la Longrais, I.A.R.; Sochirca, O.; Yu, H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 2007, 67, 10117–10122. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Yang, Y.; Feng, G.-H.; Sun, B.-F.; Chen, J.-Q.; Li, Y.-F.; Chen, Y.-S.; Zhang, X.-X.; Wang, C.-X.; Jiang, L.-Y.; et al. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017, 27, 1100–1114. [Google Scholar] [CrossRef] [PubMed]
- Bertero, A.; Brown, S.; Madrigal, P.; Osnato, A.; Ortmann, D.; Yiangou, L.; Kadiwala, J.; Hubner, N.C.; de Los Mozos, I.R.; Sadée, C.; et al. The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency. Nature 2018, 555, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.J.; Molinie, B.; Wang, J.; Qu, K.; Zhang, J.; Li, L.; Bouley, D.M.; Lujan, E.; Haddad, B.; Daneshvar, K.; et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014, 15, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.-J.; Ringeling, F.R.; Vissers, C.; Jacob, F.; Pokrass, M.; Jimenez-Cyrus, D.; Su, Y.; Kim, N.-S.; Zhu, Y.; Zheng, L.; et al. Temporal control of mammalian cortical neurogenesis by m6A Methylation. Cell 2017, 171, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.; Pan, T. Cellular dynamics of RNA modification. Acc. Chem. Res. 2011, 44, 1380–1388. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.P.; Pickering, B.F.; Cheng, Y.; Zaccara, S.; Nguyen, D.; Minuesa, G.; Chou, T.; Chow, A.; Saletore, Y.; MacKay, M.; et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017, 23, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Kudou, K.; Komatsu, T.; Nogami, J.; Maehara, K.; Harada, A.; Saeki, H.; Oki, E.; Maehara, Y.; Ohkawa, Y. The requirement of Mettl3-promoted MyoD mRNA maintenance in proliferative myoblasts for skeletal muscle differentiation. Open Biol. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chen, Y.; Sun, B.; Wang, L.; Yang, Y.; Ma, D.; Lv, J.; Heng, J.; Ding, Y.; Xue, Y.; et al. m6A modulates haematopoietic stem and progenitor cell specification. Nature 2017, 549, 273–276. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, S.; Yuan, Q. N6-Methyladenosine Methyltransferases and Demethylases: New regulators of stem cell pluripotency and differentiation. Stem Cells Dev. 2016, 25, 1050–1059. [Google Scholar] [CrossRef] [PubMed]
- Weng, H.; Huang, H.; Wu, H.; Qin, X.; Zhao, B.S.; Dong, L.; Shi, H.; Skibbe, J.; Shen, C.; Hu, C.; et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell 2018, 22, 191.e9–205.e9. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, I.; Tzelepis, K.; Pandolfini, L.; Shi, J.; Millán-Zambrano, G.; Robson, S.C.; Aspris, D.; Migliori, V.; Bannister, A.J.; Han, N.; et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 2017, 552, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Cui, Q.; Shi, H.; Ye, P.; Li, L.; Qu, Q.; Sun, G.; Sun, G.; Lu, Z.; Huang, Y.; Yang, C.-G.; et al. m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017, 18, 2622–2634. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, B.S.; Zhou, A.; Lin, K.; Zheng, S.; Lu, Z.; Chen, Y.; Sulman, E.P.; Xie, K.; Bögler, O.; et al. m6A Demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 2017, 31, 591.e6–606.e6. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Choe, J.; Du, P.; Triboulet, R.; Gregory, R.I. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 2016, 62, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Crews, L.A.; Holm, F.; Jamieson, C.H.M. RNA editing-dependent epitranscriptome diversity in cancer stem cells. Nat. Rev. Cancer 2017, 17, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhi, W.I.; Lu, H.; Samanta, D.; Chen, I.; Gabrielson, E.; Semenza, G.L. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget 2016, 7, 64527–64542. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Kurowski, A.; Nichols, J.; Watt, F.M.; Benitah, S.A.; Frye, M. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 2011, 7, e1002403. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Bandiera, R.; Popis, M.; Hussain, S.; Lombard, P.; Aleksic, J.; Sajini, A.; Tanna, H.; Cortés-Garrido, R.; Gkatza, N.; et al. Stem cell function and stress response are controlled by protein synthesis. Nature 2016, 534, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Amort, T.; Rieder, D.; Wille, A.; Khokhlova-Cubberley, D.; Riml, C.; Trixl, L.; Jia, X.-Y.; Micura, R.; Lusser, A. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017, 18, 1. [Google Scholar] [CrossRef] [PubMed]
- Fong, Y.W.; Ho, J.J.; Inouye, C.; Tjian, R. The dyskerin ribonucleoprotein complex as an OCT4/SOX2 coactivator in embryonic stem cells. eLife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Bellodi, C.; McMahon, M.; Contreras, A.; Juliano, D.; Kopmar, N.; Nakamura, T.; Maltby, D.; Burlingame, A.; Savage, S.A.; Shimamura, A.; et al. H/ACA small RNA dysfunctions in disease reveal key roles for noncoding RNA modifications in hematopoietic stem cell differentiation. Cell Rep. 2013, 3, 1493–1502. [Google Scholar] [CrossRef] [PubMed]
- Guzzi, N.; Cieśla, M.; Ngoc, P.C.T.; Lang, S.; Arora, S.; Dimitriou, M.; Pimková, K.; Sommarin, M.N.E.; Munita, R.; Lubas, M.; et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 2018, 173, 1204.e26–1216.e26. [Google Scholar] [CrossRef] [PubMed]
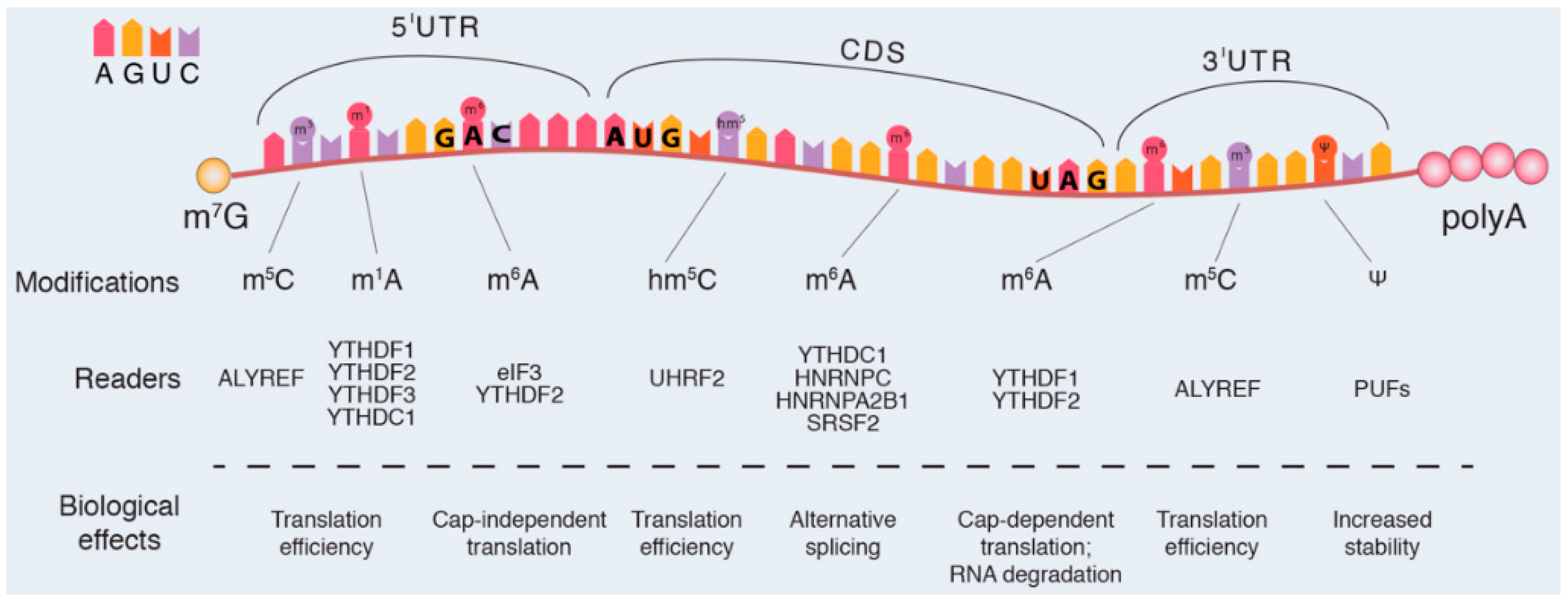
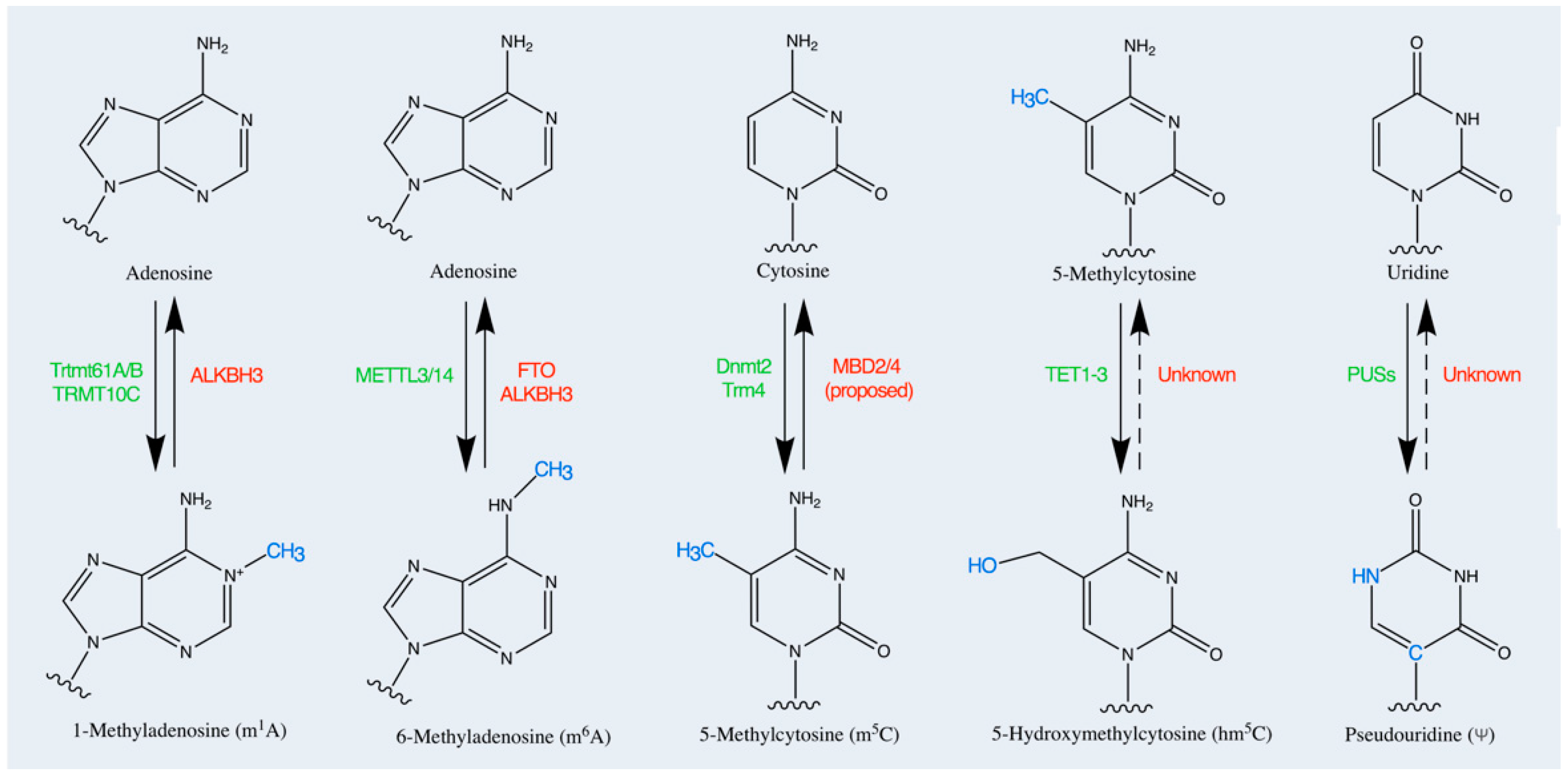
| RNA Species | Post-Transcriptional Modifications | RNA Characteristics | Ref. |
|---|---|---|---|
| mRNA | Cap 5′-end Poly-A tail 3′-end Splicing N6-methyladenosine N1-methyladenosine 5-methylcytidine Pseudouridine Adenosine to Inosine editing | Structure: The mature messenger RNA (mRNA) is a single linear polymer of ribonucleotides containing the 3′-UTR and 5′-UTR regions that border the coding sequence Function: mRNA carries the genetic information from the DNA to the ribosome where it is translated into a polymer of amino acids (protein) | [19,20,21,24,25,27,28,33] |
| tRNA | Anticodon loop: inosine, queuosine, 5-methylcytosine, 5-methoxycarbonylmethyl-2-thiouridine, threonyl-carbamoyl-adenosine and wybutosine D-loop: dihydrouridine T-loop: pseudouridine | Structure: The mature transfer RNA (tRNA) is a polymer of ribonucleotides with a characteristic three-dimensional structure. Here, the anticodon loop has the three pairing nucleotides with the codon in the mRNA; the 3′-end has the CCA sequence that allows the binding of the amino acid Function: The tRNA is the translator of the nucleotidic language to aminoacidic language | [19,20,28,29,91,92,99,106] |
| rRNA | 2′-O-methylation Uridine-to-Pseudouridine isomerization Pseudouridine | Structure: The mature ribosomal RNA (rRNA) is a polymer of ribonucleotides consisting of several hairpin clusters Function: rRNA together with riboproteins compose the ribosome. Here, rRNAs have a direct role in the formation of the peptic bond between two amino acids | [19,20,28,127,128] |
| lncRNA | N6-methyladenosine N1-methyladenosine 5-methylcytidine Pseudouridine | Structure: The mature long noncoding-RNA (lncRNA) is a polymer of ribonucleotides with secondary structure Function: lncRNA has a significant role in different biological functions such as chromatin modifications, post-transcriptional regulation | [19,20,28,72,98,99,104] |
| miRNA | Pseudouridine 5-methylcytidine Adenosine to Inosine editing | Structure: The microRNA (miRNA) is a small oligonucleotide containing about 22 nucleotides with a single hairpin structure Function: The miRNA silencing at the post-transcriptional level of regulation of gene expression | [19,20,28,98,105,137] |
| circRNA | Pseudouridine N6-methyladenosine | Structure: The circleRNA (circRNA) is a polymer of ribonucleotides with circular conformation, as the 3′-end and 5′-ends have been joined together Function: The circRNA function as a sponge of miRNA, therefore have a key role in the regulation of gene expression | [19,20,28,98,138] |
| Tools | Source | Prediction of Modifications | Description | Ref. |
|---|---|---|---|---|
| HAMR | http://www.lisanwanglab.org/hamr/ | m1A, m6A, A-to-I, Pseudouridine (Ψ), Dihydrouridine (D) | HAMR (High-throughput Annotation of Modified Ribonucleotides) is a web application that allows to detect and classify modified nucleotides in RNA-seq data | [153] |
| PAI | http://lin.uestc.edu.cn/server/PAI/ | A-to-I | Prediction of Adenosine to Inosine sitesby using pseudo nucleotide compositions | [155] |
| iRNA-AI | http://lin.uestc.edu.cn/server/iRNA-AI/ | A-to-I | Identification of Adenosine to Inosine editing sites | [156] |
| RAMPred | http://lin-group.cn/server/RAMPred/ | m1A | Identification of the N1-methyladenosine sites in eukaryotic transcriptomes | [157] |
| iRNA-3typeA | http://lin-group.cn/server/iRNA-3typeA/ | m1A, m6A, A-to-I | Identification of 3-types of modification at RNA’s Adenosine sites | [158] |
| iRNA-PseColl | http://lin.uestc.edu.cn/server/iRNA-PseColl/ | m1A, m6A, m5C | A seamless platform for identifying the occurrence sites of different RNA modifications by incorporating collective effects of nucleotides into PseKNC | [159] |
| iRNAm5C-PseDNC | http://www.jci-bioinfo.cn/iRNAm5C-PseDNC/ | m5C | Identification of RNA 5-methylcytosine sites by incorporation physical-chemical properties into pseudo dinucleotide composition | [160] |
| iRNA-Methyl | http://lin.uestc.edu.cn/server/iRNA-Methyl/ | m6A | Identification of N6-methyladenosine sites using pseudo nucleotide composition | [161] |
| m6Apred | http://lin.uestc.edu.cn/server/m6Apred/ | m6A | Identification and analysis of the N6-methyladenosine in Saccharomyces cerevisiae transcriptome | [162] |
| MethyRNA | http://lin.uestc.edu.cn/server/methyrna/ | m6A | A sequence-based tool for the identification of N6-methyladenosine sites | [163] |
| SRAMP | http://www.cuilab.cn/sramp/ | m6A | SRAMP (sequence-based RNA Adenosine methylation site predictor), a useful tool to predict m6A modification sites on the RNA sequences of interests | [164] |
| RAM-ESVM | http://server.malab.cn/RAM-ESVM/ | m6A | Identification of M6A Sites in theS. Cerevisiae Transcriptome | [165] |
| PPUS | http://lyh.pkmu.cn/ppus/ | Pseudouridine (Ψ) | PPUS is an online tool to predict Pseudouridine sites recognized by Pseudouridine synthase in RNA | [166] |
| iRNA-PseU | http://lin.uestc.edu.cn/server/iRNA-PseU/ | Pseudouridine (Ψ) | Identification of RNA Pseudouridine sites | [167] |
| tRNAMOD | http://crdd.osdd.net/raghava/trnamod/ | Pseudouridine (Ψ), Dihydrouridine (D) | Web-server for the prediction of transfer RNA (tRNA) modifications | [168] |
| Stem Cell Types | Origin | Properties | Ref. |
|---|---|---|---|
| Naïve stem cells | Zygotic stage of the mammalian embryo, immediately after the maternal redetermination. Specifically, they originate from the preimplantation epiblast | Self-renewal Pluripotency Unrestricted stem cells | [173,174,175] |
| Primed stem cells | Zygotic stage of the mammalian embryo, immediately after the maternal redetermination. Specifically, they originate from naïve stem cells that enter into a lineage commitment process | Self-renewal Pluripotency More lineage restricted stem cells compared to naive stem cells | [173,174,175] |
| Embryonic Stem cells (ESCs) | ESCs originate from the inner mass of the blastocyst | Self-renewal Pluripotent: multi-lineage differentiation through either asymmetric or symmetric division Generation of all 254 cell types originating adult tissue Generation of mouse chimeras Replacement of damaged cells with newly differentiated progenies if transplanted in degenerated tissues/organs | [170,177,196,206,207] |
| Adult stem cells (ASCs) | ASCs are created during ontogeny and persist in the adult tissues/organs within the niche | Self-renewal Multipotent: multi-lineage differentiation through either asymmetric or symmetric division Maintenance of the tissue homeostasis in the physiological condition Replacement of damaged cells with newly differentiated progenies if transplanted in degenerated tissues/organs | [170,178,196,206,207] |
| Induced pluripotentStem cells (iPSCs) | iPSCs originate in vitro from somatic differentiated cells after transduction with cMyc, Klf-4, Oct-3/4 , Sox-2 genes | Self-renewal Pluripotent Generation of patient-specific stem cells Generation of mouse chimeras Replacement of damaged cells with newly differentiated progenies if transplanted in degenerated tissues/organs | [170,179,180,196,206,207] |
| Cancer stem cells (CSCs) | CSCs originate from: (i) Malignant transformation of normal stem cells (ii) de-differentiation of cancer cell | Self-renewal through either asymmetric or symmetric division Multipotent: multi-lineage differentiation. Key role in predicting the biological aggressiveness of the cancer | [170,201,202,206,207] |
| Stem Cell Types | RNA Modification | Presence/Absence Increase/Decrease | Effect | Ref. |
|---|---|---|---|---|
| Naïve vs. primed | m6A in mRNAs | Presence | Molecular switches for differentiation and generation of EpiSCs | [215,216] |
| Naïve vs. primed | m6A in mRNAs | Absence | Molecular switches for reating the naïve status | [215,216] |
| Naïve/ESCs | pseudouridylation of tRNA | Presence | Stem cell commitment during the first stage of embryogenesis | [242,243] |
| ESCs | m6A in mRNAs | Presence | Critical steps for keeping ESCs in a stemness status | [213,217,218] |
| ESCs | m6A in mRNAs | Absence | Critical steps for keeping ESCs in a stemness status | [220,221] |
| ESCs | m5C in mRNAs | Increase | Critical steps for keeping ESCs in a stemness status | [241] |
| ESCs | m5C in mt-tRNAs | Presence | Regulator of ESCs fate | [124] |
| ASCs | m6A in mRNAs | Presence | Activation of differentiation process | [224] |
| ASCs | m6A in mRNAs | Decrease | Hamper the HSCs development | [213,227,228,229] |
| ASCs | m6A in mRNAs | Decrease | Myeloid differentiation of HSCs | [230] |
| ASCs | m5C in mRNAs | Presence | Balance of epidermis stem cell self-renewal and differentiation processes | [237,238] |
| CSCs | m6A in mRNAs | Increase | Acute myeloid leukemia | [230] |
| CSCs | m6A in mRNAs | Absence/Decrease | Growth and self-renewal of human glioblastoma stem cells | [232,234] |
| CSCs | m6A in mRNAs | Decrease | Progression of human lung cancer | [235] |
| CSCs | m6A in mRNAs | Decrease | Demethylation on NANOG mRNA in breast cancer stem cells in response to hypoxia | [81] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morena, F.; Argentati, C.; Bazzucchi, M.; Emiliani, C.; Martino, S. Above the Epitranscriptome: RNA Modifications and Stem Cell Identity. Genes 2018, 9, 329. https://doi.org/10.3390/genes9070329
Morena F, Argentati C, Bazzucchi M, Emiliani C, Martino S. Above the Epitranscriptome: RNA Modifications and Stem Cell Identity. Genes. 2018; 9(7):329. https://doi.org/10.3390/genes9070329
Chicago/Turabian StyleMorena, Francesco, Chiara Argentati, Martina Bazzucchi, Carla Emiliani, and Sabata Martino. 2018. "Above the Epitranscriptome: RNA Modifications and Stem Cell Identity" Genes 9, no. 7: 329. https://doi.org/10.3390/genes9070329
APA StyleMorena, F., Argentati, C., Bazzucchi, M., Emiliani, C., & Martino, S. (2018). Above the Epitranscriptome: RNA Modifications and Stem Cell Identity. Genes, 9(7), 329. https://doi.org/10.3390/genes9070329






