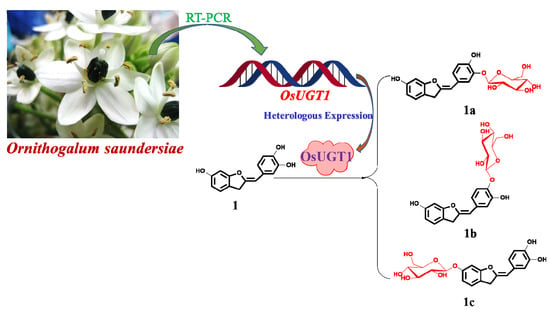
Transcriptome-Wide Identification of an Aurone Glycosyltransferase with Glycosidase Activity from Ornithogalum saundersiae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Retrieval of Unigenes encoding GT from Transcriptome Database of O. saundersiae
2.3. cDNA Isolation of the Gene Encoding OsUGT1 from O. saundersiae
2.4. Bioinformatics Analyses of OsUGT1
2.5. Heterologous Expression and Purification of OsUGT1
2.6. OsUGT1-Catalyzed Glycosylation Assays
2.7. Enzymatic Characterization
2.8. OsUGT1-Mediated Transglucosylation Assay
2.9. OsUGT1-Assisted Hydrolysis Assay
3. Results
3.1. cDNA Isolation of OsUGT1 from O. saundersiae
3.2. OsUGT1 was Predicted to Encode a Flavonoid Glycosyltransferase
3.3. OsUGT1 was Clustered Phylogenetically to a New Group of Flavonoid Glycosyltransferases
3.4. Heterologous Expression and Purification of OsUGT1
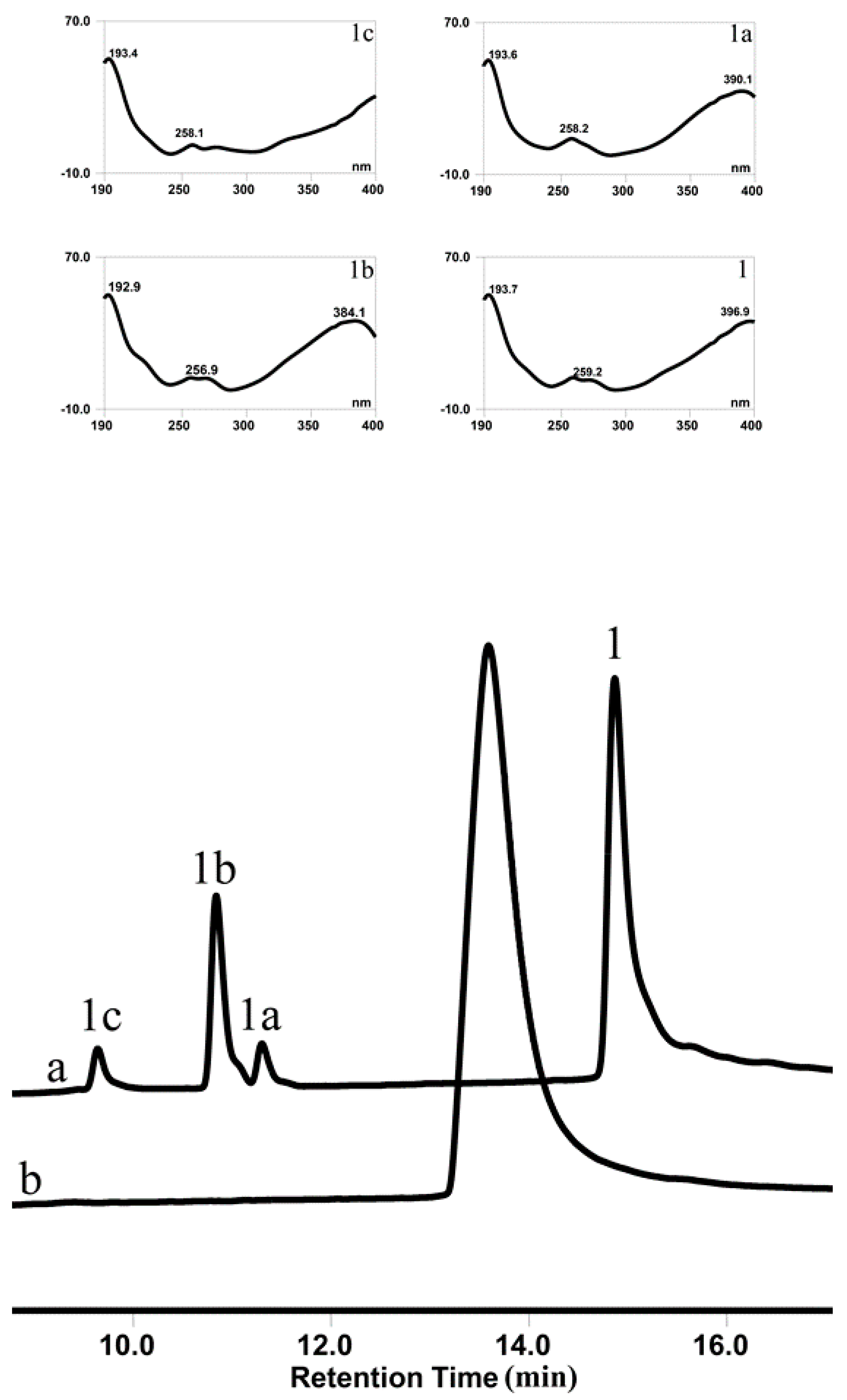
3.5. OsUGT1 is an Aurone Glycosyltransferase
3.6. Substrate Specificity of OsUGT1-Catalyzed Glycosylation
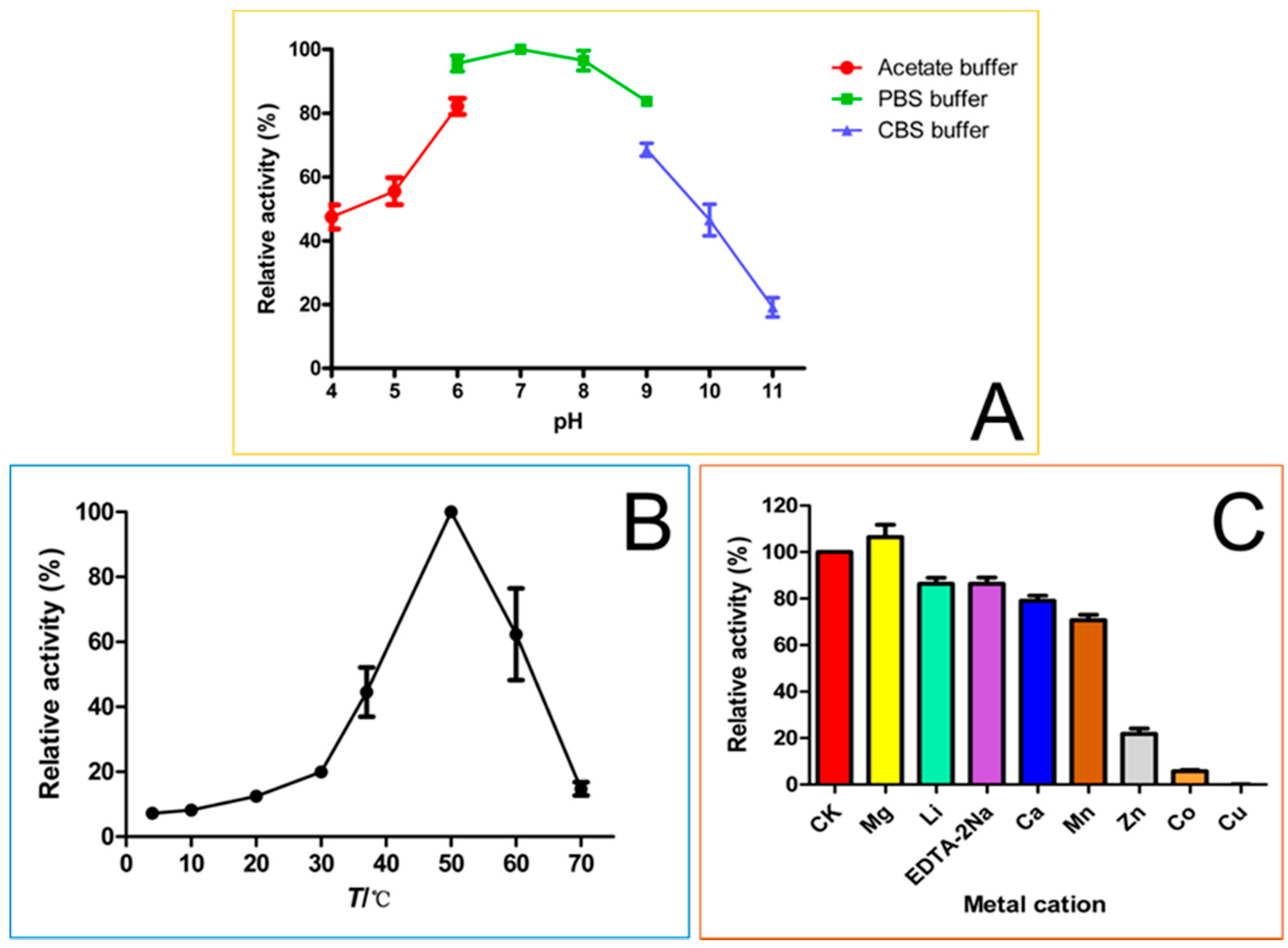
3.7. Enzymatic Properties of OsUGT1-Catalyzed Glycosylation
3.8. OsUGT1-Catalyzed Transglucosylation Action
3.9. OsUGT1-Catalyzed Hydrolysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Boucherle, B.; Peuchmaur, M.; Boumendjel, A.; Haudecoeur, R. Occurrences, biosynthesis and properties of aurones as high-end evolutionary products. Phytochemistry 2017, 142, 92–111. [Google Scholar] [CrossRef] [PubMed]
- Sutton, C.L.; Taylor, Z.E.; Farone, M.B.; Handy, S.T. Antifungal activity of substituted aurones. Bioorg. Med. Chem. Lett. 2017, 27, 901–903. [Google Scholar] [CrossRef] [PubMed]
- Haudecoeur, R.; Boumendjel, A. Recent advances in the medicinal chemistry of aurones. Curr. Med. Chem. 2012, 19, 2861–2875. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Natarajan, S.; Park, J.-H.; Lee, D.-Y.; Cho, J.-G.; Kim, G.-S.; Jeon, Y.-J.; Yeon, S.-W.; Yang, D.-C.; Baek, N.-I. Potential neuroprotective flavonoid-based inhibitors of CDK5/p25 from Rhus parviflora. Bioorg. Med. Chem. Lett. 2013, 23, 5150–5154. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hu, D.; Lam, S.; Ge, L.; Wu, D.; Zhao, J.; Long, Z.; Yang, W.; Fan, B.; Li, S. Comparison of antioxidant activities of different parts from snow chrysanthemum (Coreopsis tinctoria Nutt.) and identification of their natural antioxidants using high performance liquid chromatography coupled with diode array detection and mass spectrometry and 2, 2′-azinobis (3-ethylbenzthiazoline-sulfonic acid) diammonium salt-based assay. J. Chromatogr. A 2016, 1428, 134–142. [Google Scholar] [PubMed]
- Chand, K.; Rajeshwari; Hiremathad, A.; Singh, M.; Santos, M.A.; Keri, R.S. A review on antioxidant potential of bioactive heterocycle benzofuran: Natural and synthetic derivatives. Pharmacol. Rep. 2017, 69, 281–295. [Google Scholar] [CrossRef] [PubMed]
- Kondratyuk, T.P.; Pezzuto, J.M. Natural product polyphenols of relevance to human health. Pharm. Biol. 2004, 42, 46–63. [Google Scholar] [CrossRef]
- Westenburg, H.E.; Lee, K.-J.; Lee, S.K.; Fong, H.H.; van Breemen, R.B.; Pezzuto, J.M.; Kinghorn, A.D. Activity-guided isolation of antioxidative constituents of Cotinus coggygria. J. Nat. Prod. 2000, 63, 1696–1698. [Google Scholar] [CrossRef]
- Kumaresan, R. A comparative study on the antioxidant properties of bractein and cernuoside by the DFT method. Monatsh. Chem. 2013, 144, 1513–1524. [Google Scholar]
- Li, N.; Meng, D.; Pan, Y.; Cui, Q.; Li, G.; Ni, H.; Sun, Y.; Qing, D.; Jia, X.; Pan, Y.; et al. Anti-neuroinflammatory and NQO1 inducing activity of natural phytochemicals from Coreopsis tinctoria. J. Funct. Foods 2015, 17, 837–846. [Google Scholar] [CrossRef]
- Kayser, O.; Kiderlen, A.; Folkens, U.; Kolodziej, H. In vitro leishmanicidal activity of aurones. Planta Med. 1999, 65, 316–319. [Google Scholar] [CrossRef] [PubMed]
- Halbwirth, H.; Wimmer, G.; Wurst, F.; Forkmann, G.; Stich, K. Enzymatic glucosylation of 4-deoxyaurones and 6′-deoxychalcones with enzyme extracts of Coreopsis grandiflora, Nutt. I. Plant. Sci. 1997, 122, 125–131. [Google Scholar] [CrossRef]
- Sakakibara, K.; Fukui, Y.; Sakakibara, K.; Fukui, Y.; Tanaka, Y.; Kusumi, T.; Yoshikawa, T. Genes Encoding Protein Having Activity of Transferring Sugar onto Aurone. U.S. Patent No. 6770747, 3 August 2004. [Google Scholar]
- Yin, S.; Kong, J.-Q. Transcriptome-guided discovery and functional characterization of two UDP-sugar 4-epimerase families involved in the biosynthesis of anti-tumor polysaccharides in Ornithogalum caudatum. RSC Adv. 2016, 6, 37370–37384. [Google Scholar] [CrossRef]
- Yin, S.; Kong, J.-Q. Transcriptome-guided gene isolation and functional characterization of UDP-xylose synthase and UDP-d-apiose/UDP-d-xylose synthase families from Ornithogalum caudatum Ait. Plant. Cell Rep. 2016, 35, 2403–2421. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Yin, S.; Liu, M.; Kong, J.-Q. Isolation and characterization of a multifunctional flavonoid glycosyltransferase from Ornithogalum caudatum with glycosidase activity. Sci. Rep. 2018, 8, 5886. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Kong, J.-Q. The enzymatic biosynthesis of acylated steroidal glycosides and their cytotoxic activity. Acta Pharm. Sin. B 2018. [Google Scholar] [CrossRef]
- Li, L.-N.; Kong, J.-Q. Transcriptome-wide identification of sucrose synthase genes in Ornithogalum caudatum. RSC Adv. 2016, 6, 18778–18792. [Google Scholar] [CrossRef]
- Guo, L.; Chen, X.; Li, L.-N.; Tang, W.; Pan, Y.-T.; Kong, J.-Q. Transcriptome-enabled discovery and functional characterization of enzymes related to (2S)-pinocembrin biosynthesis from Ornithogalum caudatum and their application for metabolic engineering. Microb. Cell Fact. 2016, 15, 27. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Kubo, S.; Mimaki, Y.; Terao, M.; Sashida, Y.; Nikaido, T.; Ohmoto, T. Cholestane glycosides with potent cytostatic activities on various tumor cells from Ornithogalum saundersiae bulbs. Bioorg. Med. Chem. Lett. 1997, 7, 633–636. [Google Scholar]
- Kubo, S.; Mimaki, Y.; Terao, M.; Sashida, Y.; Nikaido, T.; Ohmoto, T. Acylated cholestane glycosides from the bulbs of Ornithogalum saundersiae. Phytochemistry 1992, 31, 3969–3973. [Google Scholar] [CrossRef]
- Vogt, T.; Jones, P. Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 2000, 5, 380–386. [Google Scholar] [CrossRef]
- Wang, L.; Han, W.; Xie, C.; Hou, J.; Fang, Q.; Gu, J.; Wang, P.G.; Cheng, J. Comparing the acceptor promiscuity of a Rosa hybrida glucosyltransferase RhGT1 and an engineered microbial glucosyltransferase OleD(PSA) toward a small flavonoid library. Carbohydr. Res. 2013, 368, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Liu, S.; Cheng, C.; Guo, R.; Chen, Y.; Xie, L.; Mao, Y.; Lin, Y.; Zhang, Z.; Lai, Z. Cloning and expression analysis of betalain biosynthesis genes in Amaranthus tricolor. Biotechnol. Lett. 2016, 38, 723–729. [Google Scholar] [CrossRef] [PubMed]
- Kamel, M.S.; Mohamed, K.M.; Hassanean, H.A.; Ohtani, K.; Kasai, R.; Yamasaki, K. Acylated flavonoid glycosides from Bassia muricata. Phytochemistry 2001, 57, 1259–1262. [Google Scholar] [CrossRef]
- Pandey, R.P. Diversifying natural products with promiscuous glycosyltransferase enzymes via a sustainable microbial fermentation approach. Front. Chem. 2017, 5, 110. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, R.; Shimabayashi, H.; Moriwaki, M. Enzymatic production of highly soluble myricitrin glycosides using β-galactosidase. Biosci. Biotechnol. Biochem. 2006, 70, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, H.; Lu, L.; Li, Z.; Xu, X.; Xiao, M. Purification and characterization of a novel β-galactosidase with transglycosylation activity from Bacillus megaterium 2-37-4-1. Appl. Biochem. Biotechnol. 2009, 158, 192–199. [Google Scholar] [CrossRef] [PubMed]




| Position | 1a | 1b | 1c |
|---|---|---|---|
| 4 | 7.60, d (8.5) | 7.62, d (8.5) | 7.70, d (8.5) |
| 5 | 6.71, m | 6.73, dd (8.5, 1.9) | 6.92, dd (8.5, 2.0) |
| 7 | 6.76, d (1.9) | 6.80, d (1.9) | 7.14, d (2.0) |
| 10 | 6.71, m | 6.69, s | 6.71, s |
| 2′ | 7.74, d (1.9) | 7.49, d (2.0) | 7.44, d (2.0) |
| 5′ | 6.94, d (8.3) | 7.20, d (8.6) | 6.86, d (8.2) |
| 6′ | 7.51, dd (8.3, 1.9) | 7.37, dd (8.6, 2.0) | 7.31, dd (8.2, 2.0) |
| Glc | Glc | Glc | |
| 1″ 1‴ | 4.82, d (7.4) | 4.84, d (7.4) | 5.16, d (7.4) |
| H of sugar | 3.0–3.8 | 3.0–3.8 | 3.0–3.8 |
| Position | 1a | 1b | 1c |
|---|---|---|---|
| 2 | 145.9, C | 146.9, C | 145.9, C |
| 3 | 181.3, C | 181.8, C | 182.0, C |
| 4 | 125.8, CH | 126.3, CH | 125.8, CH |
| 5 | 113.0, CH | 113.4, CH | 114.0, CH |
| 6 | 166.4, C | 166.9, C | 165.1, C |
| 7 | 98.6, CH | 99.0, CH | 99.8, CH |
| 8 | 167.7, C | 168.2, C | 167.5, C |
| 9 | 113.0, C | 113.5, C | 115.8, C |
| 10 | 111.5, CH | 111.4, CH | 113.4, CH |
| 1′ | 127.2, C | 126.9, C | 125.2, C |
| 2′ | 118.7, CH | 118.4, CH | 118.8, CH |
| 3′ | 145.6, C | 147.2, C | 146.1, C |
| 4′ | 148.8, C | 147.4, C | 148.8, C |
| 5′ | 116.5, CH | 116.5, CH | 116.5, CH |
| 6′ | 123.7, CH | 124.3, CH | 123.7, CH |
| Glc | Glc | Glc | |
| 1″ | 101.9, CH | 101.8, CH | 100.3, CH |
| 2″ | 73.3, CH | 73.7, CH | 73.6, CH |
| 3″ | 76.0, CH | 76.3, CH | 76.8, CH |
| 4″ | 69.6, CH | 70.3, CH | 70.0, CH |
| 5″ | 77.1, CH | 77.7, CH | 77.6, CH |
| 6″ | 60.6, CH2 | 61.2, CH2 | 61.1, CH2 |
| Substrate | Km (mM) | Vmax (mM/h) |
|---|---|---|
| sulfuretin | 0.159 ± 0.011 | 0.208 ± 0.004 |
| 7,8-dihydroxyflavone | 0.713 ± 0.069 | 1.695 ± 0.085 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, S.; Liu, M.; Yang, Y.; He, J.-M.; Wang, Y.-N.; Kong, J.-Q. Transcriptome-Wide Identification of an Aurone Glycosyltransferase with Glycosidase Activity from Ornithogalum saundersiae. Genes 2018, 9, 327. https://doi.org/10.3390/genes9070327
Yuan S, Liu M, Yang Y, He J-M, Wang Y-N, Kong J-Q. Transcriptome-Wide Identification of an Aurone Glycosyltransferase with Glycosidase Activity from Ornithogalum saundersiae. Genes. 2018; 9(7):327. https://doi.org/10.3390/genes9070327
Chicago/Turabian StyleYuan, Shuai, Ming Liu, Yan Yang, Jiu-Ming He, Ya-Nan Wang, and Jian-Qiang Kong. 2018. "Transcriptome-Wide Identification of an Aurone Glycosyltransferase with Glycosidase Activity from Ornithogalum saundersiae" Genes 9, no. 7: 327. https://doi.org/10.3390/genes9070327
APA StyleYuan, S., Liu, M., Yang, Y., He, J.-M., Wang, Y.-N., & Kong, J.-Q. (2018). Transcriptome-Wide Identification of an Aurone Glycosyltransferase with Glycosidase Activity from Ornithogalum saundersiae. Genes, 9(7), 327. https://doi.org/10.3390/genes9070327