Reorganization of the Y Chromosomes Enhances Divergence in Israeli Mole Rats Nannospalax ehrenbergi (Spalacidae, Rodentia): Comparative Analysis of Meiotic and Mitotic Chromosomes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Preparation of Mitotic Chromosomes and Fluorescent In-Situ Hybridization
2.3. Preparation of Meiotic Chromosomes
2.4. Antibodies and Immunostaining
3. Results
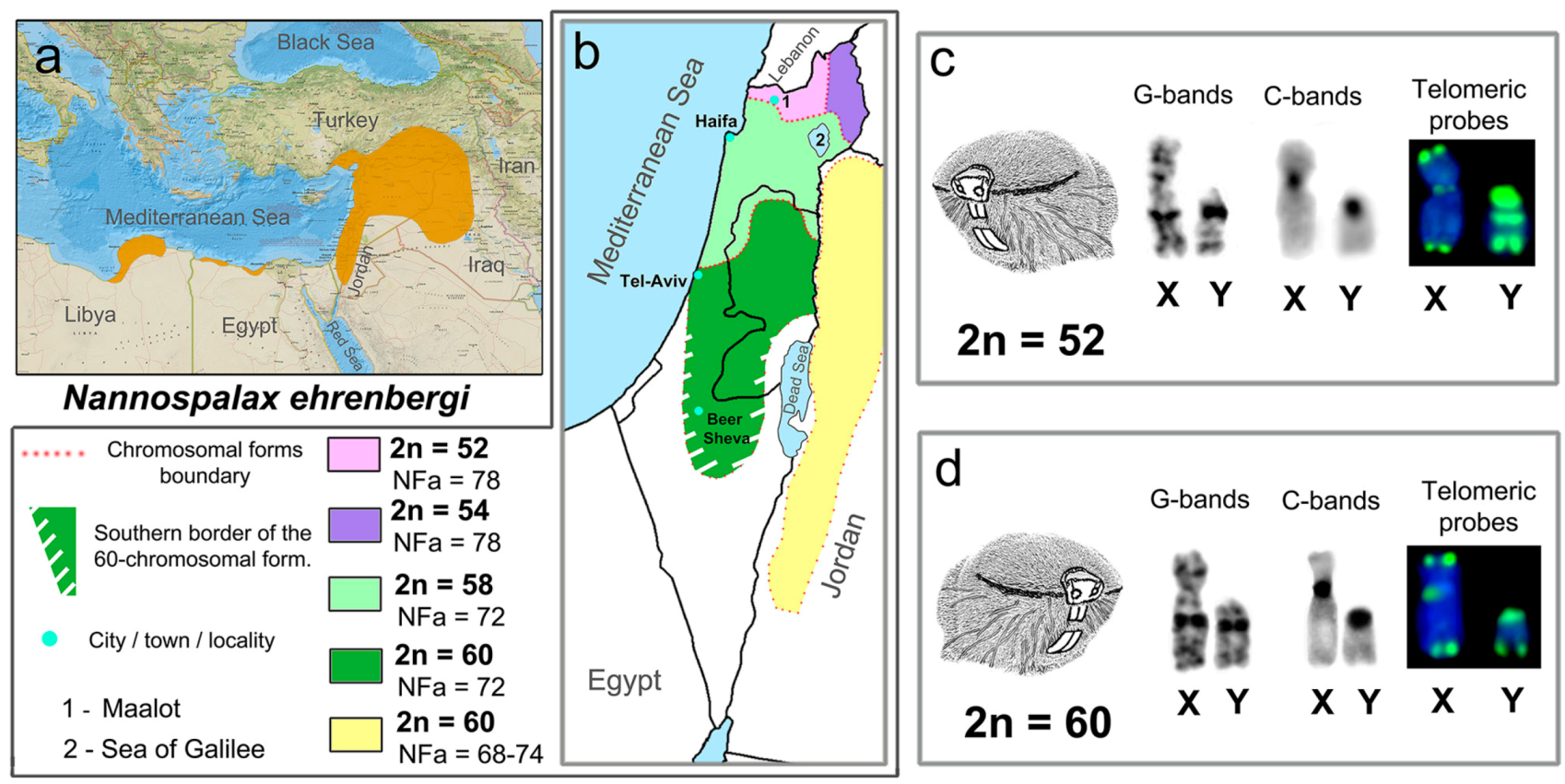
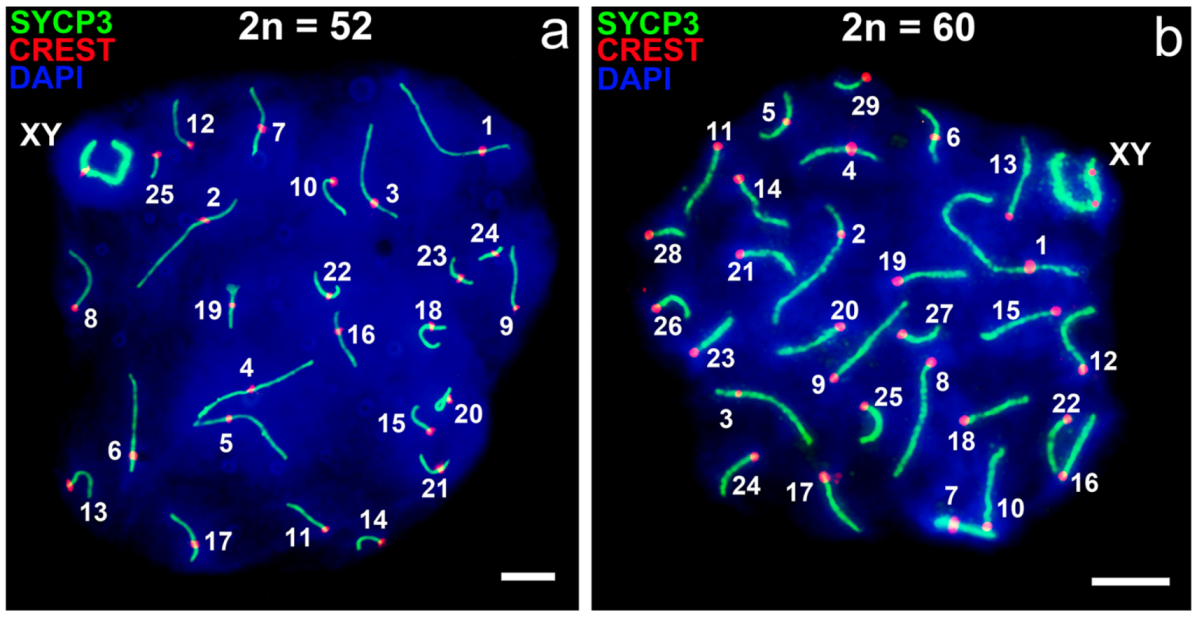
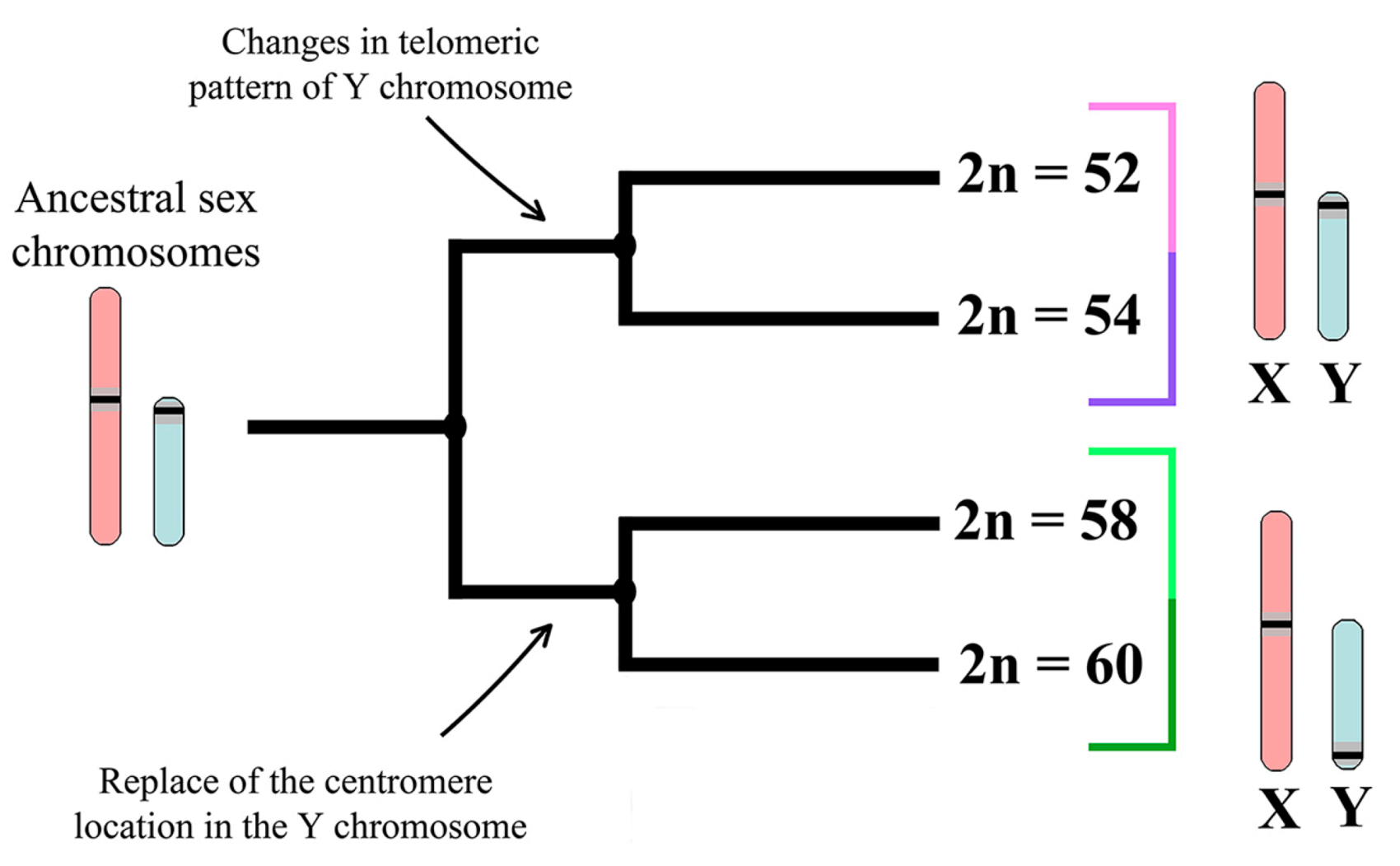
3.1. Mitotic and Meiotic Chromosomes of 52- and 60- Chromosome Mole Rats
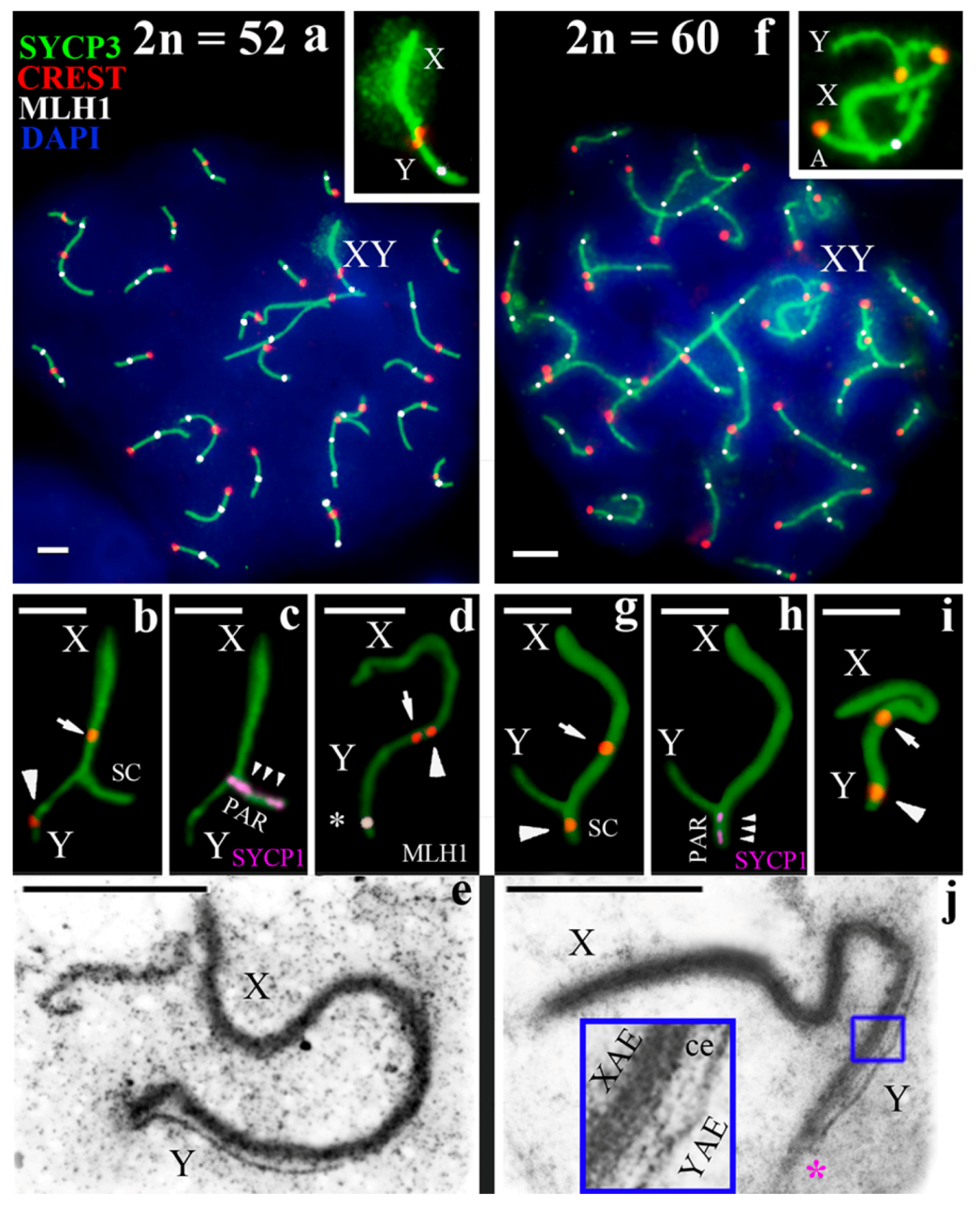
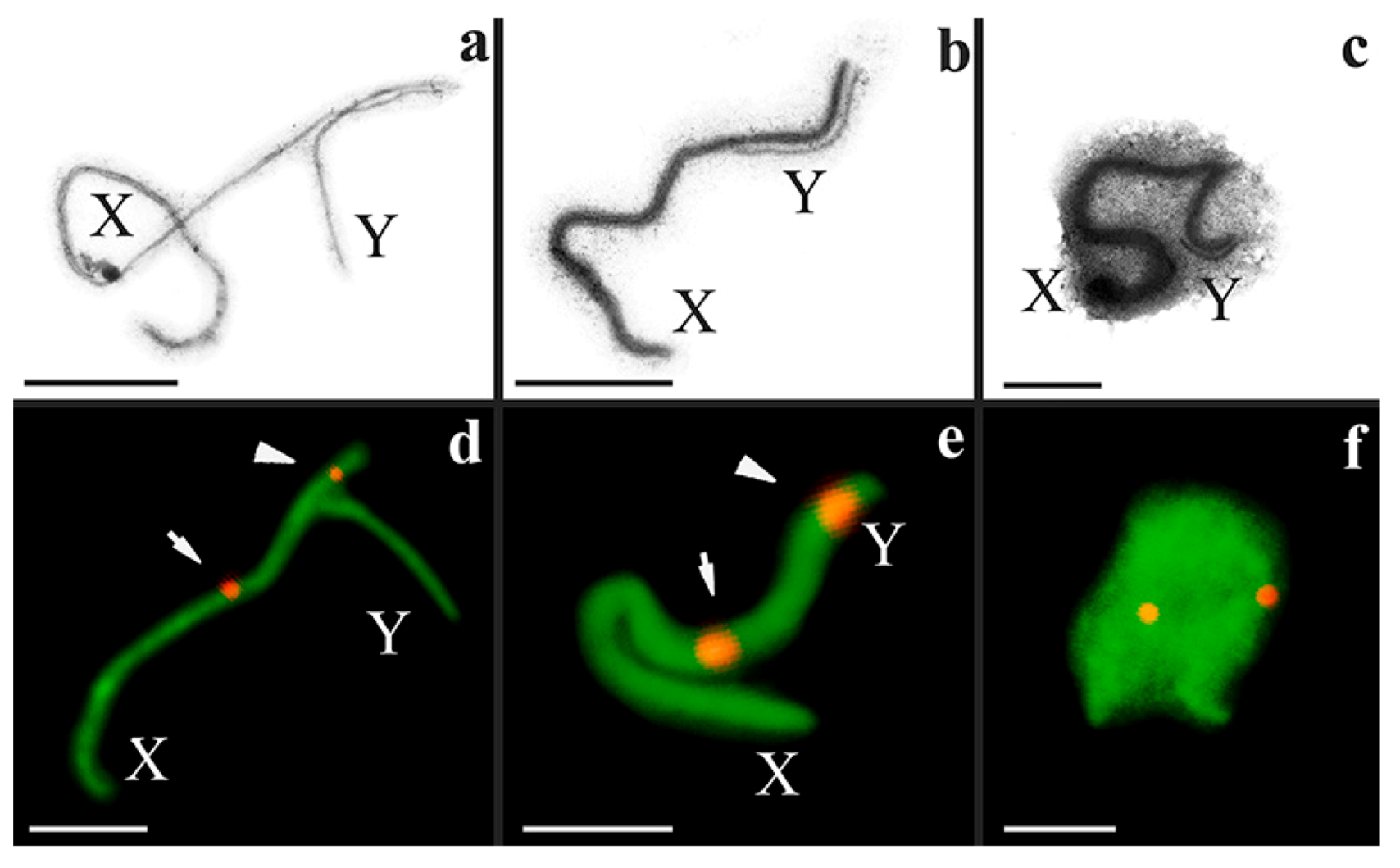
3.2. Synapsis of the Mole Rat Sex Chromosomes
3.3. Recombination of the Mole Rat Sex Chromosomes
4. Discussion
4.1. Specific Structure of the Nannospalax ehrenbergi XY Bivalent
4.2. Differences in Nannospalax ehrenbergi Sex Chromosomes
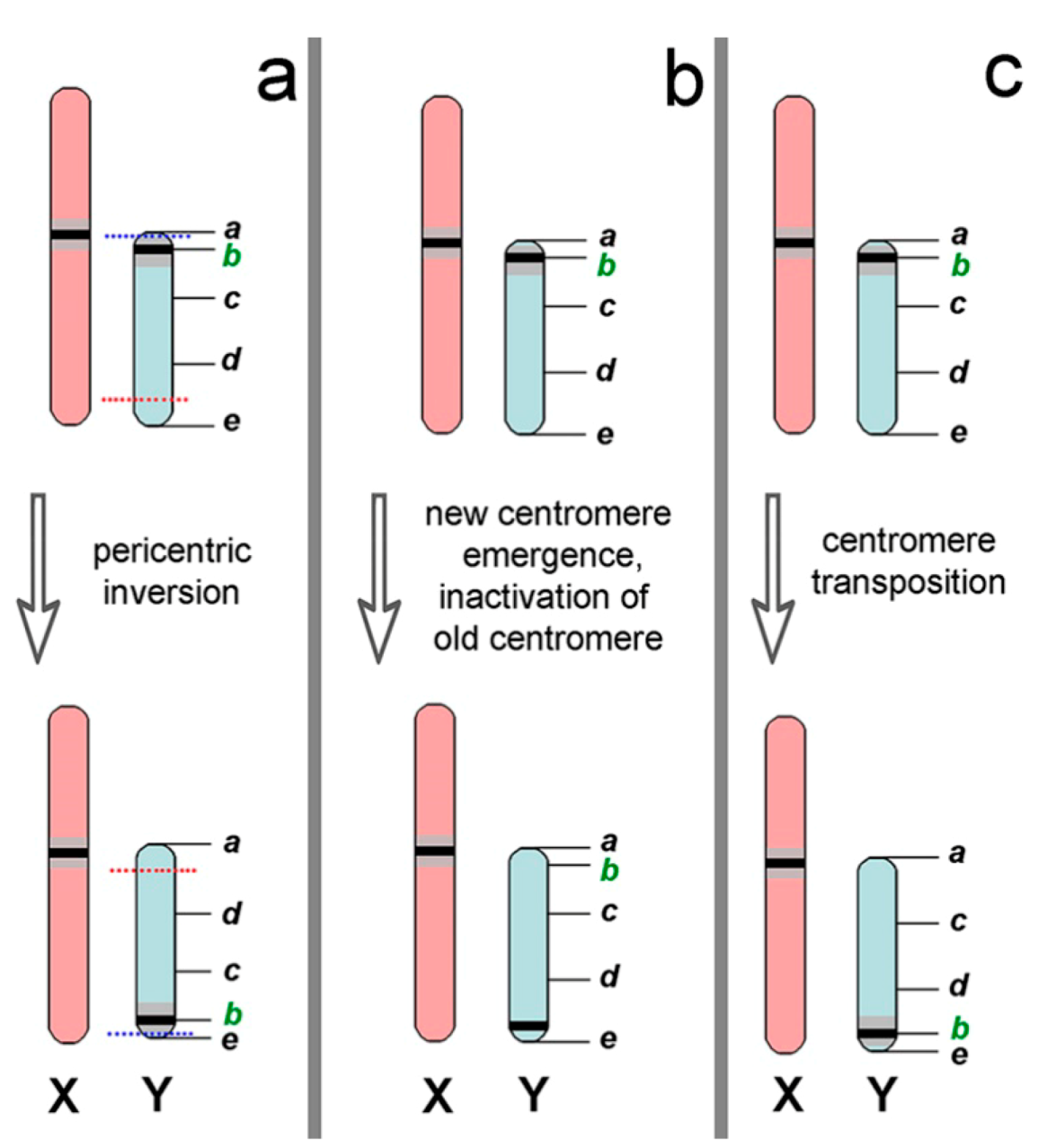
4.3. Trends and Possible Ways of Y chromosome Evolution in Nannospalax ehrenbergi
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Ezaz, T.; Srikulnath, K.; Graves, J.A.M. Origin of amniote sex chromosomes: An ancestral super-sex chromosome, or common requirements? J. Hered. 2016, 108, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A.M. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu. Rev. Genet. 2008, 42, 565–586. [Google Scholar] [CrossRef] [PubMed]
- Fredga, K. Unusual sex chromosome inheritance in mammals. Philos. Trans. R. Soc. Lond. B 1970, 259, 15–36. [Google Scholar] [CrossRef]
- Vorontsov, N.N.; Lyapunova, E.A.; Borissov, Y.M.; Dovgal, V.E. Variability of sex chromosomes in mammals. Genetica 1980, 52/53, 361–372. [Google Scholar] [CrossRef]
- Veyrunes, F.; Chevret, P.; Catalan, J.; Castiglia, R.; Watson, J.; Dobigny, G.; Robinson, T.J.; Britton-Davidian, J. A novel sex determination system in a close relative of the house mouse. Proc. R. Soc. Lond. B Biol. Sci. 2010, 277, 1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Fredga, K. Aberrant chromosomal sex-determining mechanisms in mammals, with special reference to species with XY females. Philos. Trans. R. Soc. Lond. B 1988, 322, 83–95. [Google Scholar] [CrossRef]
- Yoshida, K.; Kitano, J. The contribution of female meiotic drive to the evolution of neo-sex chromosomes. Evolution 2012, 66, 3198–3208. [Google Scholar] [CrossRef] [PubMed]
- Cortez, D.; Marin, R.; Toledo-Flores, D.; Froidevaux, L.; Liechti, A.; Waters, P.D.; Grützner, F.; Kaessmann, H. Origins and functional evolution of Y chromosomes across mammals. Nature 2014, 508, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A.M. Sex chromosome specialization and degeneration in mammals. Cell 2006, 124, 901–914. [Google Scholar] [CrossRef] [PubMed]
- Hsu, T.C.; Arrighi, F.E. Distribution of constitutive heterochromatin in mammalian chromosomes. Chromosoma 1971, 34, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.J. A gene for the fourth chromosome of Drosophila. J. Exp. Zool 1914, 17, 325–336. [Google Scholar] [CrossRef]
- Fisher, R.A. The evolution of dominance. Biol. Rev. 1931, 6, 345–368. [Google Scholar] [CrossRef]
- Nei, M. Linkage modification and sex difference in recombination. Genetics 1969, 63, 681–699. [Google Scholar] [PubMed]
- Bull, J.J. Evolution of Sex Determining Mechanisms; The Benjamin-Cummings Publishing Company: San Francisco, CA, USA, 1983; p. 316. [Google Scholar]
- Peneder, P.; Wallner, B.; Vogl, C. Exchange of genetic information between therian X and Y chromosome gametologs in old evolutionary strata. Ecol. Evol. 2017, 7, 8478–8487. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S. Sex Chromosomes and Sex-Linked Genes; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 1967; p. 191. [Google Scholar]
- Charlesworth, B.; Charlesworth, D. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000, 355, 1563–1572. [Google Scholar] [CrossRef] [PubMed]
- Bachtrog, D. Y-chromosome evolution: Emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 2013, 14, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Kolomiets, O.L.; Vorontsov, N.N.; Lyapunova, E.A.; Mazurova, T.F. Ultrastructure, meiotic behavior, and evolution of sex chromosomes of the genus Ellobius. Genetica 1991, 84, 179–189. [Google Scholar] [CrossRef]
- Matveevsky, S.; Bakloushinskaya, I.; Kolomiets, O. Unique sex chromosome systems in Ellobius: How do male XX chromosomes recombine and undergo pachytene chromatin inactivation? Sci. Rep. 2016, 6, 29949. [Google Scholar] [CrossRef] [PubMed]
- Matveevsky, S.; Kolomiets, O.; Bogdanov, A.; Hakhverdyan, M.; Bakloushinskaya, I. Chromosomal evolution in mole voles Ellobius (Cricetidae, Rodentia): Bizarre sex chromosomes, variable autosomes and meiosis. Genes 2017, 8, 306. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, Y.; Nishida-Umehara, C.; Matsuda, Y.; Sutou, S.; Suzuki, H. X-chromosomal localization of mammalian Y-linked genes in two XO species of the Ryukyu spiny rat. Cytogenet. Genome Res. 2002, 99, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A.M. The origin and function of the mammalian Y chromosome and Y-borne genes—An evolving understanding. Bioessays 1995, 17, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Saxena, R.; Brown, L.G.; Hawkins, T.; Alagappan, R.K.; Skaletsky, H.; Reeve, M.P.; Reijo, R.; Rozen, S.; Dinulos, M.B.; Disteche, C.M.; et al. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat. Genet. 1996, 14, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Skaletsky, H.; Kuroda-Kawaguchi, T.; Minx, P.J.; Cordum, H.S.; Hillier, L.; Brown, L.G.; Repping, S.; Pyntikova, T.; Ali, J.; Bieri, T.; et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 2003, 423, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Griffin, D.K. Is the Y chromosome disappearing?—Both sides of the argument. Chromosome Res. 2012, 20, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Murata, C.; Kuroki, Y.; Imoto, I.; Kuroiwa, A. Ancestral Y-linked genes were maintained by translocation to the X and Y chromosomes fused to an autosomal pair in the Okinawa spiny rat Tokudaia muenninki. Chromosome Res. 2016, 24, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Fredga, K. A new sex determining mechanism in a mammal. Chromosomes of Indian mongoose (Herpestes auropunctatus). Hereditas 1965, 52, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Murata, C.; Sawaya, H.; Nakata, K.; Yamada, F.; Imoto, I.; Kuroiwa, A. The cryptic Y-autosome translocation in the small Indian mongoose, Herpestes auropunctatus, revealed by molecular cytogenetic approaches. Chromosoma 2016, 125, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.S.F.; Elliott, M.W.; Morgan, L.; Miller, A.; Jones, T.C. Translocation of Y chromosome to an autosome in the Bolivian owl monkey, Aotus. Am. J. Phys. Anthrop. 1976, 45, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Pieczarka, J.C.; de Souza Barros, R.M.; De Faria, F.M.; Nagamachi, C.Y. Aotus from the southwestern Amazon region is geographically and chromosomally intermediate between A. azarae boliviensis and A. infulatus. Primates 1993, 34, 197–204. [Google Scholar] [CrossRef]
- Stanyon, R.; Garofalo, F.; Steinberg, E.R.; Capozzi, O.; Di Marco, S.; Nieves, M.; Archidiacono, N.; Mudry, M.D. Chromosome painting in two genera of South American monkeys: Species identification, conservation, and management. Cytogenet. Genome Res. 2011, 134, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, E.R.; Nieves, M.; Mudry, M.D. Multiple sex chromosome systems in howler monkeys (Platyrrhini, Alouatta). Comp. Cytogenet. 2014, 8, 43–69. [Google Scholar] [CrossRef] [PubMed]
- Schempp, W.; Toder, R. Evolution of the mammalian XY pairing segment. Chromosomes Today 1993, 11, 277–283. [Google Scholar]
- Burgoyne, P.S. Genetic homology and crossing over in the X and Y chromosomes of mammals. Hum. Genet. 1982, 61, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Lahn, B.T.; Page, D.C. Four evolutionary strata on the human X chromosome. Science 1999, 286, 964–967. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, R.; Wadee, A.; Rosendorff, J.; Wessels, A.; Jenkins, T. Inverted Y chromosome polymorphism in the Gujerati Muslim Indian population of South Africa. Hum. Genet. 1986, 74, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Hsu, L.Y.; Benn, P.A.; Tannenbaum, H.L.; Perlis, T.E.; Carlson, A.D.; Opitz, J.M.; Reynolds, J.F. Chromosomal polymorphisms of 1, 9, 16, and Y in 4 major ethnic groups: A large prenatal study. Am. J. Med. Genet. Part A 1987, 26, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Tomomasa, H.; Adachi, Y.; Iwabuchi, M.; Oshio, S.; Umeda, T.; Iino, Y.; Takano, T.; Nakahori, Y. Pericentric inversion of the Y chromosome of infertile male. Arch. Androl. 2000, 45, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Tyler-Smith, C.; Gimelli, G.; Giglio, S.; Floridia, G.; Pandya, A.; Terzoli, G.; Warburton, P.E.; Earnshaw, W.C.; Zuffardi, O. Transmission of a fully functional human neocentromere through three generations. Am. J. Hum. Genet. 1999, 64, 1440–1444. [Google Scholar] [CrossRef] [PubMed]
- Wahrman, J.; Goitein, R.; Nevo, E. Mole rat Spalax: Evolutionary significance of chromosome variation. Science 1969, 164, 82–84. [Google Scholar] [CrossRef] [PubMed]
- Wahrman, J.; Richler, C.; Gamperl, R.; Nevo, E. Revisiting Spalax: Mitotic and meiotic chromosome variability. Isr. J. Zool. 1985, 33, 15–38. [Google Scholar] [CrossRef]
- Shams, I.; Avivi, A.; Nevo, E. Hypoxic stress tolerance of the blind subterranean mole rat: Expression of erythropoietin and hypoxia-inducible factor 1α. Proc. Natl. Acad. Sci. USA 2004, 101, 9698–9703. [Google Scholar] [CrossRef] [PubMed]
- Brodsky, L.I.; Jacob-Hirsch, J.; Avivi, A.; Trakhtenbrot, L.; Zeligson, S.; Amariglio, N.; Paz, A.; Korol, B.; Band, M.; Rechavi, G.; et al. Evolutionary regulation of the blind subterranean mole rat, Spalax, revealed by genome-wide gene expression. Proc. Natl. Acad. Sci. USA 2005, 102, 17047–17052. [Google Scholar] [CrossRef] [PubMed]
- Gorbunova, V.; Seluanov, A.; Zhang, Z.; Gladyshev, V.N.; Vijg, J. Comparative genetics of longevity and cancer: Insights from long-lived rodents. Nat. Rev. Genet. 2014, 15, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Malik, A.; Bicker, A.; Poetzsch, G.; Avivi, A.; Shams, I.; Hankeln, T. Hypoxia tolerance, longevity and cancer-resistance in the mole rat Spalax—A liver transcriptomics approach. Sci. Rep. 2017, 7, 14348. [Google Scholar] [CrossRef] [PubMed]
- Arslan, A.; Kryštufek, B.; Matur, F.; Zima, J. Review of chromosome races in blind mole rats (Spalax and Nannospalax). Folia Zool. 2016, 65, 249–301. [Google Scholar] [CrossRef]
- Nevo, E.; Ivanitskaya, E.; Beiles, A. Adaptive Radiation of Blind Subterranean Mole Rats: Naming and Revisiting the Four Sibling Species of the Spalax ehrenbergi Superspecies in Israel: Spalax galili (2n = 52), S. golani (2n = 54), S. carmeli (2n = 58) and S. judaei (2n = 60); Backhuys Publishers: Leiden, The Netherlands, 2001; p. 190. [Google Scholar]
- Qumsiyeh, M.B. Mammals of the Holy Land; Texas Tech University Press: Lubbock, TX, USA, 1996; p. 389. [Google Scholar]
- Ivanitskaya, E.; Coskun, Y.; Nevo, E. Banded karyotypes of mole rats (Spalax, Spalacidae, Rodentia) from Turkey: A comparative analysis. J. Zool. Syst. Evol. Res. 1997, 35, 171–177. [Google Scholar] [CrossRef]
- Ivanitskaya, E.; Nevo, E. Cytogenetics of mole rats of the Spalax ehrenbergi superscpecies from Jordan (Spalacidae, Rodentia). Zeitschrift fur Saugetierkunde 1998, 63, 336–346. [Google Scholar]
- Stokes, W.S. Reducing unrelieved pain and distress in laboratory animals using humane endpoints. ILAR J. 2000, 41, 59–60. [Google Scholar] [CrossRef]
- Lee, M.R.; Elder, F.F.B. Yeast stimulation of bone marrow mitosis for cytogenetic investigations. Cytogenet. Genome Res. 1980, 26, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Seabright, M. A rapid banding technique for human chromosomes. Lancet 1971, 11, 971–972. [Google Scholar] [CrossRef]
- Navarro, J.; Vidal, F.; Guitart, M.; Egozcue, J. A method for the sequential study of synaptonemal complexes by light and electron microscopy. Hum. Genet. 1981, 59, 419–421. [Google Scholar] [CrossRef] [PubMed]
- Kolomiets, O.L.; Matveevsky, S.N.; Bakloushinskaya, I.Y. Sexual dimorphism in prophase I of meiosis in the Northern mole vole (Ellobius talpinus Pallas, 1770) with isomorphic (XX) chromosomes in males and females. Comp. Cytogenet. 2010, 4, 55–66. [Google Scholar] [CrossRef]
- Ivanitskaya, E.; Belyayev, A.; Nevo, E. Heterochromatin differentiation shows the pathways of karyotypic evolution in Israeli mole rats (Spalax, Spalacidae, Rodentia). Cytogenet. Genome Res. 2005, 111, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Ivanitskaya, E.; Rashkovetsky, L.; Nevo, E. Chromosomes in a hybrid zone of Israeli mole rats (Spalax, Radentia). Russ. J. Genet. 2010, 46, 1149–1151. [Google Scholar] [CrossRef]
- Solari, A.J.; Rahn, M.I. Asymmetry and resolution of the synaptonemal complex in the XY pair of Chinchilla laniger. Genetica 1985, 67, 63–71. [Google Scholar] [CrossRef]
- Anderson, L.K.; Reeves, A.; Webb, L.M.; Ashley, T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 1999, 151, 1569–1579. [Google Scholar] [PubMed]
- Dresser, M.E.; Moses, M.J. Synaptonemal complex karyotyping in spermatocytes of the Chinese hamster (Cricetulus griseus). IV. Light and electron microscopy of synapsis and nucleolar development by silver staining. Chromosoma 1980, 76, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Solari, A.J.; Pigozzi, M.I. Fine structure of the XY body in the XY1Y2 trivalent of the bat Artibeus lituratus. Chromosome Res. 1994, 2, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Bonvicino, C.R.; D’Andrea, P.S.; Borodin, P.M. Pericentric inversion in natural populations of Oligoryzomys nigripes (Rodentia: Sigmodontinae). Genome 2001, 44, 791–796. [Google Scholar] [CrossRef] [PubMed]
- Sciurano, R.B.; Merani, M.S.; Bustos, J.; Solari, A.J. Synaptonemal complexes and XY behavior in two species of Argentinian armadillos: Chaetophractus villosus and Dasypus hybridus (Xenarthra, Dasypodidae). Biocell 2006, 30, 57–66. [Google Scholar] [PubMed]
- Joseph, A.M.; Chandley, A.C. The morphological sequence of XY pairing in the Norway rat Rattus norvegicus. Chromosoma 1984, 89, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, D.; Leng, M.; Yang, L.; Zhong, L.; Cooke, H.J.; Shi, Q. Synapsis and meiotic recombination in male Chinese muntjac (Muntiacus reevesi). PLoS ONE 2011, 6, e19255. [Google Scholar] [CrossRef] [PubMed]
- Waters, P.D.; Ruiz-Herrera, A.; Dobigny, G.; Caldès, M.G.; Robinson, T.J. Sex chromosomes of basal placental mammals. Chromosoma 2007, 116, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Segura, J.; Ferretti, L.; Ramos-Onsins, S.; Capilla, L.; Farré, M.; Reis, F.; Oliver-Bonet, M.; Fernández-Bellón, H.; Garcia, F.; Garcia-Caldés, M.; et al. Evolution of recombination in eutherian mammals: Insights into mechanisms that affect recombination rates and crossover interference. Proc. R. Soc. Lond. B Biol. Sci. 2013, 280, 20131945. [Google Scholar] [CrossRef] [PubMed]
- Lindsley, D.L.; Sandler, L. The genetic analysis of meiosis in female Drosophila melanogaster. Philos. Trans. R. Soc. Lond. B 1977, 277, 295–312. [Google Scholar] [CrossRef]
- Charlesworth, B.; Langley, C.H.; Stephan, W. The evolution of restricted recombination and the accumulation of repeated DNA sequences. Genetics 1986, 112, 947–962. [Google Scholar] [PubMed]
- Davisson, M.T.; Akeson, E.C. Recombination suppression by heterozygous Robertsonian chromosomes in the mouse. Genetics 1993, 133, 649–667. [Google Scholar] [PubMed]
- Topp, C.N.; Dawe, R.K. Reinterpreting pericentromeric heterochromatin. Curr. Opin. Plant Biol. 2006, 9, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Froenicke, L.; Anderson, L.K.; Wienberg, J.; Ashley, T. Male mouse recombination maps for each autosome identified by chromosome painting. Am. J. Hum. Genet. 2002, 71, 1353–1368. [Google Scholar] [CrossRef] [PubMed]
- Rice, W.R. Evolution of the Y sex chromosome in animals. Bioscience 1996, 46, 331–343. [Google Scholar] [CrossRef]
- Bergero, R.; Charlesworth, D. The evolution of restricted recombination in sex chromosomes. Trends Ecol. Evol. 2009, 24, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Jackson, B.C. Recombination-suppression: How many mechanisms for chromosomal speciation? Genetica 2011, 139, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Yoshida, K.; Kitano, J. Contribution of gene flow to the evolution of recombination suppression in sex chromosomes. J. Theor. Biol. 2017, 431, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Di Meo, G.P.; Perucatti, A.; Floriot, S.; Incarnato, D.; Rullo, R.; Jambrenghi, A.C.; Ferretti, L.; Vonghia, G.; Cribiu, E.; Eggen, A.; et al. Chromosome evolution and improved cytogenetic maps of the Y chromosome in cattle, zebu, river buffalo, sheep and goat. Chromosome Res. 2005, 13, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Warburton, P.E. Chromosomal dynamics of human neocentromere formation. Chromosome Res. 2004, 12, 617. [Google Scholar] [CrossRef] [PubMed]
- Bakloushinskaya, I.Y.; Matveevsky, S.N.; Romanenko, S.A.; Serdukova, N.A.; Kolomiets, O.L.; Spangenberg, V.E.; Lyapunova, E.A.; Graphodatsky, A.S. A comparative analysis of the mole vole sibling species Ellobius tancrei and E. talpinus (Cricetidae, Rodentia) through chromosome painting and examination of synaptonemal complex structures in hybrids. Cytogenet. Genome Res. 2012, 136, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Nergadze, S.G.; Piras, F.M.; Gamba, R.; Corbo, M.; Cerutti, F.; McCarter, J.G.W.; Cappelletti, E.; Gozzo, F.; Harman, R.M.; Antczak, D.F.; et al. Birth, evolution, and transmission of satellite-free mammalian centromeric domains. Genome Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Schubert, I. What is behind “centromere repositioning”? Chromosoma 2018, 127, 229–234. [Google Scholar] [CrossRef] [PubMed]
- White, M.A.; Ikeda, A.; Payseur, B.A. A pronounced evolutionary shift of the pseudoautosomal region boundary in house mice. Mamm. Genome 2012, 23, 454–466. [Google Scholar] [CrossRef] [PubMed]
- Dumont, B.L. Meiotic consequences of genetic divergence across the murine pseudoautosomal region. Genetics 2017, 205, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- King, M. Species Evolution: The Role of Chromosome Change; Cambridge University Press: Cambridge, UK, 1993; p. 366. [Google Scholar]
- De la Fuente, R.; Sánchez, A.; Marchal, J.A.; Viera, A.; Parra, M.T.; Rufas, J.S.; Page, J. A synaptonemal complex-derived mechanism for meiotic segregation precedes the evolutionary loss of homology between sex chromosomes in arvicolid mammals. Chromosoma 2012, 121, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, B. The evolution of sex chromosomes. Science 1991, 251, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- Matveevsky, S.N. Vavilov Institute of General Genetics, Russian Academy of Sciences, Moscow, Russia. Materials and data about mole rats in Miilya hybrid zone, Israel. 2016. [Google Scholar]
- Nevo, E.; Filippucci, M.G.; Beiles, A. Genetic polymorphisms in subterranean mammals (Spalax ehrenbergi superspecies) in the near East revisited: Patterns and theory. Heredity 1994, 72, 465–487. [Google Scholar] [CrossRef] [PubMed]
- Nevo, E.; Honeycutt, R.L.; Yonekawa, H.; Nelson, K.; Hanzawa, N. Mitochondrial DNA polymorphisms in subterranean mole-rats of the Spalax ehrenbergi superspecies in Israel, and its peripheral isolates. Mol. Biol. Evol. 1993, 10, 590–604. [Google Scholar] [CrossRef] [PubMed]
- Nevo, E.; Beiles, A.; Spradling, T. Molecular evolution of cytochrome b of subterranean mole rats, Spalax ehrenbergi superspecies, in Israel. J. Mol. Evol. 1999, 49, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.J. The relation of recombination to mutational advance. Mutat. Res. Fund. Mol. Mech. Mutagen. 1964, 1, 2–9. [Google Scholar] [CrossRef]
- Gordo, I.; Charlesworth, B. The speed of Muller’s ratchet with background selection, and the degeneration of Y chromosomes. Genet. Res. 2001, 78, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Mitsainas, G.P.; Rovatsos, M.T.; Athanasopoulou, E.B.G. The HSR and heterochromatin distribution in natural populations of Mus musculus domesticus (Rodentia: Murinae) from Greece. Caryologia 2009, 62, 53–61. [Google Scholar]
- Jotterand-Bellomo, M. The African Mus genus, an example of karyotypic homogeneity: Cytogenetic study of Mus minutoides/musculoides (Ivory Coast), M. setulosus (Central African Republic), and M. mattheyi (Burkina Faso). Cytogenet. Cell Genet. 1986, 42, 99–104. [Google Scholar] [CrossRef]
- Veyrunes, F.; Catalan, J.; Sicard, B.; Robinson, T.J.; Duplantier, J.M.; Granjon, L.; Dobigny, G.; Britton-Davidian, J. Autosome and sex chromosome diversity among the African pygmy mice, subgenus Nannomys (Murinae; Mus). Chromosome Res. 2004, 12, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Veyrunes, F.; Watson, J.; Robinson, T.J.; Britton-Davidian, J. Accumulation of rare sex chromosome rearrangements in the African pygmy mouse, Mus (Nannomys) minutoides: A whole-arm reciprocal translocation (WART) involving an X-autosome fusion. Chromosome Res. 2007, 15, 223. [Google Scholar] [CrossRef] [PubMed]
- Searle, J.B.; Fedyk, S.; Fredga, K.; Hausser, J.; Volobouev, V.T. Nomenclature for the chromosomes of the common shrew Sorex araneus. Mém. Soc. Vaud. Sci. Nat. 1991, 19, 13–22. [Google Scholar] [CrossRef]
- Matveevsky, S.N.; Pavlova, S.V.; Atsaeva, M.M.; Searle, J.B.; Kolomiets, O.L. Dual mechanism of chromatin remodeling in the common shrew sex trivalent (XY1Y2). Comp. Cytogenet. 2017, 11, 727–745. [Google Scholar] [CrossRef] [PubMed]
- Guénet, J.L.; Nagamine, C.; Simon-Chazottes, D.; Montagutelli, X.; Bonhomme, F. Hst-3: An X-linked hybrid sterility gene. Genet. Res. 1990, 56, 163–165. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, Y.; Hirobe, T.; Chapman, V.M. Genetic basis of XY chromosome dissociation and male sterility in interspecific hybrids. Proc. Natl. Acad. Sci. USA 1991, 88, 4850–4854. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, Y.; Moens, P.B.; Chapman, V.M. Deficiency of X and Y chromosomal pairing at meiotic prophase in spermatocytes of sterile interspecific hybrids between laboratory mice (Mus domesticus) and Mus spretus. Chromosoma 1992, 101, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Kryštufek, B.; Ivanitskaya, E.; Arslan, A.; Arslan, E.; Buzan, E.V. Evolutionary history of mole rats (genus Nannospalax) inferred from mitochondrial cytochrome b sequence. Biol. J. Linn. Soc. 2012, 105, 446–455. [Google Scholar] [CrossRef]
- Hadid, Y.; Németh, A.; Snir, S.; Pavlíček, T.; Csorba, G.; Kázmér, M.; Major, Á.; Mezhzherin, S.; Rusin, M.; Coşkun, Y.; et al. Is evolution of blind mole rats determined by climate oscillations? PLoS ONE 2012, 7, e30043. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matveevsky, S.; Ivanitskaya, E.; Spangenberg, V.; Bakloushinskaya, I.; Kolomiets, O. Reorganization of the Y Chromosomes Enhances Divergence in Israeli Mole Rats Nannospalax ehrenbergi (Spalacidae, Rodentia): Comparative Analysis of Meiotic and Mitotic Chromosomes. Genes 2018, 9, 272. https://doi.org/10.3390/genes9060272
Matveevsky S, Ivanitskaya E, Spangenberg V, Bakloushinskaya I, Kolomiets O. Reorganization of the Y Chromosomes Enhances Divergence in Israeli Mole Rats Nannospalax ehrenbergi (Spalacidae, Rodentia): Comparative Analysis of Meiotic and Mitotic Chromosomes. Genes. 2018; 9(6):272. https://doi.org/10.3390/genes9060272
Chicago/Turabian StyleMatveevsky, Sergey, Elena Ivanitskaya, Victor Spangenberg, Irina Bakloushinskaya, and Oxana Kolomiets. 2018. "Reorganization of the Y Chromosomes Enhances Divergence in Israeli Mole Rats Nannospalax ehrenbergi (Spalacidae, Rodentia): Comparative Analysis of Meiotic and Mitotic Chromosomes" Genes 9, no. 6: 272. https://doi.org/10.3390/genes9060272
APA StyleMatveevsky, S., Ivanitskaya, E., Spangenberg, V., Bakloushinskaya, I., & Kolomiets, O. (2018). Reorganization of the Y Chromosomes Enhances Divergence in Israeli Mole Rats Nannospalax ehrenbergi (Spalacidae, Rodentia): Comparative Analysis of Meiotic and Mitotic Chromosomes. Genes, 9(6), 272. https://doi.org/10.3390/genes9060272




