Engineering the Salt-Inducible Ectoine Promoter Region of Halomonas elongata for Protein Expression in a Unique Stabilizing Environment
Abstract
1. Introduction
2. Materials and Methods
2.1. Construction of Different H. elongata Strains
2.2. Medium, Supplements and Growth Conditions
2.3. Recombinant Plasmids and Templates
2.4. Vector Construction
- Plasmid pPE, pWUB01 and pWUB02: The promoter region promA upstream of the ectoine biosynthesis cluster was amplified from genomic DNA of H. elongata DSM 2581T (pPE) and H. elongata KB1 (pWUB01), respectively, with primers for_1A and rev_1A (pPE) or rev_2B (pWUB01). For construction of pWUB02 the forward primer was changed to for_2C. Amplified fragments and the destination vector pBBR1-MCS were digested with appropriate restriction enzymes (Table 2) and ligated. Gene gfpuv was amplified from pGFPuv using primers for_3D and rev_4D, and was ligated into the corresponding vector backbone downstream of the ectoine promoter.
- Plasmid pSPE1_GFPuv and pSPE2_GFPuv: gfpuv was amplified from pGFPuv using either primers for_4E and rev_4D (pSPE1_GFPuv) or primers for_5E and rev_4D (pSPE2_GFPuv). Both forward primers for_4E and for_5E comprise a synthetic RBS adapted for expression in H. elongata DSM 2581T and E. coli, respectively (Table 2). Amplified fragments and the destination vector pPE were digested with appropriate restriction enzymes (Table 2) and ligated.
- Plasmid pWUB01_sec_mCherry, pWUB01_tat_gfpuv: Primers for_11H and rev_8H were used to amplify a fragment of pcoA encoding the entire signal sequence and the first four amino acid residues of the mature protein, and, in addition, an overlap extension complementary to the gfpuv gene. Primers for_13H and rev_4D were used to amplify gfpuv gene from pGFPuv without start codon ATG, but an additional overlap extension complementary to the pcoA leader. Both of the fragments were fused in an overlap extension polymerase chain reaction using primers for_12H and rev_4D. The resulting product contained PscI and HindIII restriction sites. Primers for_8G and rev_6G were used to amplify a fragment of teaA encoding the entire signal sequence and the first four amino acid residues of the mature protein, and, in addition, an overlap extension complementary to the mCherry gene. Primers for_10G and rev_7G were used to amplify mCherry gene from pCQ11ftsZmCh without start codon but an additional overlap extension complementary to the teaA leader. Both of the fragments were fused in an overlap extension polymerase chain reaction using primers for_9G and rev_7G. The resulting product contained PagI and HindIII restriction sites.
2.5. Cultivation for Fluorescence Measurements
2.6. Cultivation for Fluorescence Microscopy
2.7. Fluorescence Microscopy
2.8. Purification of Soluble Protein Fractions
2.9. Protein Quantification
2.10. Polyacrylamide Gel Electrophoresis
2.11. Fluorescence Measurements
2.12. Calculation of Optimized Ribosomal Binding Sites
3. Results
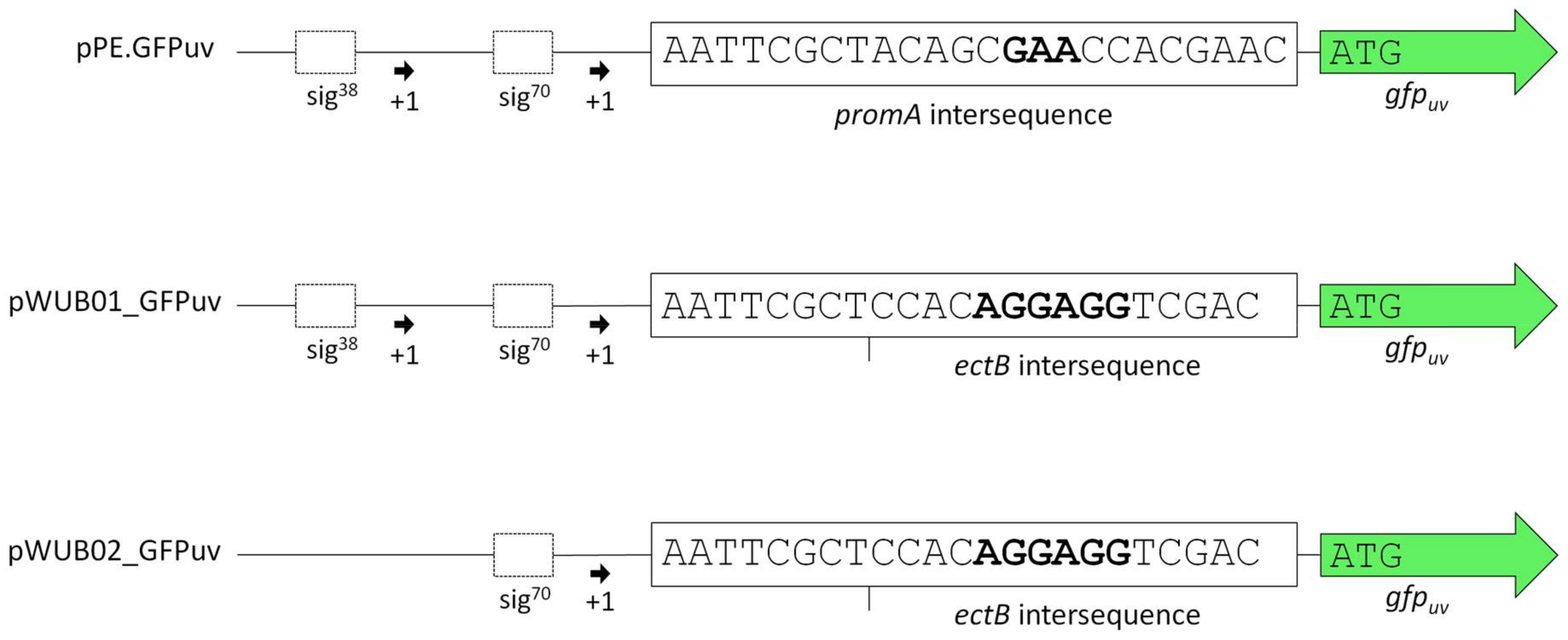
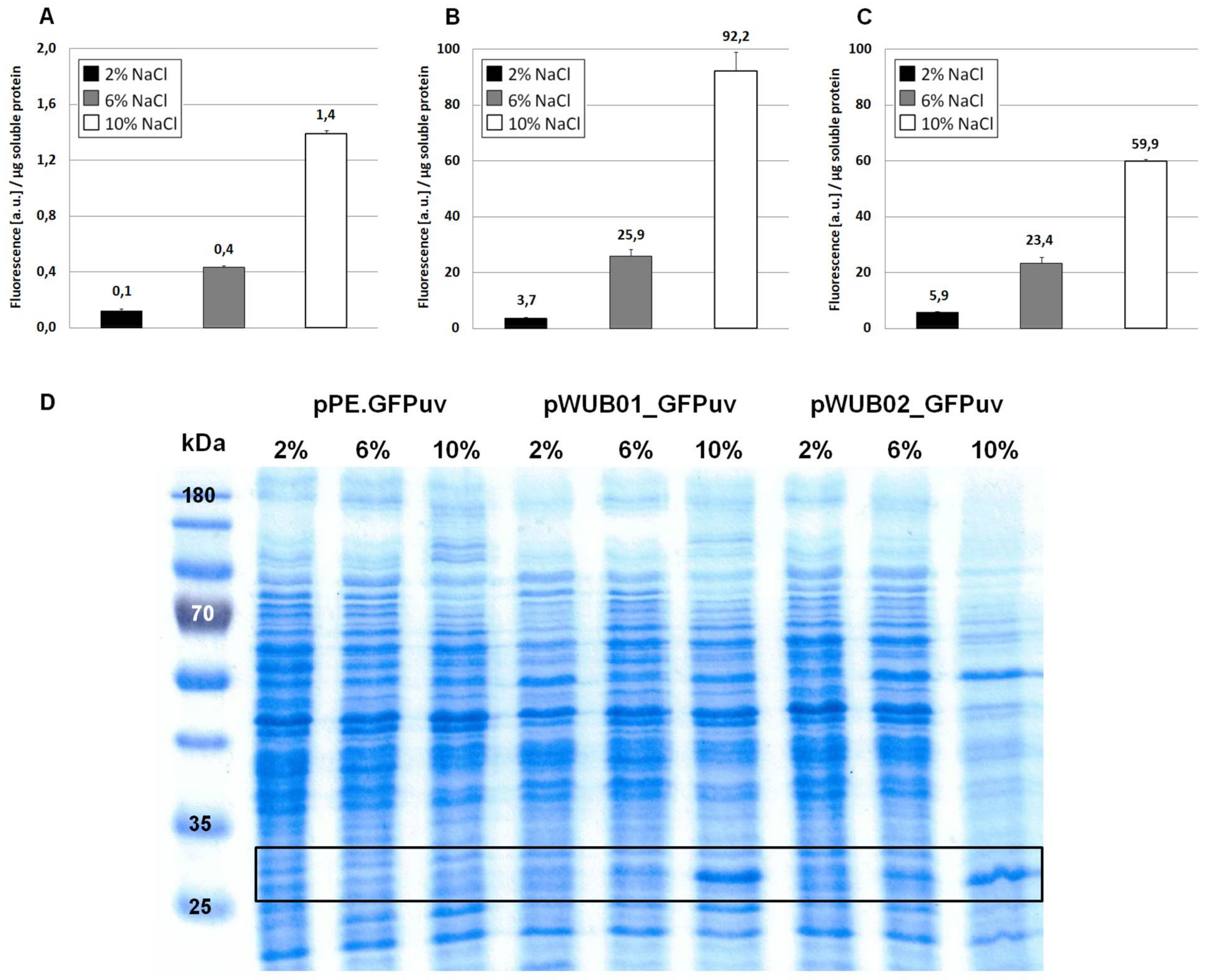
3.1. Construction of a Suitable Expression Vector for H. elongata
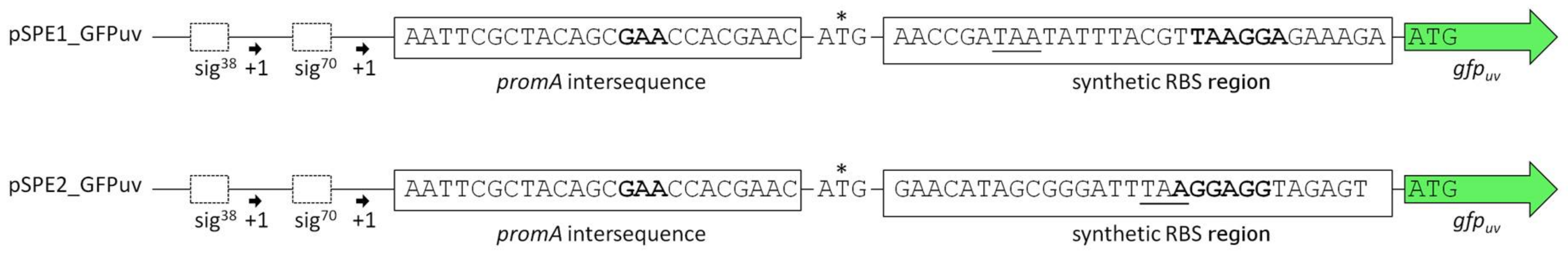
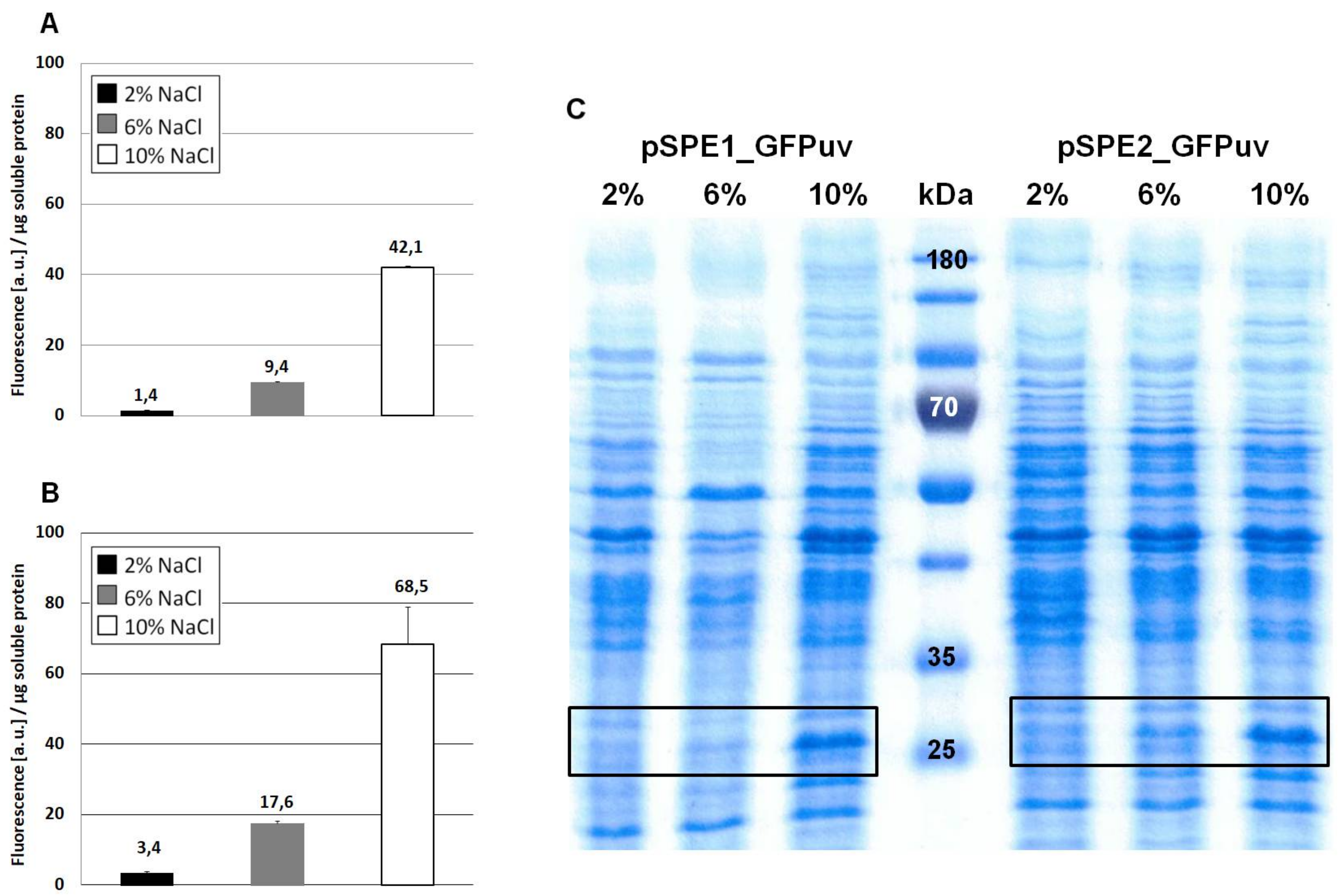
3.2. Optimization of Translation Based on Messenger RNA Folding Energies
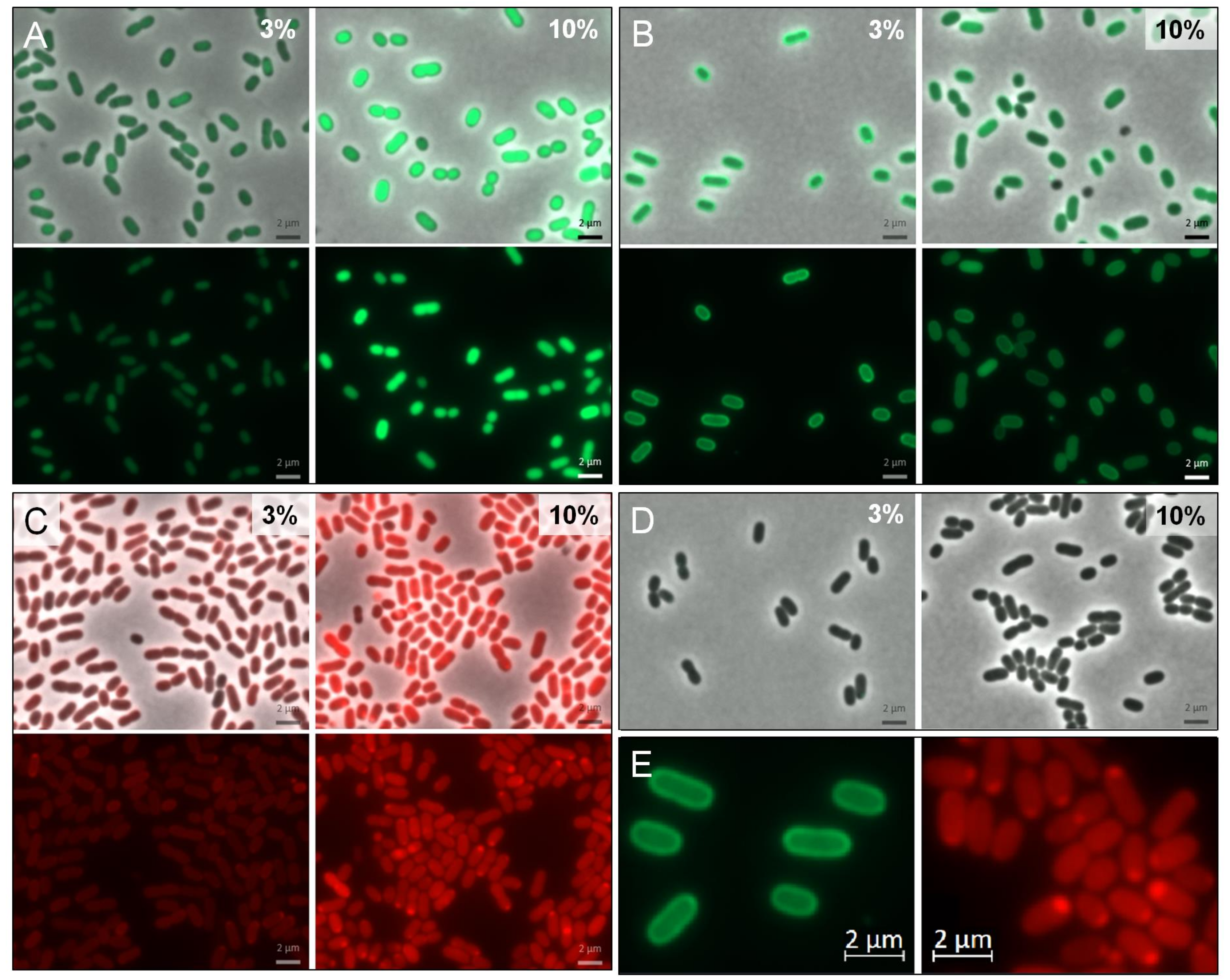
3.3. Periplasmatic Expression of GFPuv and mCherry
4. Discussion
4.1. Characterization of the Promoter Region Upstream of ectA
4.2. Optimizing Translation Initiation Using the RBS Calculator v2.0
4.3. Directing Proteins into the Periplasmic Environment
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Vreeland, R.H.; Litchfield, C.D.; Martin, E.L.; Elliot, E. Halomonas elongata, a new genus and species of extremely salt-tolerant bacteria. Int. J. Syst. Bacteriol. 1980, 30, 485–495. [Google Scholar] [CrossRef]
- Ventosa, A.; Nieto, J.J.; Oren, A. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 1998, 62, 504–544. [Google Scholar] [PubMed]
- Dötsch, A.; Severin, J.; Alt, W.; Galinski, E.A.; Kreft, J.U. A mathematical model for growth and osmoregulation in halophilic bacteria. Microbiology 2008, 154, 2956–2969. [Google Scholar] [CrossRef] [PubMed]
- Schwibbert, K.; Marin-Sanguino, A.; Bagyan, I.; Heidrich, G.; Lentzen, G.; Seitz, H.; Rampp, M.; Schuster, S.C.; Klenk, H.P.; Pfeiffer, F.; et al. A blueprint of ectoine metabolism from the genome of the industrial producer Halomonas elongata DSM 2581T. Environ. Microbiol. 2011, 13, 1973–1994. [Google Scholar] [CrossRef] [PubMed]
- Peters, P.; Galinski, E.A.; Truper, H.G. The biosynthesis of ectoine. Fems Microbiol. Lett. 1990, 71, 157–162. [Google Scholar] [CrossRef]
- Reuter, K.; Pittelkow, M.; Bursy, J.; Heine, A.; Craan, T.; Bremer, E. Synthesis of 5-hydroxyectoine from ectoine: Crystal structure of the non-heme iron(II) and 2-oxoglutarate-dependent dioxygenase EctD. PLoS ONE 2010, 5, e10647. [Google Scholar] [CrossRef] [PubMed]
- Widderich, N.; Kobus, S.; Hoppner, A.; Riclea, R.; Seubert, A.; Dickschat, J.S.; Heider, J.; Smits, S.H.; Bremer, E. Biochemistry and crystal structure of ectoine synthase: A metal-containing member of the cupin superfamily. PLoS ONE 2016, 11, e0151285. [Google Scholar] [CrossRef] [PubMed]
- Severin, J.; Wohlfarth, A.; Galinski, E.A. The predominant role of recently discovered tetrahydropyrimidines for the osmoadaptation of halophilic eubacteria. J. Gen. Microbiol. 1992, 138, 1629–1638. [Google Scholar] [CrossRef]
- Kindzierski, V.; Raschke, S.; Knabe, N.; Siedler, F.; Scheffer, B.; Pfluger-Grau, K.; Pfeiffer, F.; Oesterhelt, D.; Marin-Sanguino, A.; Kunte, H.J. Osmoregulation in the halophilic bacterium Halomonas elongata: A case study for integrative systems biology. PLoS ONE 2017, 12, e0168818. [Google Scholar] [CrossRef] [PubMed]
- Pastor, J.M.; Bernal, V.; Salvador, M.; Argandona, M.; Vargas, C.; Csonka, L.; Sevilla, A.; Iborra, J.L.; Nieto, J.J.; Canovas, M. Role of central metabolism in the osmoadaptation of the halophilic bacterium Chromohalobacter salexigens. J. Biol. Chem. 2013, 288, 17769–17781. [Google Scholar] [CrossRef] [PubMed]
- Sauer, T.; Galinski, E.A. Bacterial milking: A novel bioprocess for production of compatible solutes. Biotechnol. Bioeng. 1998, 59, 128. [Google Scholar] [CrossRef]
- Kunte, H.J.; Lentzen, G.; Galinski, E.A. Industrial production of the cell protectant ectoine: Protection mechanisms, processes, and products. Curr. Biotechnol. 2014, 3, 10–25. [Google Scholar] [CrossRef]
- Kanapathipillai, M.; Ku, S.H.; Girigoswami, K.; Park, C.B. Small stress molecules inhibit aggregation and neurotoxicity of prion peptide 106–126. Biochem. Biophys. Res. Commun. 2008, 365, 808–813. [Google Scholar] [CrossRef] [PubMed]
- Arora, A.; Ha, C.; Park, C.B. Inhibition of insulin amyloid formation by small stress molecules. FEBS Lett. 2004, 564, 121–125. [Google Scholar] [CrossRef]
- Hahn, M.B.; Meyer, S.; Schroter, M.A.; Kunte, H.J.; Solomun, T.; Sturm, H. DNA protection by ectoine from ionizing radiation: Molecular mechanisms. Phys. Chem. Chem. Phys. 2017, 19, 25717–25722. [Google Scholar] [CrossRef] [PubMed]
- Kunte, H.J.; Galinski, E.A. Transposon mutagenesis in halophilic eubacteria: Conjugal transfer and insertion of transposon Tn5 and Tn1732 in Halomonas elongata. FEMS Microbiol. Lett. 1995, 128, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Grammann, K.; Volke, A.; Kunte, H.J. New type of osmoregulated solute transporter identified in halophilic members of the bacteria domain: TRAP transporter TeaABC mediates uptake of ectoine and hydroxyectoine in Halomonas elongata DSM 2581T. J. Bacteriol. 2002, 184, 3078–3085. [Google Scholar] [CrossRef] [PubMed]
- Vargas, C.; Nieto, J.J. Genetic tools for the manipulation of moderately halophilic bacteria of the family Halomonadaceae. Methods Mol. Biol. 2004, 267, 183–208. [Google Scholar] [PubMed]
- Simon, R.; Priefer, U.; Puhler, A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Chung, C.T.; Niemela, S.L.; Miller, R.H. One-step preparation of competent Escherichia coli: Transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. USA 1989, 86, 2172–2175. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Nojima, H.; Okayama, H. High efficiency transformation of Escherichia coli with plasmids. Gene 1990, 96, 23–28. [Google Scholar] [CrossRef]
- Kovach, M.E.; Phillips, R.W.; Elzer, P.H.; Roop, R.M., 2nd.; Peterson, K.M. pBBR1MCS: A broad-host-range cloning vector. Biotechniques 1994, 16, 800–802. [Google Scholar] [PubMed]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop, R.M., 2nd.; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef]
- Hardt, P.; Engels, I.; Rausch, M.; Gajdiss, M.; Ulm, H.; Sass, P.; Ohlsen, K.; Sahl, H.-G.; Bierbaum, G.; Schneider, T.; et al. The cell wall precursor lipid II acts as a molecular signal for the Ser/Thr kinase PknB of Staphylococcus aureus. Int. J. Med. Microbiol. 2017, 307, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Brünig, A. Molekulargenetische und physiologische Studien zur Entwicklung eines Expressionssystems in Halomonas elongata DSMZ 2581T. Diploma Thesis, Rheinische Friedrich-Wilhelms-Universität Bonn, Bonn, Germany, 2005. [Google Scholar]
- Kurz, M.; Brunig, A.N.; Galinski, E.A. NhaD type sodium/proton-antiporter of Halomonas elongata: A salt stress response mechanism in marine habitats? Saline Syst. 2006, 2, 10. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Salis, H.M.; Mirsky, E.A.; Voigt, C.A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009, 27, 946–950. [Google Scholar] [CrossRef] [PubMed]
- Espah Borujeni, A.; Channarasappa, A.S.; Salis, H.M. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 2014, 42, 2646–2659. [Google Scholar] [CrossRef] [PubMed]
- Durfee, T.; Nelson, R.; Baldwin, S.; Plunkett, G., 3rd.; Burland, V.; Mau, B.; Petrosino, J.F.; Qin, X.; Muzny, D.M.; Ayele, M.; et al. The complete genome sequence of Escherichia coli DH10B: Insights into the biology of a laboratory workhorse. J. Bacteriol. 2008, 190, 2597–2606. [Google Scholar] [CrossRef] [PubMed]
- Politi, N.; Pasotti, L.; Zucca, S.; Casanova, M.; Micoli, G.; Cusella De Angelis, M.G.; Magni, P. Half-life measurements of chemical inducers for recombinant gene expression. J. Biol. Eng. 2014, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Kurz, M.; Burch, A.Y.; Seip, B.; Lindow, S.E.; Gross, H. Genome-driven investigation of compatible solute biosynthesis pathways of Pseudomonas syringae pv. syringae and their contribution to water stress tolerance. Appl. Environ. Microbiol. 2010, 76, 5452–5462. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Gralla, J.D. Osmo-regulation of bacterial transcription via poised RNA polymerase. Mol. Cell 2004, 14, 153–162. [Google Scholar] [CrossRef]
- Rosenthal, A.Z.; Hu, M.; Gralla, J.D. Osmolyte-induced transcription: -35 region elements and recognition by sigma38 (rpoS). Mol. Microbiol. 2006, 59, 1052–1061. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Zhou, T.; Wilke, C.O. A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput. Biol. 2010, 6, e1000664. [Google Scholar] [CrossRef] [PubMed]
- Tuller, T.; Waldman, Y.Y.; Kupiec, M.; Ruppin, E. Translation efficiency is determined by both codon bias and folding energy. Proc. Natl. Acad. Sci. USA 2010, 107, 3645–3650. [Google Scholar] [CrossRef] [PubMed]
- Salis, H.M. Chapter Two—The Ribosome Binding Site Calculator. In Methods in Enzymology; Voigt, C., Ed.; Academic Press, Elsevier Inc.: Cambridge, MA, USA, 2011; Volume 498, pp. 19–42. [Google Scholar]
- Bagos, P.G.; Nikolaou, E.P.; Liakopoulos, T.D.; Tsirigos, K.D. Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinformatics 2010, 26, 2811–2817. [Google Scholar] [CrossRef] [PubMed]
- Bendtsen, J.D.; Nielsen, H.; Widdick, D.; Palmer, T.; Brunak, S. Prediction of twin-arginine signal peptides. BMC Bioinform. 2005, 6, 167. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Calderon, M.I.; Vargas, C.; Rojo, F.; Iglesias-Guerra, F.; Csonka, L.N.; Ventosa, A.; Nieto, J.J. Complex regulation of the synthesis of the compatible solute ectoine in the halophilic bacterium Chromohalobacter salexigens DSM 3043T. Microbiology 2004, 150, 3051–3063. [Google Scholar] [CrossRef] [PubMed]
- Salvador, M.; Argandona, M.; Pastor, J.M.; Bernal, V.; Canovas, M.; Csonka, L.N.; Nieto, J.J.; Vargas, C. Contribution of RpoS to metabolic efficiency and ectoines synthesis during the osmo- and heat-stress response in the halophilic bacterium Chromohalobacter salexigens. Environ. Microbiol. Rep. 2015, 7, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Typas, A.; Stella, S.; Johnson, R.C.; Hengge, R. The −35 sequence location and the Fis-sigma factor interface determine σS selectivity of the proP (P2) promoter in Escherichia coli. Mol. Microbiol. 2007, 63, 780–796. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Liu, J.; Han, R.; Shen, G.; Long, Q.; Wei, X.; Liu, D. Identification and characterization of ectoine biosynthesis genes and heterologous expression of the ectABC gene cluster from Halomonas sp. QHL1, a moderately halophilic bacterium isolated from Qinghai Lake. J. Microbiol. 2014, 52, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Typas, A.; Becker, G.; Hengge, R. The molecular basis of selective promoter activation by the σS subunit of RNA polymerase. Mol. Microbiol. 2007, 63, 1296–1306. [Google Scholar] [CrossRef] [PubMed]
- Czech, L.; Poehl, S.; Hub, P.; Stoeveken, N.; Bremer, E. Tinkering with osmotically controlled transcription allows enhanced production and excretion of ectoine and hydroxyectoine from a microbial cell factory. Appl. Environ. Microbiol. 2018, 84, e01772-17. [Google Scholar] [CrossRef] [PubMed]
- Omotajo, D.; Tate, T.; Cho, H.; Choudhary, M. Distribution and diversity of ribosome binding sites in prokaryotic genomes. BMC Genom. 2015, 16, 604. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Niimura, Y.; Miura, K.; Gojobori, T. Dynamic evolution of translation initiation mechanisms in prokaryotes. Proc. Natl. Acad. Sci. USA 2010, 107, 6382–6387. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H.; Chang, B.C.; Chiang, P.W.; Tang, S.L. Questionable 16S ribosomal RNA gene annotations are frequent in completed microbial genomes. Gene 2008, 416, 44–47. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, F.; Bagyan, I.; Alfaro-Espinoza, G.; Zamora-Lagos, M.A.; Habermann, B.; Marin-Sanguino, A.; Oesterhelt, D.; Kunte, H.J. Revision and reannotation of the Halomonas elongata DSM 2581T genome. Microbiologyopen 2017, 6, e465. [Google Scholar] [CrossRef] [PubMed]
- Feilmeier, B.J.; Iseminger, G.; Schroeder, D.; Webber, H.; Phillips, G.J. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 2000, 182, 4068–4076. [Google Scholar] [CrossRef] [PubMed]
- Dammeyer, T.; Timmis, K.N.; Tinnefeld, P. Broad host range vectors for expression of proteins with (Twin-) Strep-tag, His-tag and engineered, export optimized yellow fluorescent protein. Microb. Cell Fact. 2013, 12, 49. [Google Scholar] [CrossRef] [PubMed]
- Panahandeh, S.; Maurer, C.; Moser, M.; DeLisa, M.P.; Muller, M. Following the path of a twin-arginine precursor along the TatABC translocase of Escherichia coli. J. Biol. Chem. 2008, 283, 33267–33275. [Google Scholar] [CrossRef] [PubMed]
- Di Cola, A.; Robinson, C. Large-scale translocation reversal within the thylakoid Tat system in vivo. J. Cell. Biol. 2005, 171, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Branston, S.D.; Matos, C.F.; Freedman, R.B.; Robinson, C.; Keshavarz-Moore, E. Investigation of the impact of Tat export pathway enhancement on E. coli culture, protein production and early stage recovery. Biotechnol. Bioeng. 2012, 109, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Barrett, C.M.; Ray, N.; Thomas, J.D.; Robinson, C.; Bolhuis, A. Quantitative export of a reporter protein, GFP, by the twin-arginine translocation pathway in Escherichia coli. Biochem. Biophys. Res. Commun. 2003, 304, 279–284. [Google Scholar] [CrossRef]
- Santini, C.L.; Bernadac, A.; Zhang, M.; Chanal, A.; Ize, B.; Blanco, C.; Wu, L.F. Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J. Biol. Chem. 2001, 276, 8159–8164. [Google Scholar] [CrossRef] [PubMed]
- Quan, S.; Hiniker, A.; Collet, J.F.; Bardwell, J.C. Isolation of bacteria envelope proteins. Methods Mol. Biol. 2013, 966, 359–366. [Google Scholar] [PubMed]
| Relevant Characteristics/Description | Source/Reference | |
|---|---|---|
| Strains | ||
| Halomonas elongata | ||
| DSM 2581T | wild type | [1] |
| KB1 | ΔectA | [17] |
| Escherichia coli | ||
| S17.1 | RP4-2 (Tc::Mu) (Km::Tn7); SmR, pro, thi, recA | [19] |
| Plasmids | ||
| pBBR1-MCS | Broad host range cloning vector | [22,23] |
| pGFPuv | Expression vector containing the gene sequence of GFPuv | Clontech Laboratories, Inc. (Mountain View, CA, USA) |
| pCQ11-ftsZmCh | Contains the gene sequence of mCherry, a DsRed derivative | [24] |
| pPE | Cloning vector; pBBR1MCS derivative equipped with the salt-dependent H. elongata promoter region approx. 470 bp upstream of ectA (HELO_2588) | [25,26] |
| pPE.GFPuv | pBBR1MCS derivative equipped with the promoter region upstream of ectA (HELO_2588), followed by gfpuv | [25] |
| pSPE1_GFPuv | pBBR1MCS derivative equipped with the promoter region upstream of ectA (HELO_2588), followed by a synthetic RBS region optimized for H. elongata DSM 2581T and the gene gfpuv | This work |
| pSPE2_GFPuv | pBBR1MCS derivative equipped with the promoter region upstream of ectA (HELO_2588), followed by a synthetic RBS region optimized for E. coli K-12 substr. DH10B and the gene gfpuv | This work |
| pWUB01_GFPuv | pBBR1MCS derivative equipped with the promoter region upstream of ectA (HELO_2588) from H. elongata DSM 2581T and the RBS upstream of ectB (HELO_2589) from H. elongata KB1 (promKB1), followed by gfpuv | This work |
| pWUB02_GFPuv | pBBR1MCS derivative equipped with part of the promoter region upstream of ectA (HELO_2588) (including only the σ70 sequence) from H. elongata DSM 2581T and the RBS upstream of ectB (HELO_2589) from H. elongata KB1 (promKB1), followed by gfpuv | This work |
| Primer | Sequence (5′ → 3′) | Restriction Site |
|---|---|---|
| for_1A | GGAGGCCGTCTAGATCATCCAGG | XbaI |
| rev_1A | CTCTGTGGATCCGTACATGTTCGTGGT | BamHI, PscI |
| rev_2B | AATGGATCCCTACATGTCGACCTCCTGT | BamHI |
| for_2C | TATTCTAGAGGAATTCAGCAAGCAAGAT | XbaI |
| rev_3C | ATAACAATTTCACACAGGAAACAGCTA | - |
| for_3D | CCGGTAGAAATCATGAGTAAAGGAGAAG | PagI |
| rev_4D | GGCCGACTAGTAAGCTTATTATTTTTGACAC | HindIII |
| for_4E | ATTCTGCAGAACCGATAATATTTACGTTAAGGAGAAAGAATGAGTAAAGGAGAAGAACTT | PstI |
| for_5E | ATTCTGCAGGAACATAGCGGGATTTAAGGAGGTAGAGTATGAGTAAAGGAGAAGAACTT | PstI |
| for_6F | ATTTCTAGAGGGCGCGAAGCCTGCCCGTC | XbaI |
| rev_5F | ATTGGATCCTTGCAATCTTCCTTATGACT | BamHI |
| for_7F | ATTAAGCTTGAACATAGCGGGATTTAAGG | HindIII |
| for_8G | CAAGCAAGCCGAACTGGACGCCGAACGC | - |
| for_9G | ACAATCATGAAGGCATACAAGCTGCTGAC | PagI |
| rev_6G | TCGCCCTTGCTCACACGCCAGTTGTCGG | - |
| for_10G | CCGACAACTGGCGTGTGAGCAAGGGCGA | - |
| rev_7G | ATTAAGCTTATAGGCGCGCCTTACTT | HindIII |
| for_11H | TTCGATCCTGATCCAGTTGCTTGATCA | - |
| for_12H | ATTACATGTCAATGCCCCAGAAGCCTTTA | PscI |
| rev_8H | TTCTTCTCCTTTACTTCCCCAGGGACTGG | - |
| for_13H | CCAGTCCCTGGGGAAGTAAAGGAGAAGAA | - |
| Vector | H. elongata DSM 2581T | E. coli DH10B |
|---|---|---|
| pSPE1_GFPuv | 365,234.33 | 65,308.16 |
| pSPE2_GFPuv | 95,871.18 | 1,231,096.69 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stiller, L.M.; Galinski, E.A.; Witt, E.M.H.J. Engineering the Salt-Inducible Ectoine Promoter Region of Halomonas elongata for Protein Expression in a Unique Stabilizing Environment. Genes 2018, 9, 184. https://doi.org/10.3390/genes9040184
Stiller LM, Galinski EA, Witt EMHJ. Engineering the Salt-Inducible Ectoine Promoter Region of Halomonas elongata for Protein Expression in a Unique Stabilizing Environment. Genes. 2018; 9(4):184. https://doi.org/10.3390/genes9040184
Chicago/Turabian StyleStiller, Lisa M., Erwin A. Galinski, and Elisabeth M. H. J. Witt. 2018. "Engineering the Salt-Inducible Ectoine Promoter Region of Halomonas elongata for Protein Expression in a Unique Stabilizing Environment" Genes 9, no. 4: 184. https://doi.org/10.3390/genes9040184
APA StyleStiller, L. M., Galinski, E. A., & Witt, E. M. H. J. (2018). Engineering the Salt-Inducible Ectoine Promoter Region of Halomonas elongata for Protein Expression in a Unique Stabilizing Environment. Genes, 9(4), 184. https://doi.org/10.3390/genes9040184




