The Unexplored Diversity of Pleolipoviruses: The Surprising Case of Two Viruses with Identical Major Structural Modules
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Media
2.2. Virus Isolation and Preparation of Virus Stocks
2.3. Sensitivity of Virus Infectivity
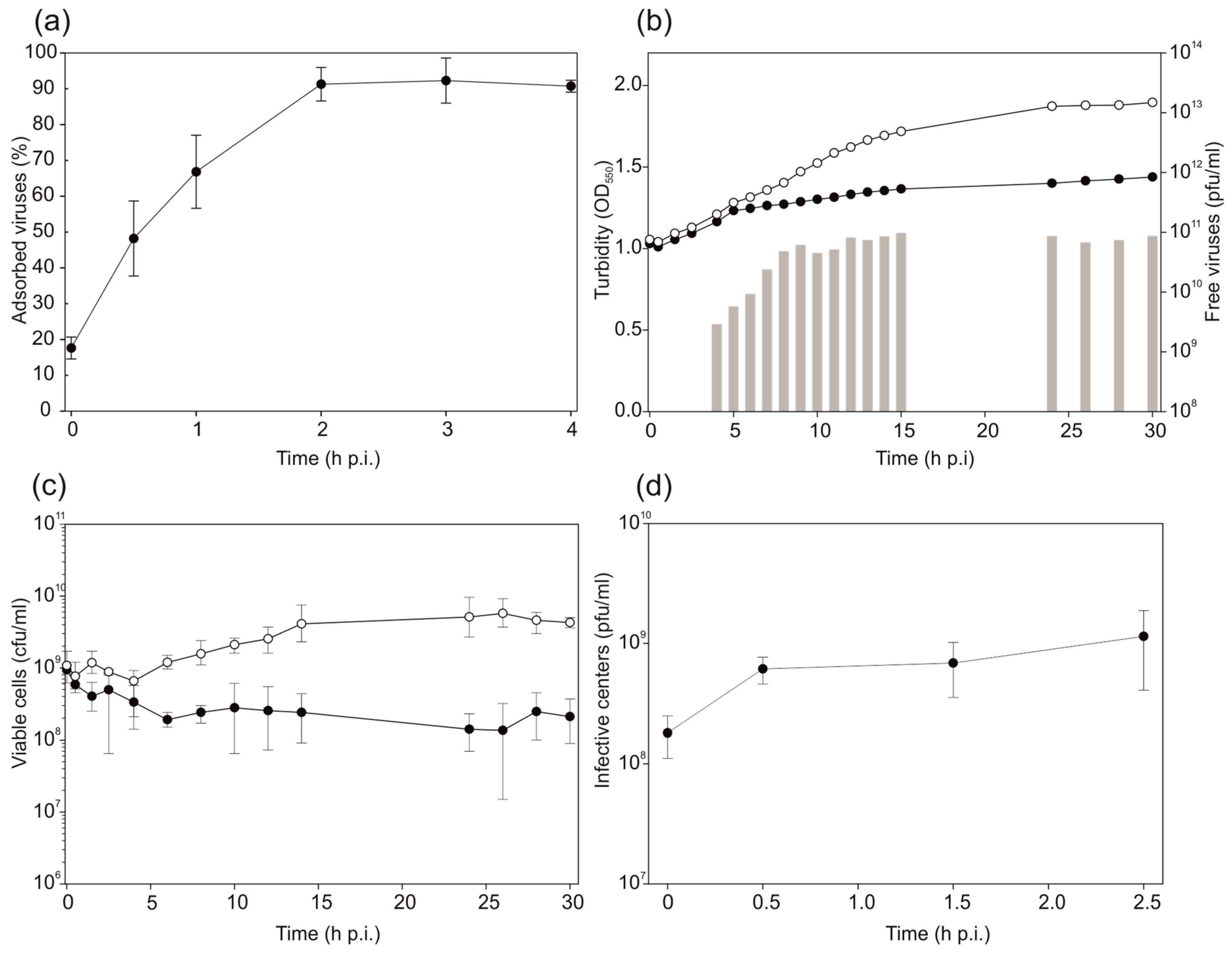
2.4. Virus Adsorption and Life Cycle
2.5. Virus Purification
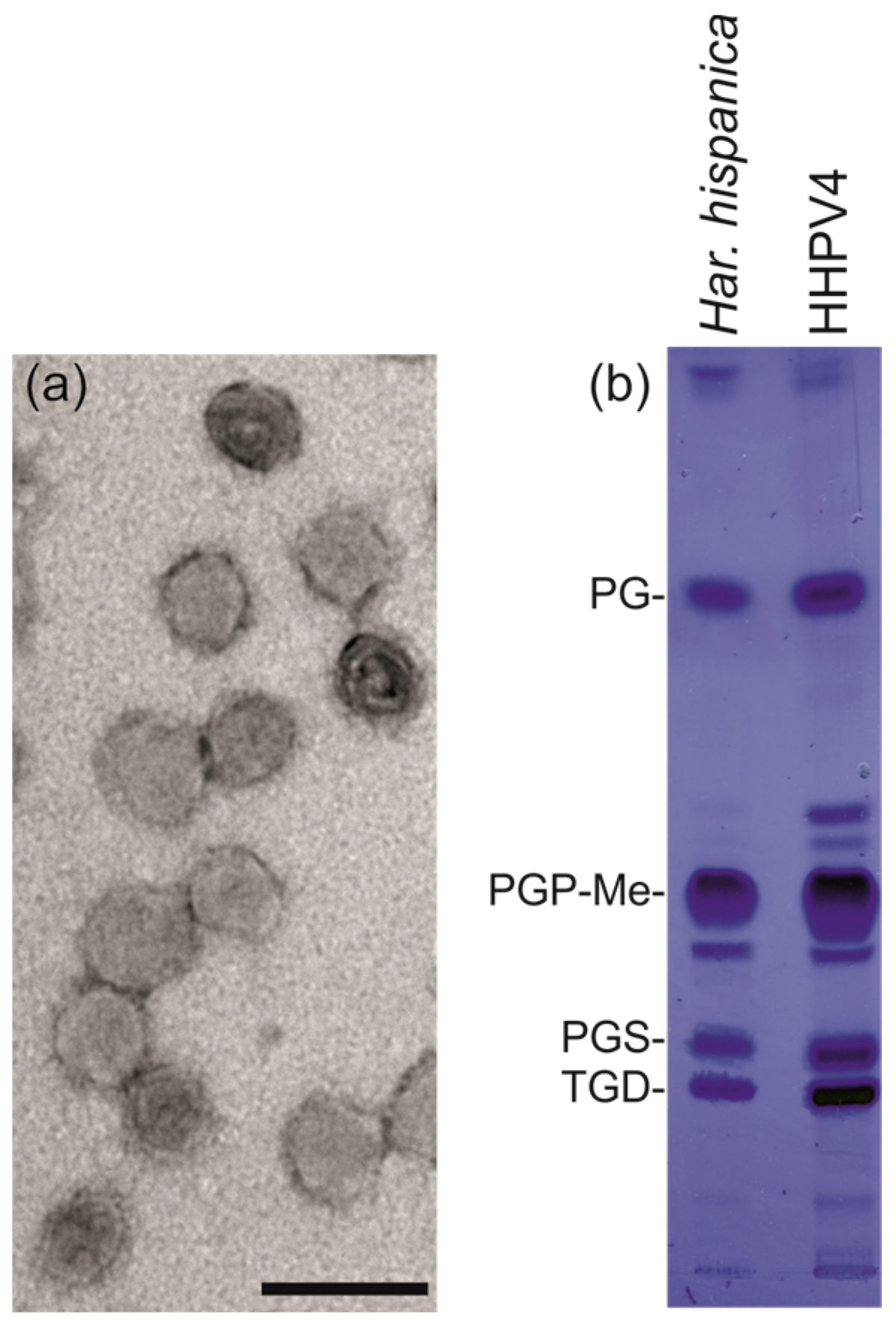
2.6. Analysis of Extracted Lipids
2.7. Negative Stain Electron Microscopy
2.8. Genome Isolation and Sequencing
2.9. Genome Annotation
2.10. Plaque Purification and PCR
2.11. Phylogenomic Analysis
3. Results
3.1. HHPV4 Was Isolated From Spot Assay Plates of H. hispanica
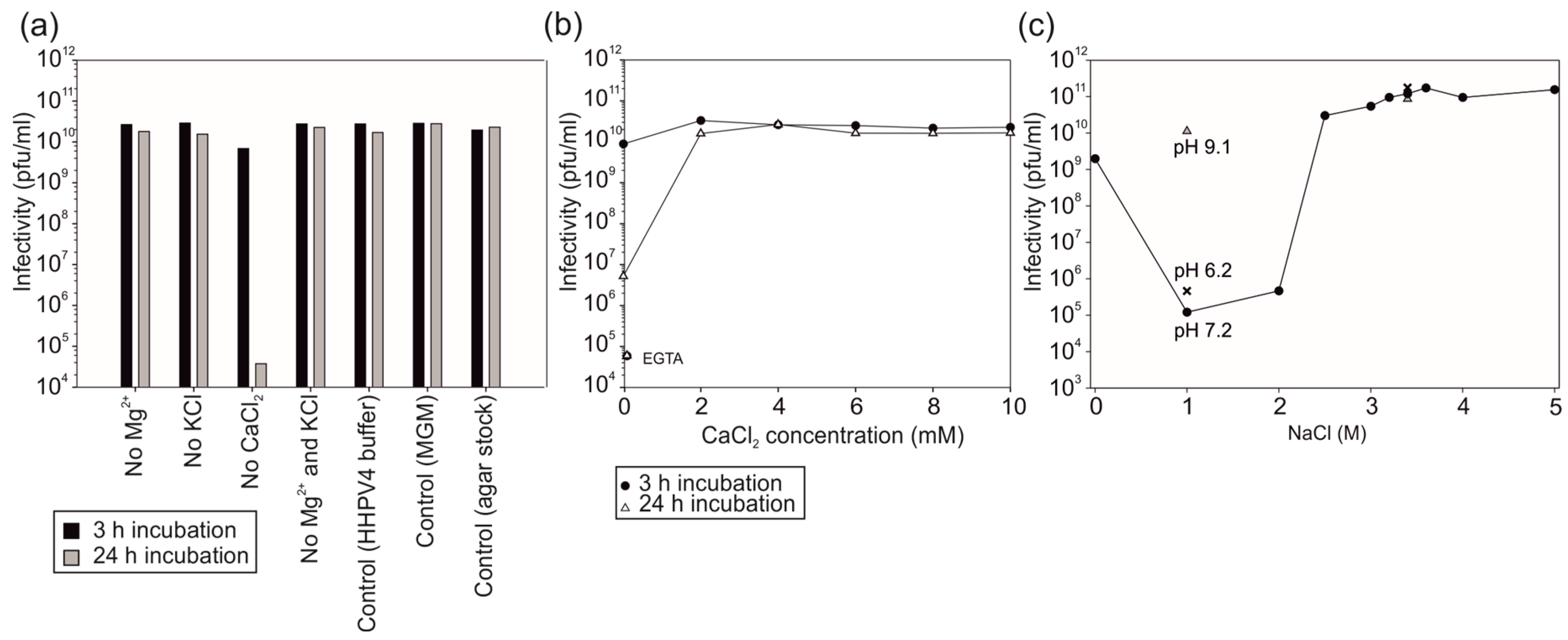
3.2. Virus Infectivity Depends on Salt Concentrations
3.3. HHPV4 Infection Leads to High Progeny Production
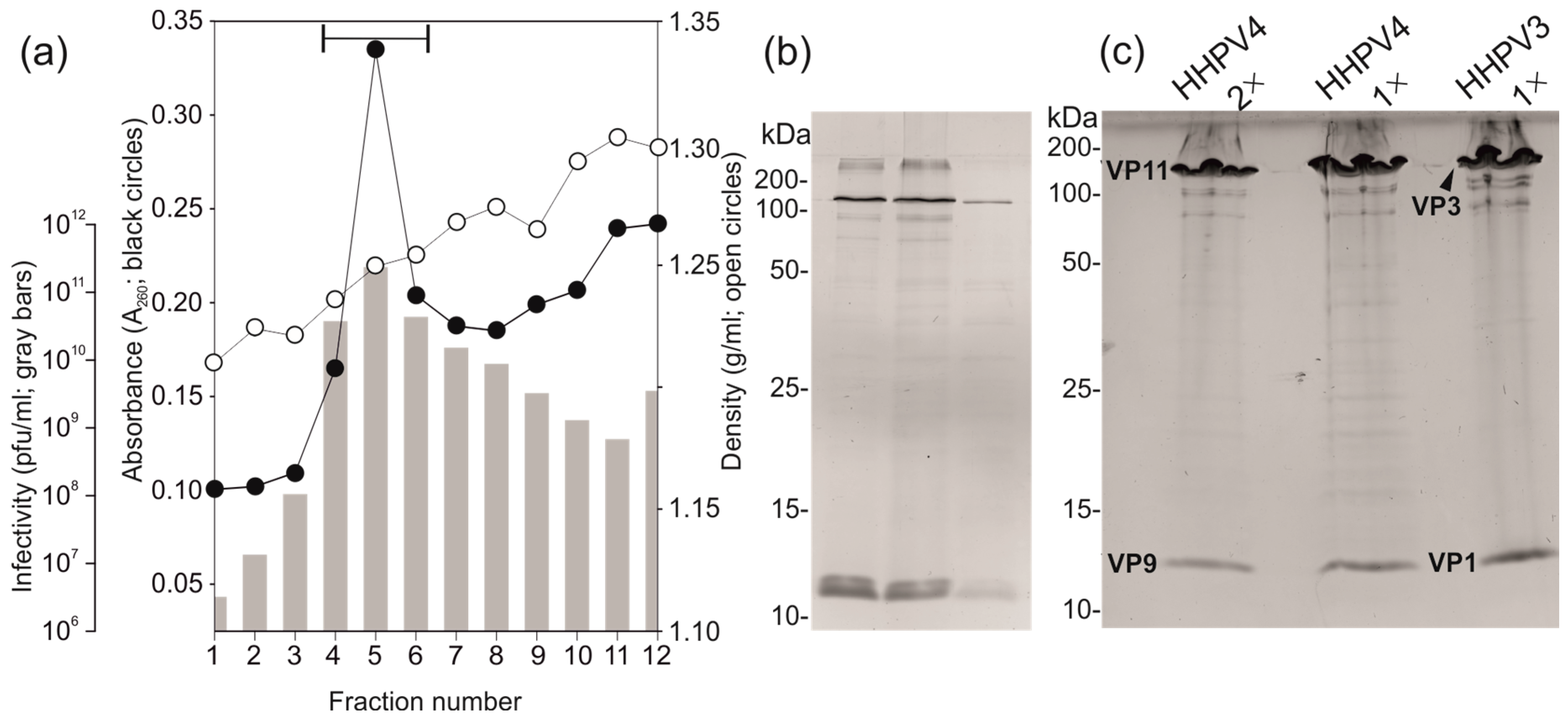
3.4. Two-Step Virus Purification Yielded a High Number of Pure Infectious Particles
3.5. HHPV4 is a Pleomorphic, Membrane-Containing Virus
3.6. The HHPV4 Genome Is a Circular dsDNA Molecule
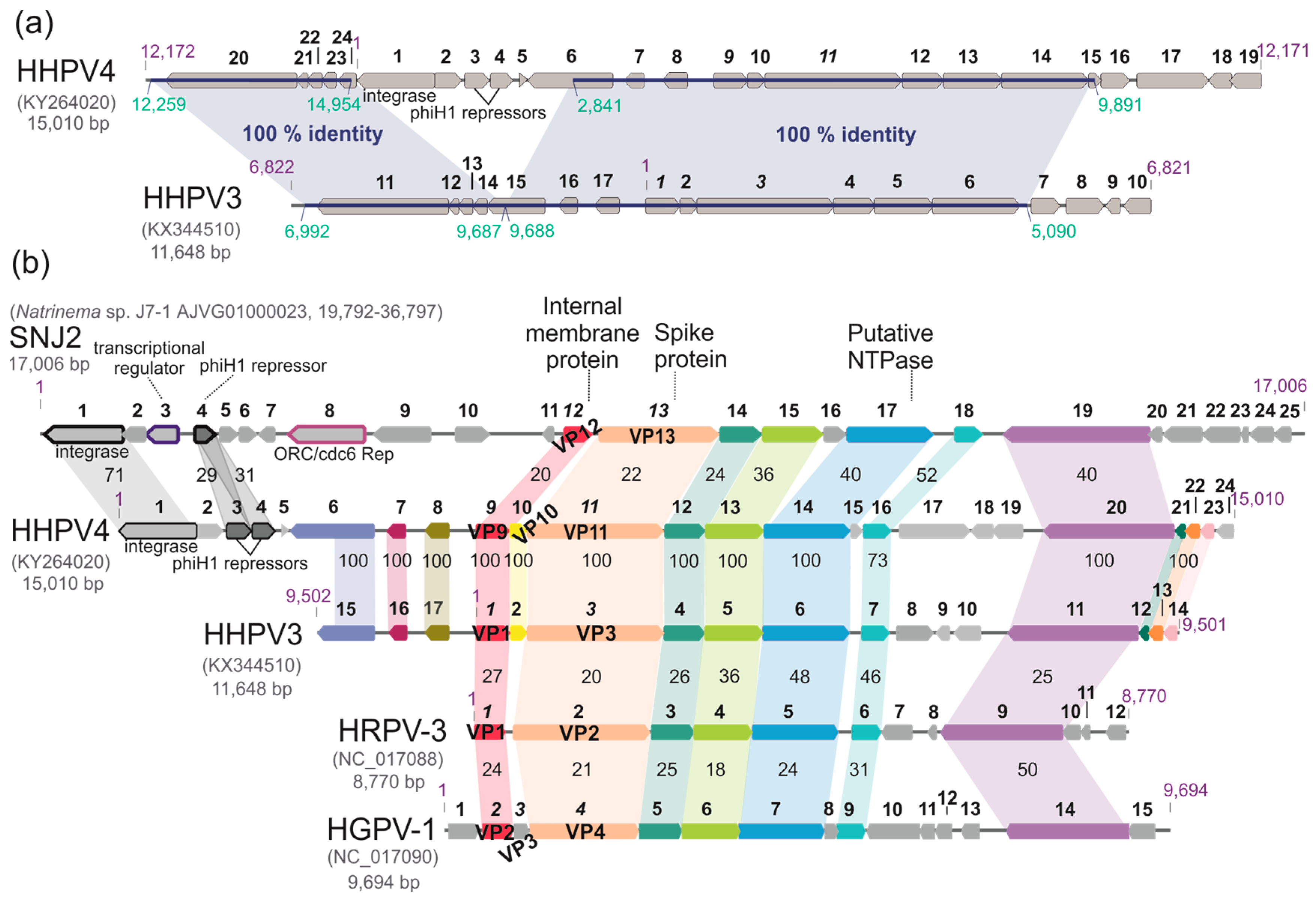
3.7. The HHPV4 Genome Sequence Is Largely Identical to That of HHPV3 and Contains a Conserved Pleolipoviral Block
3.8. HHPV4 Genome Differs from That of HHPV3 by the Presence of an Integrase-Encoding Gene
3.9. HHPV4-Like Proviruses Are Detected in Haloarchaeal Genomes
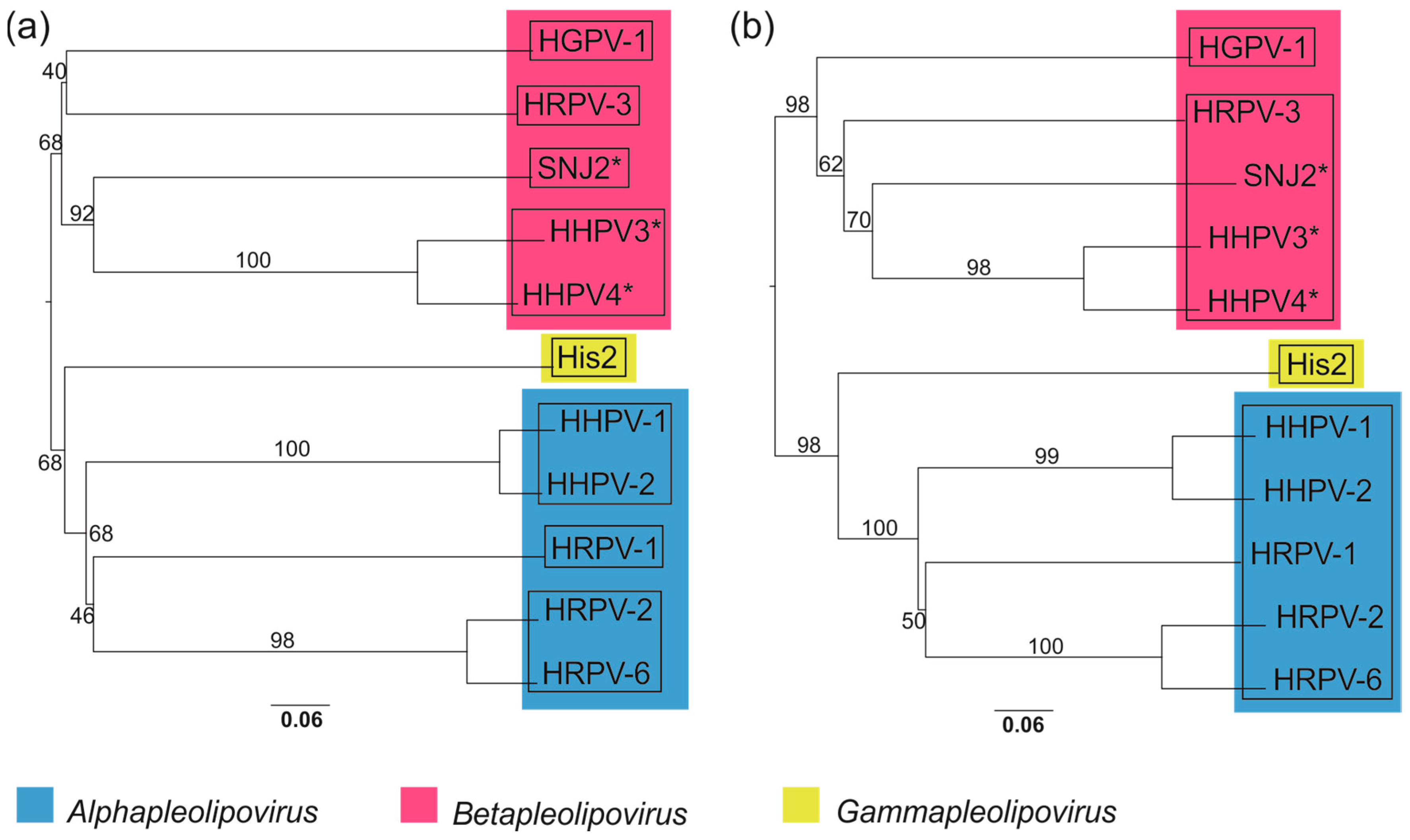
3.10. Whole Genome-Based Analysis Reinforces the Current Classification of Pleolipoviruses
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bamford, D.H.; Pietilä, M.K.; Roine, E.; Atanasova, N.S.; Dienstbier, A.; Oksanen, H.M. ICTV Report Consortium. ICTV Virus Taxonomy Profile: Pleolipoviridae. J. Gen. Virol. 2017, 98, 2916–2917. [Google Scholar] [CrossRef] [PubMed]
- Atanasova, N.S.; Demina, T.A.; Buivydas, A.; Bamford, D.H.; Oksanen, H.M. Archaeal viruses multiply: Temporal screening in a solar saltern. Viruses 2015, 7, 1902–1926. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, J.; Liu, Y.; Wang, Y.; Zhang, Z.; Oksanen, H.M.; Bamford, D.H.; Chen, X. Identification and characterization of SNJ2, the first temperate pleolipovirus integrating into the genome of the SNJ1-lysogenic archaeal strain. Mol. Microbiol. 2015, 98, 1002–1020. [Google Scholar] [CrossRef] [PubMed]
- Demina, T.A.; Atanasova, N.S.; Pietilä, M.K.; Oksanen, H.M.; Bamford, D.H. Vesicle-like virion of Haloarcula hispanica pleomorphic virus 3 preserves high infectivity in saturated salt. Virology 2016, 499, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Senčilo, A.; Paulin, L.; Kellner, S.; Helm, M.; Roine, E. Related haloarchaeal pleomorphic viruses contain different genome types. Nucleic Acids Res. 2012, 40, 5523–5534. [Google Scholar] [CrossRef] [PubMed]
- Pietilä, M.K.; Atanasova, N.S.; Manole, V.; Liljeroos, L.; Butcher, S.J.; Oksanen, H.M.; Bamford, D.H. Virion architecture unifies globally distributed pleolipoviruses infecting halophilic Archaea. J. Virol. 2012, 86, 5067–5079. [Google Scholar] [CrossRef] [PubMed]
- Pietilä, M.K.; Laurinavičius, S.; Sund, J.; Roine, E.; Bamford, D.H. The single-stranded DNA genome of novel archaeal virus Halorubrum pleomorphic virus 1 is enclosed in the envelope decorated with glycoprotein spikes. J. Virol. 2010, 84, 788–798. [Google Scholar] [CrossRef] [PubMed]
- Pietilä, M.K.; Roine, E.; Paulin, L.; Kalkkinen, N.; Bamford, D.H. An ssDNA virus infecting Archaea: a new lineage of viruses with a membrane envelope. Mol. Microbiol. 2009, 72, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Bath, C.; Cukalac, T.; Porter, K.; Dyall-Smith, M.L. His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus group, Salterprovirus. Virology 2006, 350, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, Y.; Liu, Y.; Du, K.; Xu, S.; Wang, Y.; Krupovic, M.; Chen, X. A novel family of tyrosine integrases encoded by the temperate pleolipovirus SNJ2. Nucleic Acids Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Messina, E.; Sorokin, D.Y.; Kublanov, I.V.; Toshchakov, S.; Lopatina, A.; Arcadi, E.; Smedile, F.; La Spada, G.; La Cono, V.; Yakimov, M.M. Complete genome sequence of ‘Halanaeroarchaeum sulfurireducens’ M27-SA2, a sulfur-reducing and acetate-oxidizing haloarchaeon from the deep-sea hypersaline anoxic lake Medee. Stand. Genomic Sci. 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Cvirkaite-Krupovic, V.; Iranzo, J.; Prangishvili, D.; Koonin, E.V. Viruses of Archaea: Structural, functional, environmental and evolutionary genomics. Virus Res. 2017, 244, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Dybvig, K.; Nowak, J.A.; Sladek, T.L.; Maniloff, J. Identification of an enveloped phage, mycoplasma virus L172, that contains a 14-kilobase single-stranded DNA genome. J. Virol. 1985, 53, 384–390. [Google Scholar] [PubMed]
- Dybvig, K.; Maniloff, J. Integration and lysogeny by an enveloped mycoplasma virus. J. Gen. Virol. 1983, 64 Pt 8, 1781–1785. [Google Scholar] [CrossRef] [PubMed]
- Nolte-’t Hoen, E.; Cremer, T.; Gallo, R.C.; Margolis, L.B. Extracellular vesicles and viruses: Are they close relatives? Proc. Natl. Acad. Sci. USA 2016, 113, 9155–9161. [Google Scholar] [CrossRef] [PubMed]
- Soler, N.; Krupovic, M.; Marguet, E.; Forterre, P. Membrane vesicles in natural environments: A major challenge in viral ecology. ISME J. 2015, 9, 793–796. [Google Scholar] [CrossRef] [PubMed]
- Deatherage, B.L.; Cookson, B.T. Membrane vesicle release in bacteria, eukaryotes, and Archaea: A conserved yet underappreciated aspect of microbial life. Infect. Immun. 2012, 80, 1948–1957. [Google Scholar] [CrossRef] [PubMed]
- Gaudin, M.; Gauliard, E.; Schouten, S.; Houel-Renault, L.; Lenormand, P.; Marguet, E.; Forterre, P. Hyperthermophilic Archaea produce membrane vesicles that can transfer DNA. Environ. Microbiol. Rep. 2013, 5, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Ellen, A.F.; Albers, S.V.; Huibers, W.; Pitcher, A.; Hobel, C.F.; Schwarz, H.; Folea, M.; Schouten, S.; Boekema, E.J.; Poolman, B.; et al. Proteomic analysis of secreted membrane vesicles of archaeal Sulfolobus species reveals the presence of endosome sorting complex components. Extremophiles 2009, 13, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Prangishvili, D.; Holz, I.; Stieger, E.; Nickell, S.; Kristjansson, J.K.; Zillig, W. Sulfolobicins, specific proteinaceous toxins produced by strains of the extremely thermophilic archaeal genus Sulfolobus. J. Bacteriol. 2000, 182, 2985–2988. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, S.; Tschitschko, B.; Zhong, L.; Raftery, M.J.; Cavicchioli, R. A plasmid from an Antarctic haloarchaeon uses specialized membrane vesicles to disseminate and infect plasmid-free cells. Nat. Microbiol. 2017, 2, 1446–1455. [Google Scholar] [CrossRef] [PubMed]
- Atanasova, N.S.; Roine, E.; Oren, A.; Bamford, D.H.; Oksanen, H.M. Global network of specific virus-host interactions in hypersaline environments. Environ. Microbiol. 2012, 14, 426–440. [Google Scholar] [CrossRef] [PubMed]
- Juez, G.; Rodriguez-Valera, F.; Ventosa, A.; Kushner, D.J. Haloarcula hispanica spec. nov. and Haloferax gibbonsii spec. nov., two new species of extemely halophilic archaebacteria. Syst. Appl. Microbiol. 1986, 8, 75–79. [Google Scholar] [CrossRef]
- Nuttall, S.D.; Dyall-Smith, M.L. HF1 and HF2: Novel bacteriophages of halophilic Archaea. Virology 1993, 197, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Dyall-Smith, M. The Halohandbook: Protocols for Haloarchaeal Genetics, Version 7.2; 2009. 144 pages. Available online: http://www.haloarchaea.com/resources/halohandbook/ (accessed on 7 August 2017).
- Adams, M.H. Bacteriophages; Interscience Publishers: New York, NY, USA, 1959; pp. 1–592. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Olkkonen, V.M.; Bamford, D.H. Quantitation of the adsorption and penetration stages of bacteriophage φ6 infection. Virology 1989, 171, 229–238. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [PubMed]
- Kates, M. Techniques of Lipidology: Isolation, Analysis and Identification of Lipids; North-Holland Publishing Co.: Amsterdam, The Netherlands, 1972; pp. 1–464. [Google Scholar]
- Arnold, H.P.; Zillig, W.; Ziese, U.; Holz, I.; Crosby, M.; Utterback, T.; Weidmann, J.F.; Kristjanson, J.K.; Klenk, H.P.; Nelson, K.E.; et al. A novel lipothrixvirus, SIFV, of the extremely thermophilic crenarchaeon Sulfolobus. Virology 2000, 267, 252–266. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Zhang, C.T.; Gao, F. Ori-Finder 2, an integrated tool to predict replication origins in the archaeal genomes. Front. Microbiol. 2014, 5, 482. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Lupas, A.; Van Dyke, M.; Stock, J. Predicting coiled coils from protein sequences. Science 1991, 252, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Boratyn, G.M.; Schaffer, A.A.; Agarwala, R.; Altschul, S.F.; Lipman, D.J.; Madden, T.L. Domain enhanced lookup time accelerated BLAST. Biol. Direct 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics (Oxford, England) 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Arahal, D.R.; Dewhirst, F.E.; Paster, B.J.; Volcani, B.E.; Ventosa, A. Phylogenetic analyses of some extremely halophilic Archaea isolated from Dead Sea water, determined on the basis of their 16S rRNA sequences. Appl. Environ. Microbiol. 1996, 62, 3779–3786. [Google Scholar] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Goker, M. VICTOR: Genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 2017, 33, 3396–3404. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [PubMed]
- Farris, J.S. Estimating Phylogenetic Trees from Distance Matrices. Am. Naturalist 1972, 106, 645–668. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree 1.4.0—A Graphical Viewer of Phylogenetic Trees and a Program for Producing Publication-Ready Figures; Institute of Evolutionary Biology, University of Edinburgh, Ashworth Laboratories: Edinburgh, UK, 2006. [Google Scholar]
- Göker, M.; Garcia-Blazquez, G.; Voglmayr, H.; Telleria, M.T.; Martin, M.P. Molecular taxonomy of phytopathogenic fungi: A case study in Peronospora. PLoS ONE 2009, 4, e6319. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A. Complete genome sequence of DSM 30083 T, the type strain (U5/41 T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genomic Sci. 2014, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Bamford, D.H.; Ravantti, J.J.; Rönnholm, G.; Laurinavičius, S.; Kukkaro, P.; Dyall-Smith, M.; Somerharju, P.; Kalkkinen, N.; Bamford, J.K. Constituents of SH1, a novel lipid-containing virus infecting the halophilic euryarchaeon Haloarcula hispanica. J. Virol. 2005, 79, 9097–9107. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, C.; Xu, J.P.; Yang, Z.L. Molecular characterization of pHRDV1, a new virus-like mobile genetic element closely related to pleomorphic viruses in haloarchaea. Extremophiles 2014, 18, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Dyall-Smith, M.L.; Pfeiffer, F.; Klee, K.; Palm, P.; Gross, K.; Schuster, S.C.; Rampp, M.; Oesterhelt, D. Haloquadratum walsbyi: Limited diversity in a global pond. PLoS ONE 2011, 6, e20968. [Google Scholar] [CrossRef] [PubMed]
- Roine, E.; Kukkaro, P.; Paulin, L.; Laurinavičius, S.; Domanska, A.; Somerharju, P.; Bamford, D.H. New, closely related haloarchaeal viral elements with different nucleic acid types. J. Virol. 2010, 84, 3682–3689. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wu, Z.; Li, M.; Zhang, F.; Zheng, H.; Han, J.; Liu, J.; Zhou, J.; Wang, S.; Xiang, H. Complete genome sequence of Haloarcula hispanica, a Model Haloarchaeon for studying genetics, metabolism, and virus-host interaction. J. Bacteriol. 2011, 193, 6086–6087. [Google Scholar] [CrossRef] [PubMed]
- Abelson, J.; Thomas, C. The anatomy of the T5 bacteriophage DNA molecule. J. Mol. Biol. 1966, 18, 262–288. [Google Scholar] [CrossRef]
- Khan, S.A.; Hayes, S.J.; Wright, E.T.; Watson, R.H.; Serwer, P. Specific single-stranded breaks in mature bacteriophage T7 DNA. Virology 1995, 211, 329–331. [Google Scholar] [CrossRef] [PubMed][Green Version]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atanasova, N.S.; Heiniö, C.H.; Demina, T.A.; Bamford, D.H.; Oksanen, H.M. The Unexplored Diversity of Pleolipoviruses: The Surprising Case of Two Viruses with Identical Major Structural Modules. Genes 2018, 9, 131. https://doi.org/10.3390/genes9030131
Atanasova NS, Heiniö CH, Demina TA, Bamford DH, Oksanen HM. The Unexplored Diversity of Pleolipoviruses: The Surprising Case of Two Viruses with Identical Major Structural Modules. Genes. 2018; 9(3):131. https://doi.org/10.3390/genes9030131
Chicago/Turabian StyleAtanasova, Nina S., Camilla H. Heiniö, Tatiana A. Demina, Dennis H. Bamford, and Hanna M. Oksanen. 2018. "The Unexplored Diversity of Pleolipoviruses: The Surprising Case of Two Viruses with Identical Major Structural Modules" Genes 9, no. 3: 131. https://doi.org/10.3390/genes9030131
APA StyleAtanasova, N. S., Heiniö, C. H., Demina, T. A., Bamford, D. H., & Oksanen, H. M. (2018). The Unexplored Diversity of Pleolipoviruses: The Surprising Case of Two Viruses with Identical Major Structural Modules. Genes, 9(3), 131. https://doi.org/10.3390/genes9030131





