Gene Variations in Cis-Acting Elements between the Taiwan and Prototype Strains of Porcine Epidemic Diarrhea Virus Alter Viral Gene Expression
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Amplification of the Genome and sgmRNA Using RT-PCR and Sequencing Analyses
2.3. Plasmid Constructs
2.4. Analysis of DI RNA and sgm DI RNA Synthesis Using RT-qPCR
2.5. Western Blot Analysis for DI RNA Translation in Cells
2.6. Statistical Analysis and Sequence Alignment
3. Results and Discussion
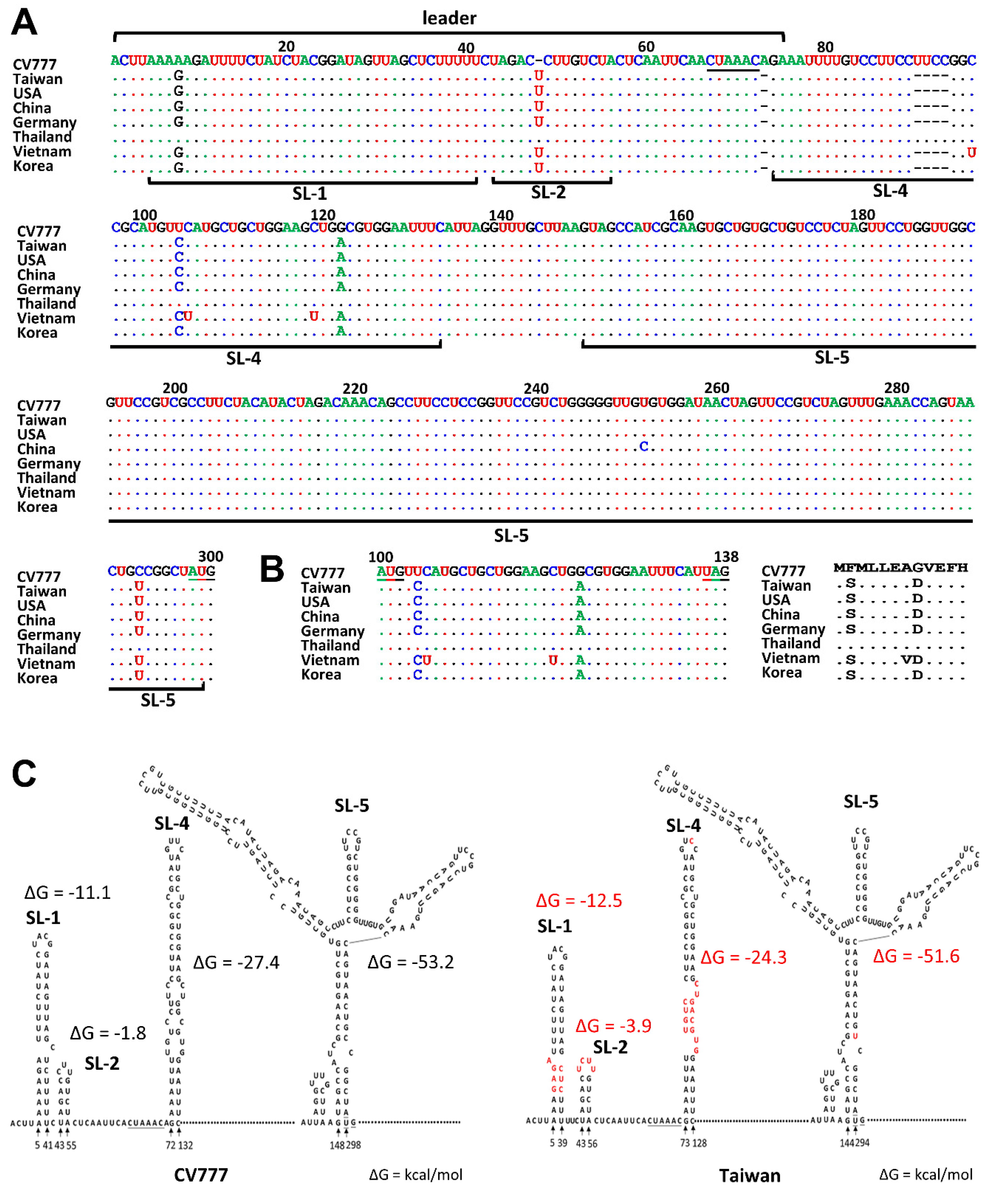
3.1. Comparison of Sequence and Structure in the 5′ UTR between CV777 and TW Strain
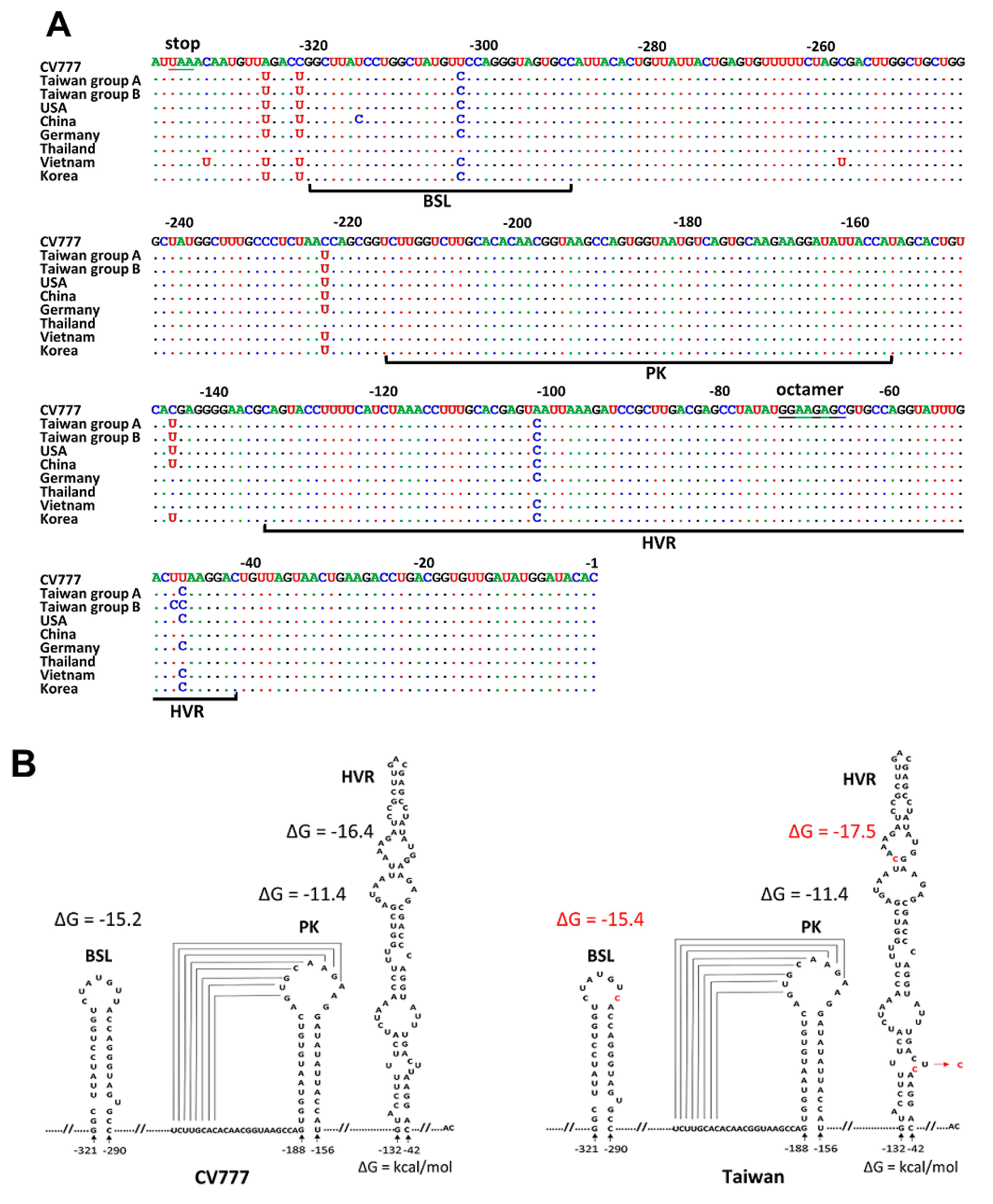
3.2. Comparison of 3′ UTR Sequence and Structure between CV777 and TW Strains
3.3. Identification of a Novel Subgenomic mRNA Species Derived from the 3′ UTR
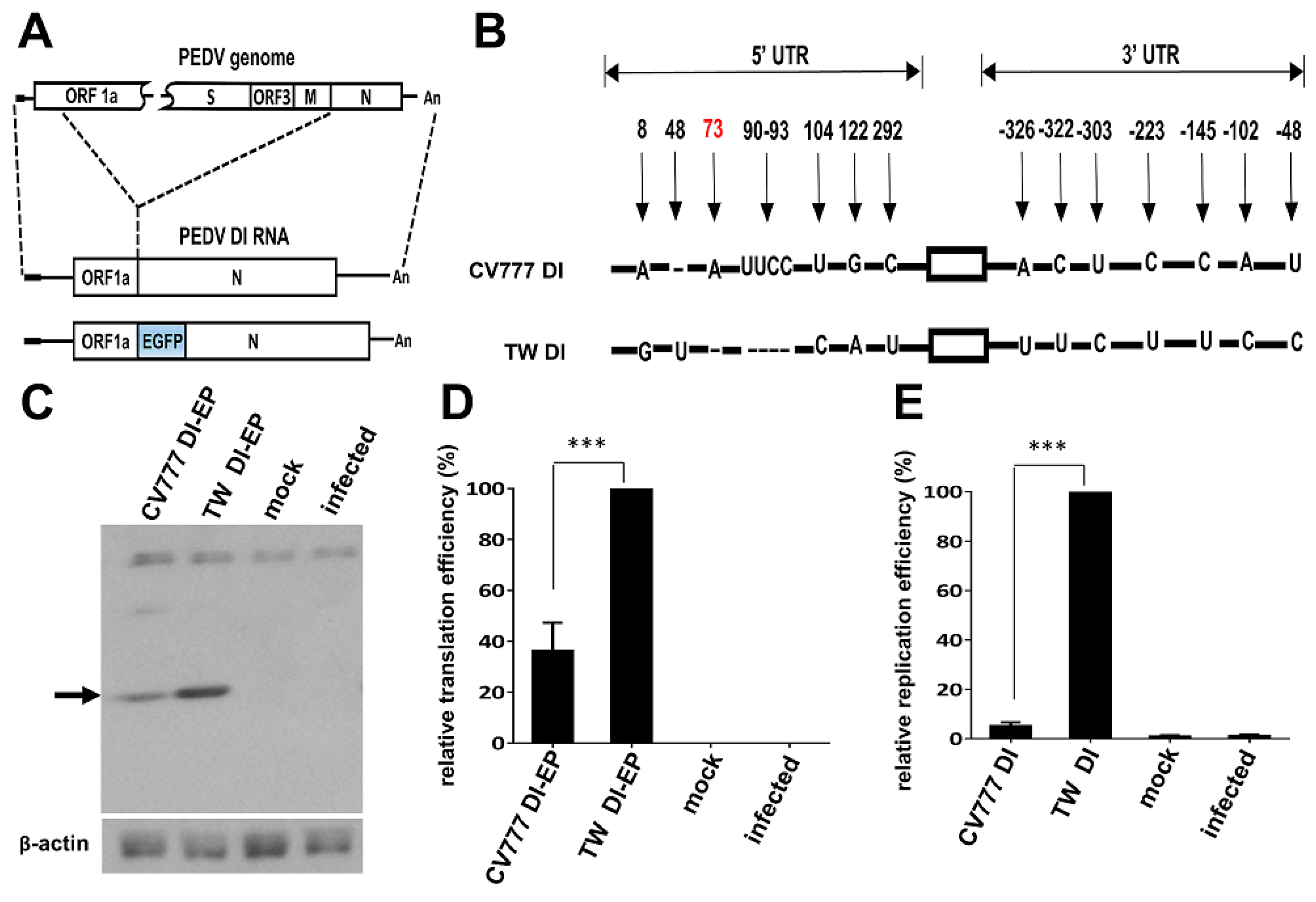
3.4. Effects of the Cis-Acting Element Variations between CV777 and TW Strains on Gene Expression
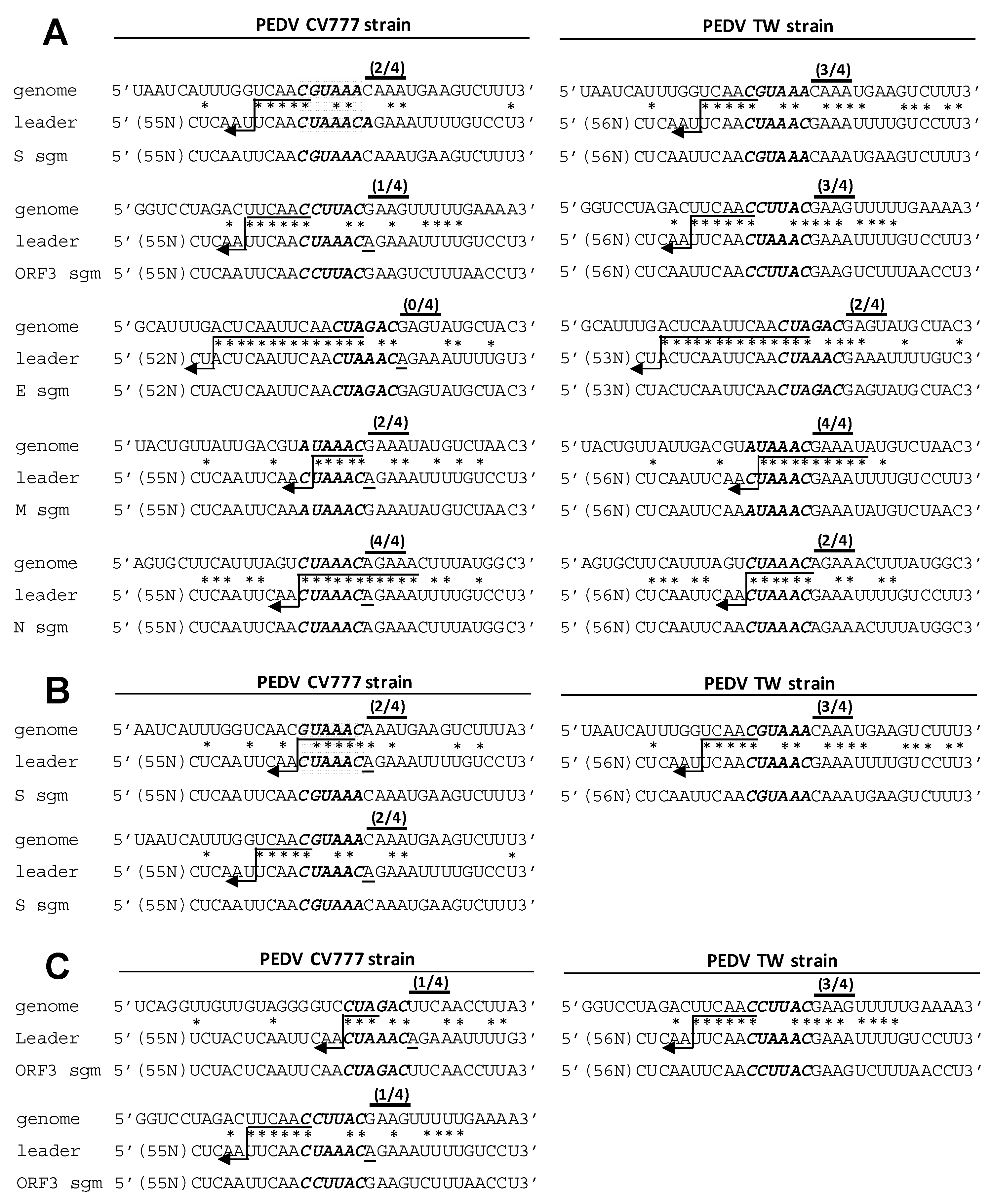
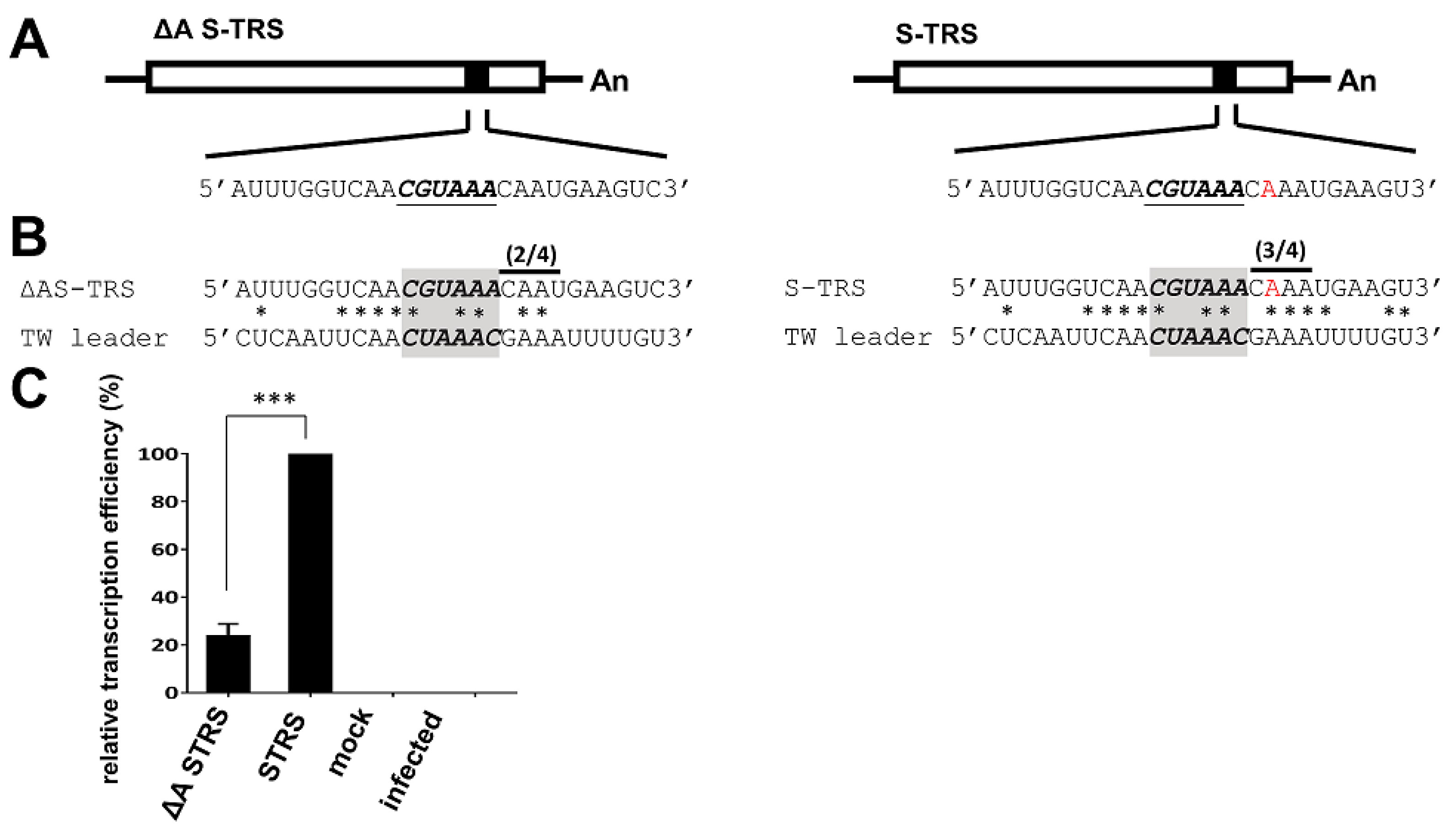
3.5. Analyses of the TRS for CV777 and TW Strain
3.6. Evaluation of the Effect of Variations in TRS between CV777 and TW Strains on sgmRNA Synthesis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- International Committee on Taxonomy of Viruses; King, A.M.Q. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses; Academic Press: London, UK; Waltham, MA, USA, 2012; pp. 1326–1327. [Google Scholar]
- Brian, D.A.; Baric, R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005, 287, 1–30. [Google Scholar] [PubMed]
- Huang, Y.W.; Dickerman, A.W.; Pineyro, P.; Li, L.; Fang, L.; Kiehne, R.; Opriessnig, T.; Meng, X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. Mbio 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Madhugiri, R.; Fricke, M.; Marz, M.; Ziebuhr, J. Coronavirus cis-acting RNA elements. Adv. Virus Res. 2016, 96, 127–163. [Google Scholar] [PubMed]
- Sola, I.; Almazan, F.; Zuniga, S.; Enjuanes, L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015, 2, 265–288. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.Y.; Guan, B.J.; Su, Y.P.; Fan, Y.H.; Brian, D.A. Reselection of a genomic upstream open reading frame in mouse hepatitis coronavirus 5′-untranslated-region mutants. J. Virol. 2014, 88, 846–858. [Google Scholar] [CrossRef] [PubMed]
- Madhugiri, R.; Fricke, M.; Marz, M.; Ziebuhr, J. RNA structure analysis of Alphacoronavirus terminal genome regions. Virus Res. 2014, 194, 76–89. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Leibowitz, J.L. The structure and functions of coronavirus genomic 3′ and 5′ ends. Virus Res. 2015, 206, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Hsue, B.; Hartshorne, T.; Masters, P.S. Characterization of an essential RNA secondary structure in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 2000, 74, 6911–6921. [Google Scholar] [CrossRef] [PubMed]
- Hsue, B.; Masters, P.S. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J. Virol. 1997, 71, 7567–7578. [Google Scholar] [PubMed]
- Williams, G.D.; Chang, R.Y.; Brian, D.A. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J. Virol. 1999, 73, 8349–8355. [Google Scholar] [PubMed]
- Goebel, S.J.; Miller, T.B.; Bennett, C.J.; Bernard, K.A.; Masters, P.S. A hypervariable region within the 3′ cis-acting element of the murine coronavirus genome is nonessential for RNA synthesis but affects pathogenesis. J. Virol. 2007, 81, 1274–1287. [Google Scholar] [CrossRef] [PubMed]
- Sola, I.; Mateos-Gomez, P.A.; Almazan, F.; Zuniga, S.; Enjuanes, L. RNA-RNA and RNA-protein interactions in coronavirus replication and transcription. RNA Biol. 2011, 8, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, S.G.; Sawicki, D.L. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 1998, 440, 215–219. [Google Scholar] [PubMed]
- Sawicki, S.G.; Sawicki, D.L.; Siddell, S.G. A contemporary view of coronavirus transcription. J. Virol. 2007, 81, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Alonso, S.; Izeta, A.; Sola, I.; Enjuanes, L. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J. Virol. 2002, 76, 293–1308. [Google Scholar] [CrossRef]
- Sola, I.; Moreno, J.L.; Zuniga, S.; Alonso, S.; Enjuanes, L. Role of nucleotides immediately flanking, the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis. J. Virol. 2005, 79, 2506–2516. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, T.B.; Boniotti, M.B.; Papetti, A.; Grasland, B.; Frossard, J.P.; Dastjerdi, A.; Hulst, M.; Hanke, D.; Pohlmann, A.; Blome, S.; et al. Full-length genome sequences of porcine epidemic diarrhoea virus strain CV777; Use of NGS to analyse genomic and sub-genomic RNAs. PLoS ONE 2018, 13. [Google Scholar] [CrossRef] [PubMed]
- Song, D.P.; Huang, D.Y.; Peng, Q.; Huang, T.; Chen, Y.J.; Zhang, T.S.; Nie, X.W.; He, H.J.; Wang, P.; Liu, Q.L.; et al. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea viruses associated with outbreaks of severe diarrhea in piglets in Jiangxi, China 2013. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Debouck, P.; Pensaert, M. Experimental infection of pigs with a new porcine enteric coronavirus, CV 777. Am. J. Vet. Res. 1980, 41, 219–223. [Google Scholar] [PubMed]
- Pensaert, M.B.; de Bouck, P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978, 58, 243–247. [Google Scholar] [CrossRef]
- Zimmerman, J.J.; Karriker, L.A.; Ramirez, A.; Schwartz, K.J.; Stevenson, G.W. Diseases of Swine, 10th ed.; John Wiley & Sons: Chichester, West Sussex, UK, 1958. [Google Scholar]
- Lee, C. Porcine epidemic diarrhea virus: An emerging and re-emerging epizootic swine virus (vol 12, 193, 2015). Virol. J. 2016, 13. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.N.; Chung, W.B.; Chang, S.W.; Wen, C.C.; Liu, H.; Chien, C.H.; Chiou, M.T. US-like strain of porcine epidemic diarrhea virus outbreaks in Taiwan, 2013–2014. J. Vet. Med. Sci. 2014, 76, 1297–1299. [Google Scholar] [CrossRef] [PubMed]
- Li, B.X.; Ge, J.W.; Li, Y.J. Porcine aminopeptidase N is a functional receptor for the PEDV coronavirus. Virology 2007, 365, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Nam, E.; Lee, C. Contribution of the porcine aminopeptidase N (CD13) receptor density to porcine epidemic diarrhea virus infection. Vet. Microbiol. 2010, 144, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.H.; Bae, J.L.; Kang, T.J.; Kim, J.; Chung, G.H.; Lim, C.W.; Laude, H.; Yang, M.S.; Jang, Y.S. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol. Cells 2002, 14, 295–299. [Google Scholar] [PubMed]
- Kang, T.J.; Kang, K.H.; Kim, J.A.; Kwon, T.H.; Jang, Y.S.; Yang, M.S. High-level expression of the neutralizing epitope of porcine epidemic diarrhea virus by a tobacco mosaic virus-based vector. Protein Expres Purif. 2004, 38, 129–135. [Google Scholar] [CrossRef]
- Oszvald, M.; Kang, T.J.; Tomoskozi, S.; Tamas, C.; Tamas, L.; Kim, T.G.; Yang, M.S. Expression of a synthetic neutralizing epitope of porcine epidemic diarrhea virus fused with synthetic B subunit of Escherichia coli heat labile enterotoxin in rice endosperm. Mol. Biotechnol. 2007, 35, 215–223. [Google Scholar] [CrossRef]
- Chiou, H.Y.; Huang, Y.L.; Deng, M.C.; Chang, C.Y.; Jeng, C.R.; Tsai, P.S.; Yang, C.; Pang, V.F.; Chang, H.W. Phylogenetic analysis of the spike (S) gene of the new variants of porcine epidemic diarrhoea virus in Taiwan. Transbound. Emerg. Dis. 2017, 64, 157–166. [Google Scholar] [CrossRef]
- Chen, Q.; Li, G.W.; Stasko, J.; Thomas, J.T.; Stensland, W.R.; Pillatzki, A.E.; Gauger, P.C.; Schwartz, K.J.; Madson, D.; Yoon, K.J.; et al. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2014, 52, 234–243. [Google Scholar] [CrossRef]
- Liao, W.Y.; Ke, T.Y.; Wu, H.Y. The 3′-terminal 55 nucleotides of bovine coronavirus defective interfering RNA harbor cis-acting elements required for both negative- and positive-strand RNA synthesis. PLoS ONE 2014, 9, e98422. [Google Scholar] [CrossRef]
- Chang, Y.C.; Kao, C.F.; Chang, C.Y.; Jeng, C.R.; Tsai, P.S.; Pang, V.F.; Chiou, H.Y.; Peng, J.Y.; Cheng, I.C.; Chang, H.W. Evaluation and comparison of the pathogenicity and host immune responses induced by a G2b Taiwan porcine epidemic diarrhea virus (Strain Pintung 52) and its highly cell-culture passaged strain in conventional 5-week-old pigs. Viruses 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, M.; Wyler, R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 1988, 26, 2235–2239. [Google Scholar] [PubMed]
- Kweon, C.H.; Kwon, B.J.; Lee, J.G.; Kwon, G.O.; Kang, Y.B. Derivation of attenuated porcine epidemic diarrhea virus (PEDV) as vaccine candidate. Vaccine 1999, 17, 2546–2553. [Google Scholar] [CrossRef]
- Zuker, M. Prediction of RNA secondary structure by energy minimization. Methods Mol. Biol. 1994, 25, 267–294. [Google Scholar] [PubMed]
- Cruz, J.L.G.; Sola, I.; Becares, M.; Alberca, B.; Plana, J.; Enjuanes, L.; Zuniga, S. Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog. 2011, 7. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.G.; Nixon, K.S.; Senanayake, S.D.; Brian, D.A. An RNA stem-loop within the bovine coronavirus nsp1 coding region is a cis-acting element in defective interfering RNA replication. J. Virol. 2007, 81, 7716–7724. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.Y.; Hofmann, M.A.; Sethna, P.B.; Brian, D.A. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J. Virol. 1994, 68, 8223–8231. [Google Scholar]
- Raman, S.; Brian, D.A. Stem-loop IV in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 2005, 79, 12434–12446. [Google Scholar] [CrossRef]
- Spagnolo, J.F.; Hogue, B.G. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 2000, 74, 5053–5065. [Google Scholar] [CrossRef]
- Wu, H.Y.; Brian, D.A. 5′-proximal hot spot for an inducible positive-to-negative-strand template switch by coronavirus RNA-dependent RNA polymerase. J. Virol. 2007, 81, 3206–3215. [Google Scholar] [CrossRef]
- Chang, R.Y.; Krishnan, R.; Brian, D.A. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J. Virol. 1996, 70, 2720–2729. [Google Scholar] [PubMed]
- Wu, H.Y.; Guy, J.S.; Yoo, D.; Vlasak, R.; Urbach, E.; Brian, D.A. Common RNA replication signals exist among group 2 coronaviruses: Evidence for in vivo recombination between animal and human coronavius molecules. Virology 2003, 315, 174–183. [Google Scholar] [CrossRef]
- Johnson, R.F.; Feng, M.; Liu, P.H.; Millership, J.J.; Yount, B.; Baric, R.S.; Leibowitz, J.L. Effect of mutations in the mouse hepatitis virus 3‘(+)42 protein binding element on RNA replication. J. Virol. 2005, 79, 14570–14585. [Google Scholar] [CrossRef] [PubMed]
- Tobler, K.; Ackermann, M. PEDV leader sequence and junction sites. Adv. Exp. Med. Biol. 1995, 380, 541–542. [Google Scholar] [PubMed]
- Li, C.; Li, Z.; Zou, Y.; Wicht, O.; van Kuppeveld, F.J.; Rottier, P.J.; Bosch, B.J. Manipulation of the porcine epidemic diarrhea virus genome using targeted RNA recombination. PLoS ONE 2013, 8, e69997. [Google Scholar] [CrossRef] [PubMed]
- Ozdarendeli, A.; Ku, S.; Rochat, S.; Williams, G.D.; Senanayake, S.D.; Brian, D.A. Downstream sequences influence the choice between a naturally occurring noncanonical and closely positioned upstream canonical heptameric fusion motif during bovine coronavirus subgenomic mRNA synthesis. J. Virol. 2001, 75, 7362–7374. [Google Scholar] [CrossRef] [PubMed]
- Pasternak, A.O.; Gultyaev, A.P.; Spaan, W.J.; Snijder, E.J. Genetic manipulation of arterivirus alternative mRNA leader-body junction sites reveals tight regulation of structural protein expression. J. Virol. 2000, 74, 11642–11653. [Google Scholar] [CrossRef]
- Pasternak, A.O.; Spaan, W.J.; Snijder, E.J. Regulation of relative abundance of arterivirus subgenomic mRNAs. J. Virol. 2004, 78, 8102–8113. [Google Scholar] [CrossRef]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, T.-L.; Su, C.-C.; Hsieh, C.-C.; Lin, C.-N.; Chang, H.-W.; Lo, C.-Y.; Lin, C.-H.; Wu, H.-Y. Gene Variations in Cis-Acting Elements between the Taiwan and Prototype Strains of Porcine Epidemic Diarrhea Virus Alter Viral Gene Expression. Genes 2018, 9, 591. https://doi.org/10.3390/genes9120591
Tsai T-L, Su C-C, Hsieh C-C, Lin C-N, Chang H-W, Lo C-Y, Lin C-H, Wu H-Y. Gene Variations in Cis-Acting Elements between the Taiwan and Prototype Strains of Porcine Epidemic Diarrhea Virus Alter Viral Gene Expression. Genes. 2018; 9(12):591. https://doi.org/10.3390/genes9120591
Chicago/Turabian StyleTsai, Tsung-Lin, Chen-Chang Su, Ching-Chi Hsieh, Chao-Nan Lin, Hui-Wen Chang, Chen-Yu Lo, Ching-Houng Lin, and Hung-Yi Wu. 2018. "Gene Variations in Cis-Acting Elements between the Taiwan and Prototype Strains of Porcine Epidemic Diarrhea Virus Alter Viral Gene Expression" Genes 9, no. 12: 591. https://doi.org/10.3390/genes9120591
APA StyleTsai, T.-L., Su, C.-C., Hsieh, C.-C., Lin, C.-N., Chang, H.-W., Lo, C.-Y., Lin, C.-H., & Wu, H.-Y. (2018). Gene Variations in Cis-Acting Elements between the Taiwan and Prototype Strains of Porcine Epidemic Diarrhea Virus Alter Viral Gene Expression. Genes, 9(12), 591. https://doi.org/10.3390/genes9120591





