Functional Evolution of Avian RIG-I-Like Receptors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Collection and Alignment
2.2. Six-Class Assessment of Sequence Face Quality
2.3. Datasets Preparation
2.4. Molecular Evolutionary Analyses
2.5. Correlation Analysis of Species-to-Ancestor dN/dS of Avian RLRs vs. ERV Abundance
3. Results
3.1. Face Quality of Predicted CDSs
3.2. Gene-Wide Conservation and dN/dS Levels of Avian RLRs
3.3. Positive Selection and Positively Selected Sites in Avian RLRs
3.4. Changes of Selection Intensity in Avian RLR Evolution
3.5. Correlation between Functional Constraint of Avian RLR Genes and ERV Abundance
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Medzhitov, R.; Janeway, C.A. Innate immunity: The virtues of a nonclonal system of recognition. Cell 1997, 91, 295–298. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Loo, Y.M.; Gale, M. Immune Signaling by RIG-I-like Receptors. Immunity 2011, 34, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, M.; Fujita, T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 2009, 227, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Hirai, R.; Loo, Y.-M.; Owen, D.; Johnson, C.L.; Sinha, S.C.; Akira, S.; Fujita, T.; Gale, M. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. USA 2007, 104, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Takeuchi, O.; Mikamo-Satoh, E.; Hirai, R.; Kawai, T.; Matsushita, K.; Hiiragi, A.; Dermody, T.S.; Fujita, T.; Akira, S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid–inducible gene-I and melanoma differentiation–associated gene 5. J. Exp. Med. 2008, 205, 1601–1610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bamming, D.; Horvath, C.M. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I, and LGP2. J. Biol. Chem. 2009, 284, 9700–9712. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.; Kato, H.; Kumagai, Y.; Yoneyama, M.; Sato, S.; Matsushita, K.; Tsujimura, T.; Fujita, T.; Akira, S.; Takeuchi, O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc. Natl. Acad. Sci. USA 2010, 107, 1512–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venkataraman, T.; Valdes, M.; Elsby, R.; Kakuta, S.; Caceres, G.; Saijo, S.; Iwakura, Y.; Barber, G.N. Loss of DExD/H Box RNA Helicase LGP2 Manifests Disparate Antiviral Responses. J. Immunol. 2007, 178, 6444–6455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Ranjith-Kumar, C.T.; Brooks, M.T.; Dharmaiah, S.; Herr, A.B.; Kao, C.; Li, P. The RIG-I-like receptor LGP2 recognizes the termini of double-stranded RNA. J. Biol. Chem. 2009, 284, 13881–13891. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Chang, M.; Nie, P.; Secombes, C.J. Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol. Biol. 2009, 9, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barber, M.R.W.; Aldridge, J.R.; Webster, R.G.; Magor, K.E. Association of RIG-I with innate immunity of ducks to influenza. Proc. Natl. Acad. Sci. USA 2010, 107, 5913–5918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Ding, N.; Ding, S.S.; Yu, S.; Meng, C.; Chen, H.; Qiu, X.; Zhang, S.; Yu, Y.; Zhan, Y.; et al. Goose RIG-I functions in innate immunity against Newcastle disease virus infections. Mol. Immunol. 2013, 53, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Cheng, A.; Wang, M. Innate sensing of viruses by pattern recognition receptors in birds. Vet. Res. 2013, 44, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liniger, M.; Summerfield, A.; Zimmer, G.; McCullough, K.C.; Ruggli, N. Chicken Cells Sense Influenza A Virus Infection through MDA5 and CARDIF-signaling Involving LGP2. J. Virol. 2012, 86, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Watanabe, C.; Suzuki, Y.; Tanikawa, T.; Uchida, Y.; Saito, T. Chicken MDA5 senses short double-stranded RNA with implications for antiviral response against avian influenza viruses in chicken. J. Innate Immun. 2014, 6, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Xu, W.; Yan, L.; Liu, J.; Rui, L.; Xiao, X.; Yu, X.; Lu, Y.; Li, Z. Function of duck RIG-I in induction of antiviral response against IBDV and avian influenza virus on chicken cells. Virus Res. 2014, 191, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Shao, Q.; Zang, Y.; Guo, Q.; Zhang, Y.; Li, Z. Pigeon RIG-I function in innate immunity against H9N2 IAV and IBDV. Viruses 2015, 7, 4131–4151. [Google Scholar] [CrossRef] [PubMed]
- Hurst, T.P.; Magiorkinis, G. Activation of the innate immune response by endogenous retroviruses. J. Gen. Virol. 2015, 96, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Kassiotis, G.; Stoye, J.P. Immune responses to endogenous retroelements: Taking the bad with the good. Nat. Rev. Immunol. 2016, 16, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Hayward, A.; Katzourakis, A. Endogenous retroviruses. Curr. Biol. 2015, 25, R644–R646. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.K.; Chitti, S. V Molecular Biology and Pathogenesis of Retroviruses. In Advances in Molecular Retrovirology; InTech: London, UK, 2016; ISBN 978-953-51-2261-6. [Google Scholar]
- Bannert, N.; Kurth, R. The Evolutionary Dynamics of Human Endogenous Retroviral Families. Annu. Rev. Genom. Hum. Genet. 2006, 7, 149–173. [Google Scholar] [CrossRef] [PubMed]
- Dewannieux, M.; Harper, F.; Richaud, A.; Letzelter, C.; Ribet, D.; Pierron, G.; Heidmann, T. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 2006, 16, 1548–1556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magiorkinis, G.; Gifford, R.J.; Katzourakis, A.; De Ranter, J.; Belshaw, R. Env-less endogenous retroviruses are genomic superspreaders. Proc. Natl. Acad. Sci. USA 2012, 109, 7385–7390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belshaw, R.; Pereira, V.; Katzourakis, A.; Talbot, G.; Paces, J.; Burt, A.; Tristem, M. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. USA 2004, 101, 4894–4899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. International Human Genome Sequencing, C. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, E.M.; McDonald, J.F. Long terminal repeat retrotransposons of Mus musculus. Genome Biol. 2004, 5, R14. [Google Scholar] [CrossRef] [PubMed]
- Berg, R.K.; Melchjorsen, J.; Rintahaka, J.; Diget, E.; Søby, S.; Horan, K.A.; Gorelick, R.J.; Matikainen, S.; Larsen, C.S.; Ostergaard, L.; et al. Genomic HIV RNA induces innate immune responses through RIG-I-dependent sensing of secondary-structured RNA. PLoS ONE 2012, 7, e29291. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Zhao, W.; Huang, Z.; Jarvis, E.D.; Gilbert, M.T.P.; Walker, P.J.; Holmes, E.C.; Zhang, G. Low frequency of paleoviral infiltration across the avian phylogeny. Genome Biol. 2014, 15, 539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.; Blackshields, G.; Brown, N.; Chenna, R.; McGettigan, P.; McWilliam, H.; Valentin, F.; Wallace, I.; Wilm, A.; Lopez, R.; et al. ClustalW and ClustalX version 2. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Valdar, W.S.J. Scoring residue conservation. Proteins Struct. Funct. Genet. 2002, 48, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Kosakovsky Pond, S.L.; Frost, S.D.W. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005, 22, 1208–1222. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [PubMed]
- Scheffler, K.; Martin, D.P.; Seoighe, C. Robust inference of positive selection from recombining coding sequences. Bioinformatics 2006, 22, 2493–2499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosakovsky Pond, S.L.; Murrell, B.; Fourment, M.; Frost, S.D.W.; Delport, W.; Scheffler, K. A random effects branch-site model for detecting episodic diversifying selection. Mol. Biol. Evol. 2011, 28, 3033–3043. [Google Scholar] [CrossRef] [PubMed]
- Holm, S. A Simple Sequentially Rejective Multiple Test Procedure. Scand. J Stat. 1979, 6, 65–70. [Google Scholar] [CrossRef]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Kosakovsky Pond, S.L. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef] [PubMed]
- Wertheim, J.O.; Murrell, B.; Smith, M.D.; Pond, S.L.K.; Scheffler, K. RELAX: Detecting relaxed selection in a phylogenetic framework. Mol. Biol. Evol. 2015, 32, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, E.D.; Mirarab, S.; Aberer, A.J.; Li, B.; Houde, P.; Li, C.; Ho, S.Y.W.; Faircloth, B.C.; Nabholz, B.; Howard, J.T.; et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 2014, 346, 1320–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- BirdTree. Available online: https://birdtree.org (accessed on 4 January 2018).
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Meredith, R.W.; Janečka, J.E.; Gatesy, J.; Ryder, O.A.; Fisher, C.A.; Teeling, E.C.; Goodbla, A.; Eizirik, E.; Simão, T.L.L.; Stadler, T.; et al. Impacts of the Cretaceous Terrestrial Revolution and KPg Extinction on Mammal Diversification. Science 2011, 334, 521–524. [Google Scholar] [CrossRef] [PubMed]
- Kosakovsky Pond, S.L.; Frost, S.D.W.; Muse, S.V. HyPhy: Hypothesis testing using phylogenies. Bioinformatics 2005, 21, 676–679. [Google Scholar] [CrossRef] [PubMed]
- Delport, W.; Poon, A.F.Y.; Frost, S.D.W.; Kosakovsky Pond, S.L. Datamonkey 2010: A suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 2010, 26, 2455–2457. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Phylogenies and the Comparative Method. Am. Nat. 1985, 125, 1–15. [Google Scholar] [CrossRef]
- Sukumaran, J.; Holder, M.T. DendroPy: A Python library for phylogenetic computing. Bioinformatics 2010, 26, 1569–1571. [Google Scholar] [CrossRef] [PubMed]
- Miranzo-Navarro, D.; Magor, K.E. Activation of duck RIG-I by TRIM25 is independent of anchored ubiquitin. PLoS ONE 2014, 9, e86968. [Google Scholar] [CrossRef]
- Zhang, G.; Li, C.; Li, Q.; Li, B.; Larkin, D.M.; Lee, C.; Storz, J.F.; Antunes, A.; Greenwold, M.J.; Meredith, R.W.; et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 2014, 346, 1311–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cagliani, R.; Forni, D.; Tresoldi, C.; Pozzoli, U.; Filippi, G.; Rainone, V.; De Gioia, L.; Clerici, M.; Sironi, M. RIG-I-like receptors evolved adaptively in mammals, with parallel evolution at LGP2 and RIG-I. J. Mol. Biol. 2014, 426, 1351–1365. [Google Scholar] [CrossRef] [PubMed]
- Dayhoff, M.O. Observed frequencies of amino acid replacements between closely related proteins. In Atlas of Protein Sequence and Structure; National Biomedical Research Foundation: Washington, DC, USA, 1978; Volume 5. [Google Scholar]
- Uchikawa, E.; Lethier, M.; Malet, H.; Brunel, J.; Gerlier, D.; Cusack, S. Structural Analysis of dsRNA Binding to Anti-viral Pattern Recognition Receptors LGP2 and MDA5. Mol. Cell 2016, 62, 586–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kowalinski, E.; Lunardi, T.; McCarthy, A.A.; Louber, J.; Brunel, J.; Grigorov, B.; Gerlier, D.; Cusack, S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 2011, 147, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Gack, M.U.; Kirchhofer, A.; Shin, Y.C.; Inn, K.-S.; Liang, C.; Cui, S.; Myong, S.; Ha, T.; Hopfner, K.-P.; Jung, J.U. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc. Natl. Acad. Sci. USA 2008, 105, 16743–16748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 2016, 351, 1083–1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
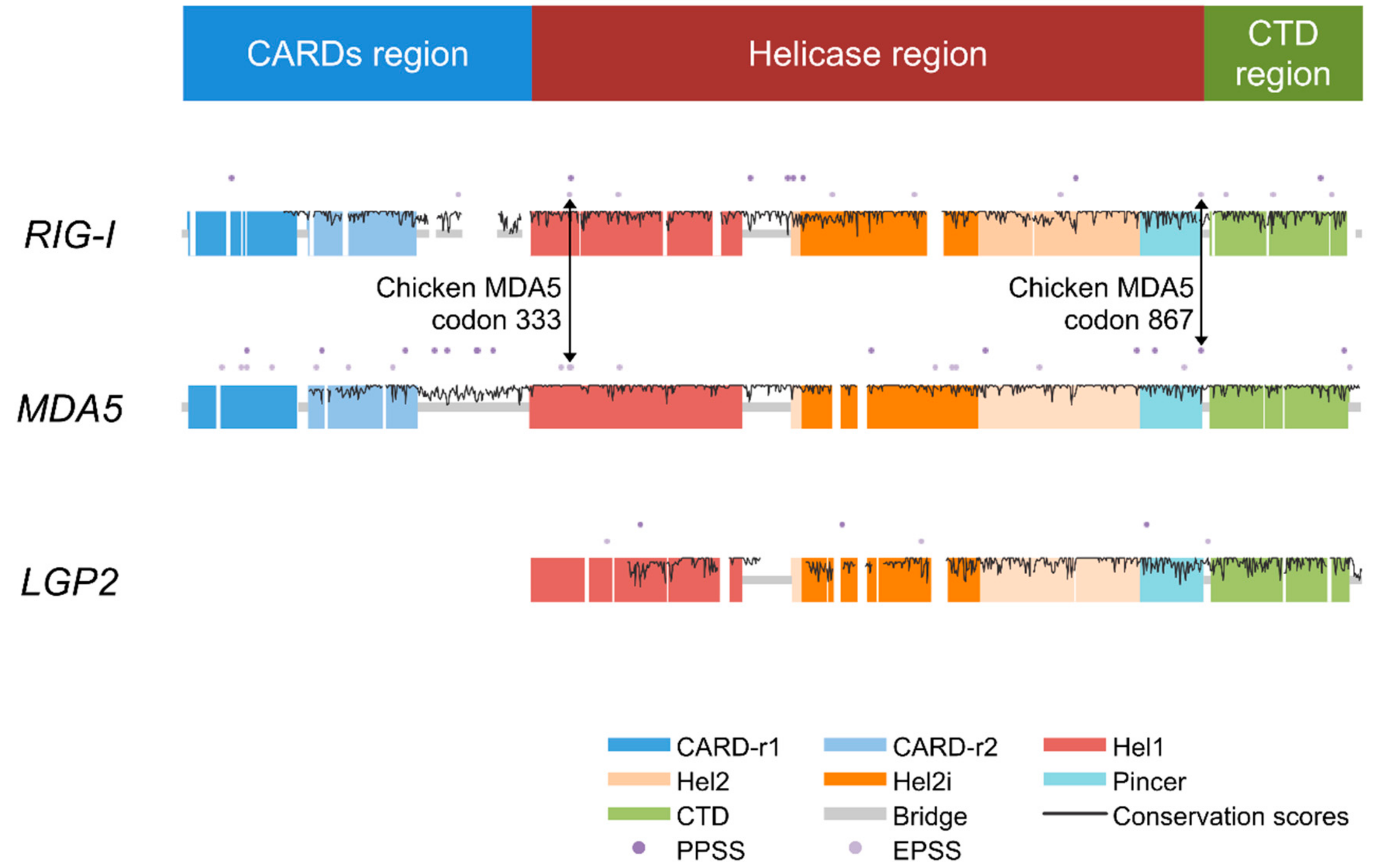
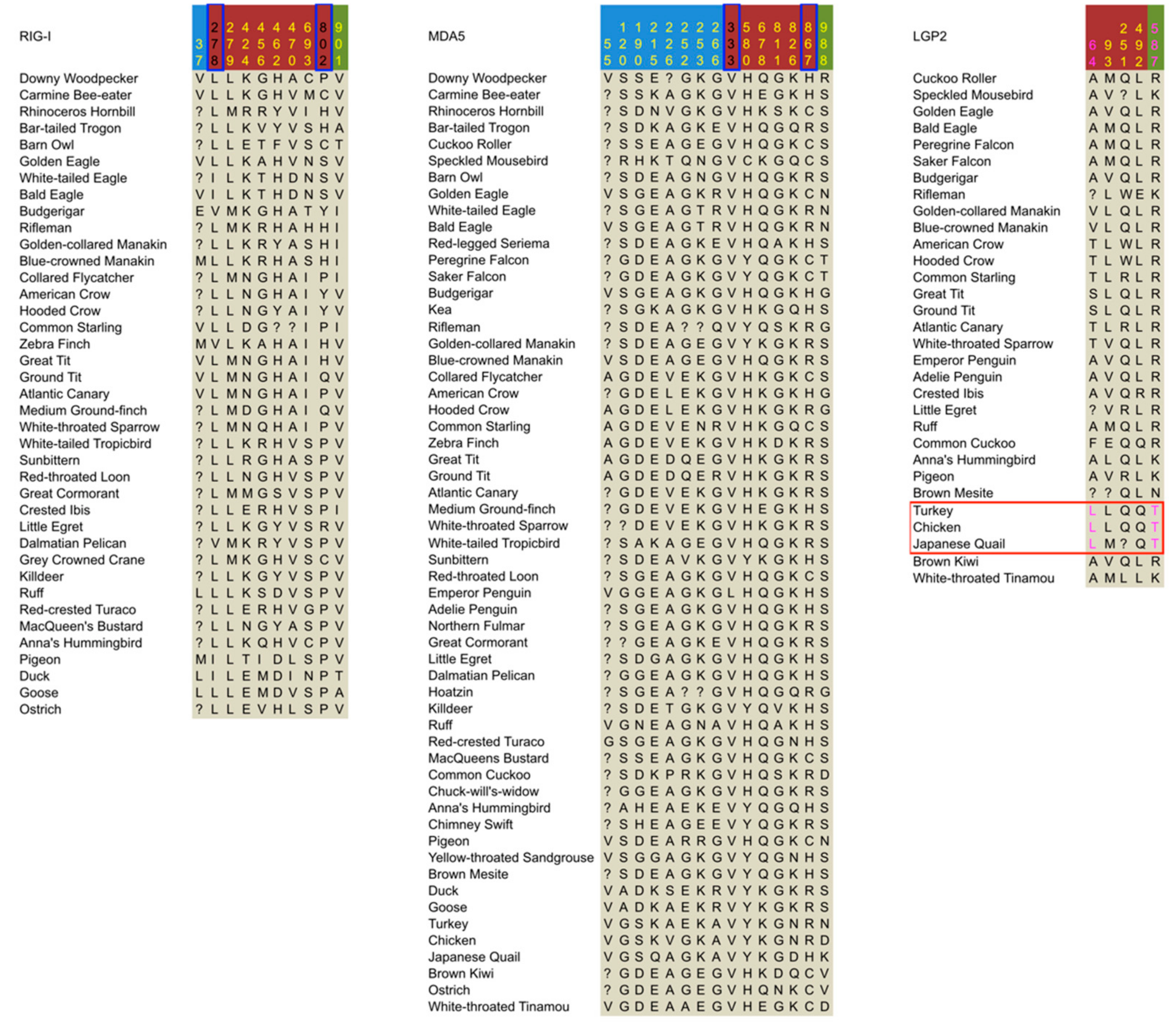
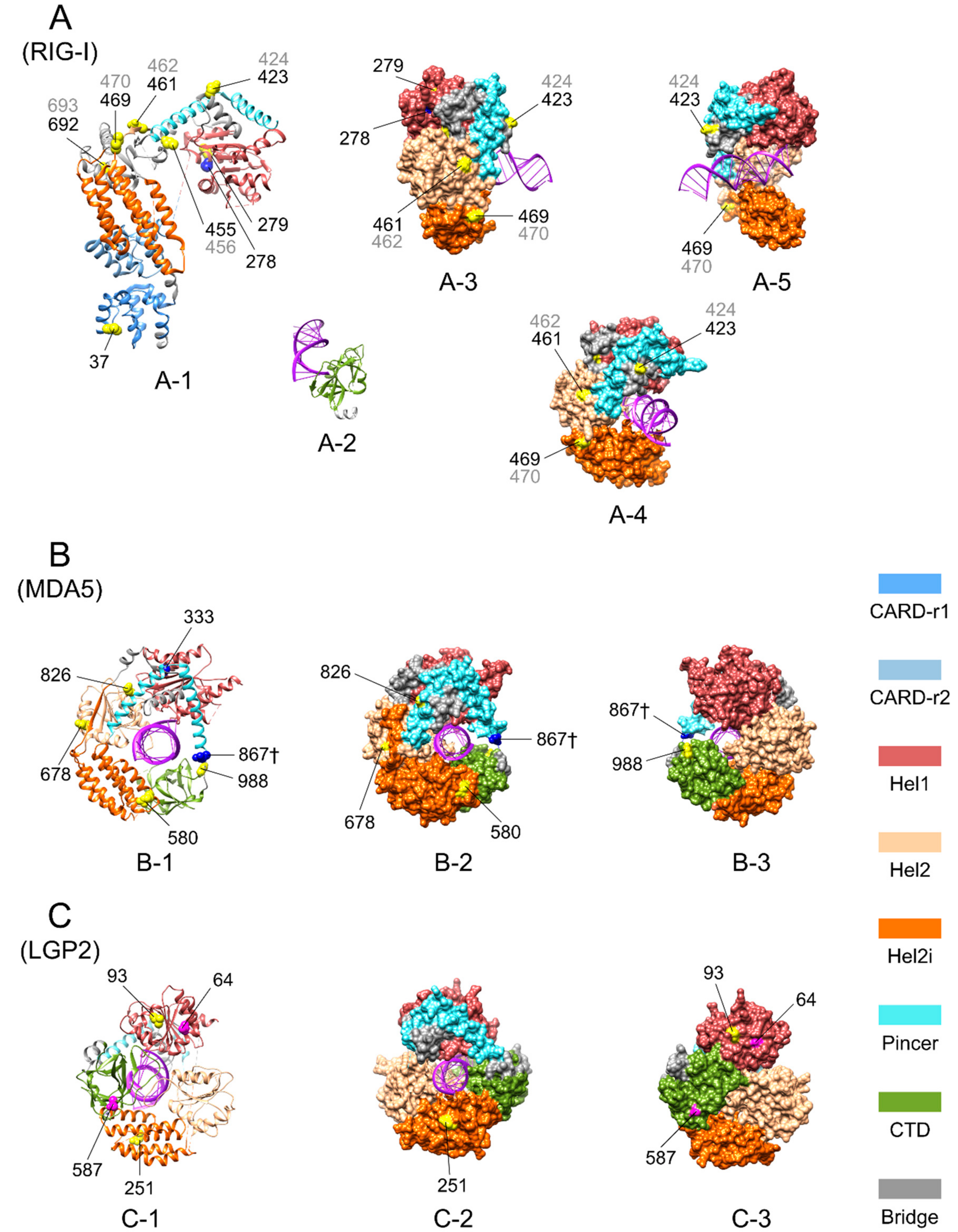

| Domain Regions | RIG-I | MDA5 | LGP2 |
|---|---|---|---|
| Three regions | |||
| Avg. ± S.D. | 0.913 ± 0.114 | 0.933 ± 0.096 | N/A |
| Invariant/all | 37% | 43% | |
| Without CARDs region | |||
| Avg. ± S.D. | 0.915 ± 0.112 | 0.953 ± 0.080 | 0.881 ± 0.139 |
| Invariant/all | 37% | 50% | 36% |
| CARDs region | |||
| Avg. ± S.D. | 0.903 ± 0.121 | 0.861 ± 0.113 | N/A |
| Invariant/all | 35% | 15% | |
| Helicase region | |||
| Avg. ± S.D. | 0.914 ± 0.111 | 0.955 ± 0.079 | 0.883 ± 0.138 |
| Invariant/all | 36% | 53% | 35% |
| CTD region | |||
| Avg. ± S.D. | 0.916 ± 0.116 | 0.942 ± 0.085 | 0.877 ± 0.141 |
| Invariant/all | 41% | 39% | 39% |
| Domain Regions and Datasets | No. of Species | dN/dS (95% Confidence Interval) | ||||
|---|---|---|---|---|---|---|
| RIG-I/MDA5/LGP2 | RIG-I | MDA5 | LGP2 | |||
| Dataset 2 and literature | ||||||
| All regions | Birds (dataset 2) | 39/57/31 | † | 0.385 | 0.369 | 0.222 |
| (0.368, 0.401) | (0.355, 0.383) | (0.211, 0.234) | ||||
| Mammals [54] | 42/46/46 | ‡ | 0.403 | 0.293 | 0.221 | |
| (0.390, 0.416) | (0.284, 0.302) | (0.213, 0.230) | ||||
| Dataset 3 | ||||||
| Three regions | Birds | 9/12/7 | † | 0.350 | 0.384 | — |
| (0.325, 0.376) | (0.357, 0.411) | |||||
| ‡ | 0.237 | 0.237 | — | |||
| Mammals | 7/10/8 | † | 0.408 | 0.330 | — | |
| (0.374, 0.443) | (0.306, 0.356) | |||||
| ‡ | 0.283 | 0.225 | — | |||
| Without CARDs region | Birds | 9/12/7 | † | 0.352 | 0.284 | 0.200 |
| (0.323, 0.383) | (0.257, 0.312) | (0.180, 0.220) | ||||
| ‡ | 0.237 | 0.185 | 0.253 | |||
| Mammals | 7/10/8 | † | 0.370 | 0.260 | 0.240 | |
| (0.334, 0.410) | (0.234, 0.288) | (0.219, 0.264) | ||||
| ‡ | 0.262 | 0.180 | 0.254 | |||
| Sites | No. Codons (Proportion to the Alignment) | |||||
|---|---|---|---|---|---|---|
| RIG-I | MDA5 | LGP2 | ||||
| Alignment | 933 | (100%) | 1025 | (100%) | 677 | (100%) |
| PPSS | 8 | (0.9%) | 14 | (1.4%) | 3 | (0.4%) |
| EPSS | 10 | (1.1%) | 17 | (1.7%) | 3 | (0.4%) |
| Total | 18 | (1.9%) | 31 | (3.0%) | 6 | (0.9%) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, W.; Satta, Y. Functional Evolution of Avian RIG-I-Like Receptors. Genes 2018, 9, 456. https://doi.org/10.3390/genes9090456
Zheng W, Satta Y. Functional Evolution of Avian RIG-I-Like Receptors. Genes. 2018; 9(9):456. https://doi.org/10.3390/genes9090456
Chicago/Turabian StyleZheng, Wanjing, and Yoko Satta. 2018. "Functional Evolution of Avian RIG-I-Like Receptors" Genes 9, no. 9: 456. https://doi.org/10.3390/genes9090456
APA StyleZheng, W., & Satta, Y. (2018). Functional Evolution of Avian RIG-I-Like Receptors. Genes, 9(9), 456. https://doi.org/10.3390/genes9090456





