Advances on Aptamers against Protozoan Parasites
Abstract
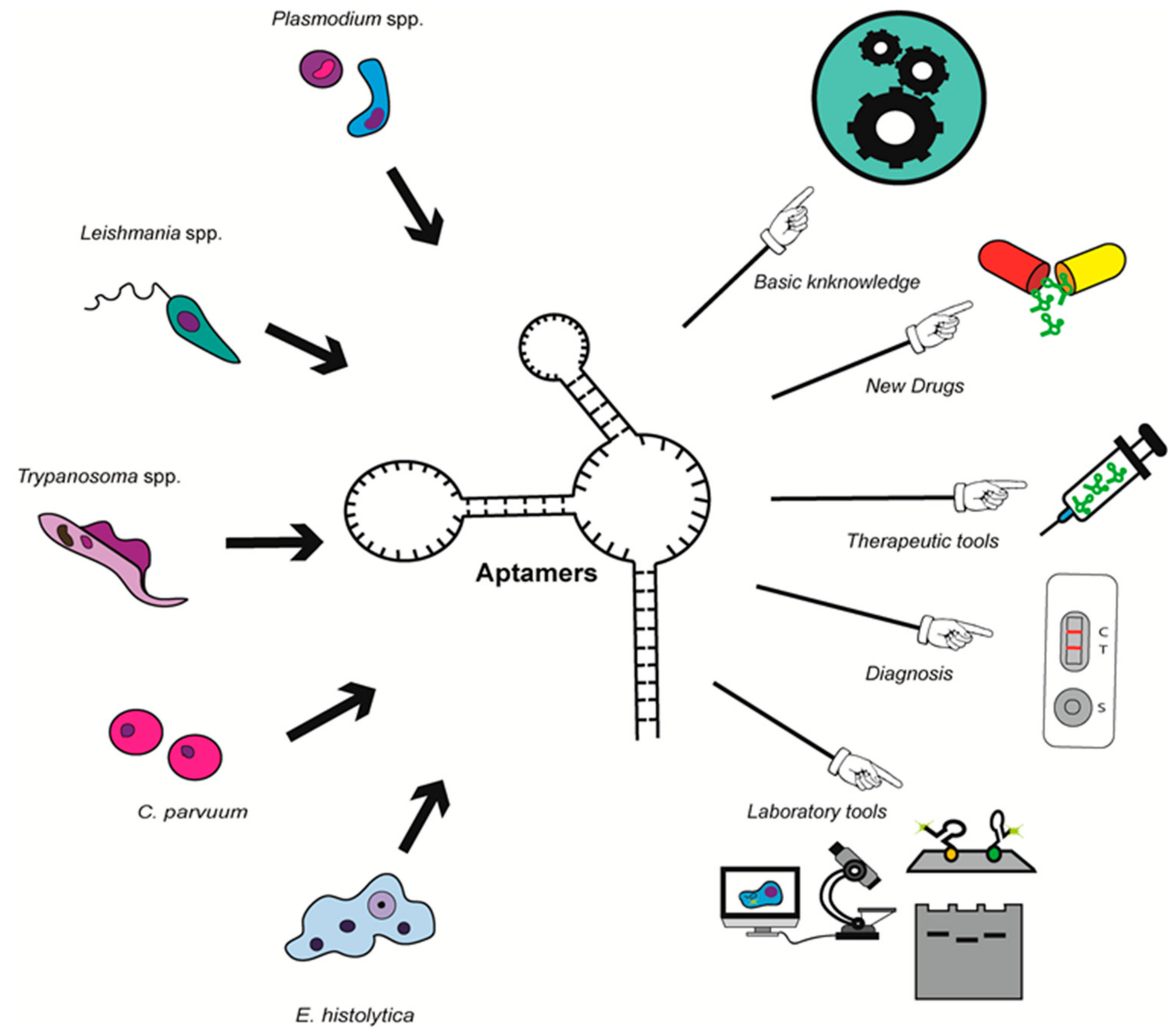
1. Introduction
2. Generation and Modification of DNA/RNA Aptamers
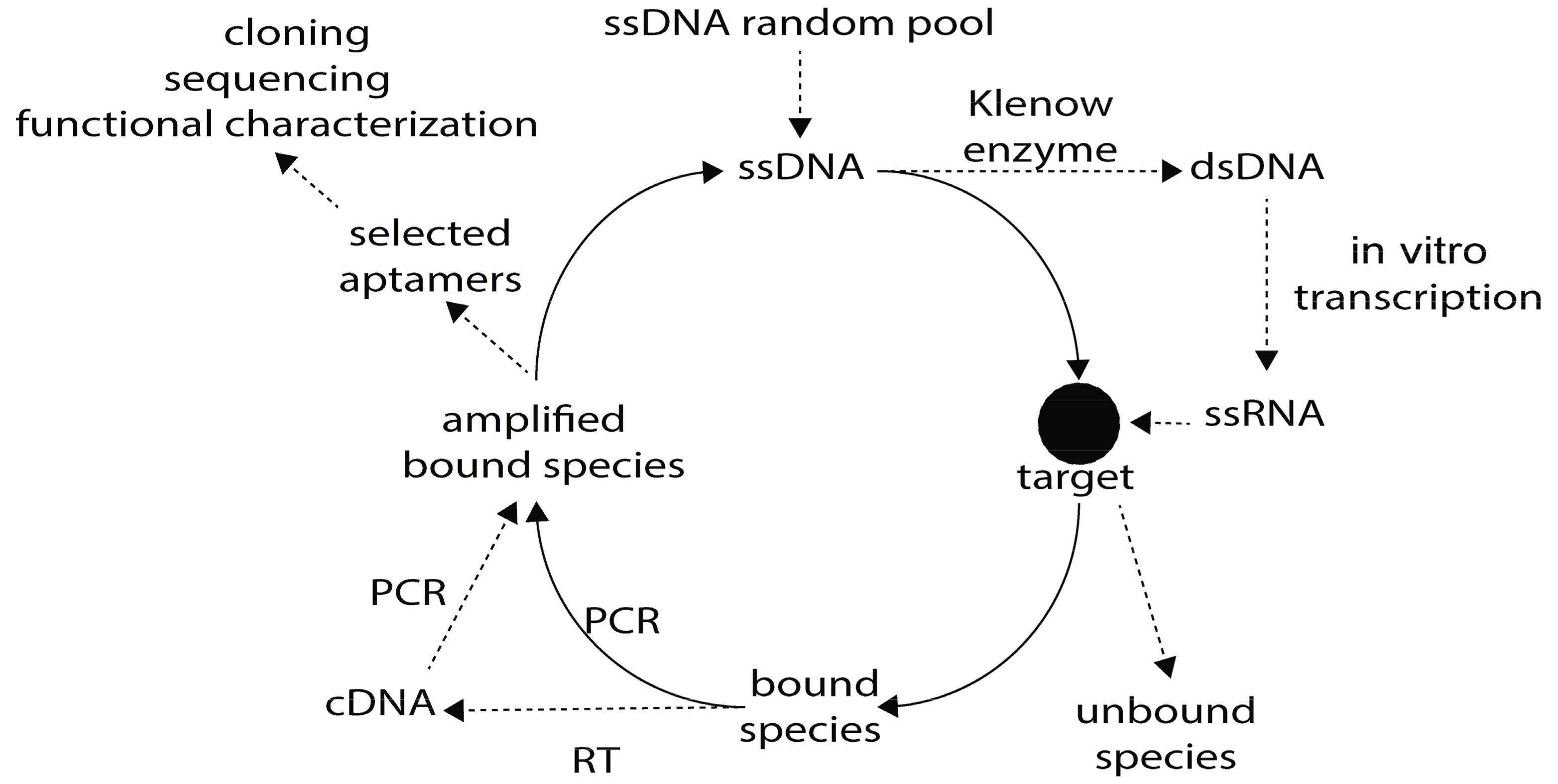
2.1. SELEX for DNA/RNA Parasite-Specific Aptamer Selection
2.2. Modifications of Parasite-Specific Aptamers to Increase their Efficacy
3. A Few Words about Peptide Aptamers
4. Development of DNA/RNA Aptamers in Parasitology
4.1. Aptamers against Trypanosoma spp.
4.2. Aptamers against Leishmania spp.
4.3. Aptamers against Plasmodium spp.
4.4. Aptamers against Cryptosporidium parvuum
4.5. Aptamers against Entamoeba histolytica
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Robertson, D.L.; Joyce, G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990, 344, 467–468. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.L.; Sooter, L.J. Single-stranded DNA aptamers against pathogens and toxins: Identification and biosensing applications. Biomed. Res. Int. 2015, 2015, 419318. [Google Scholar] [CrossRef] [PubMed]
- McKeague, M.; McConnell, E.M.; Cruz-Toledo, J.; Bernard, E.D.; Pach, A.; Mastronardi, E.; Zhang, X.; Beking, M.; Francis, T.; Giamberardino, A.; et al. Analysis of in vitro aptamer selection parameters. J. Mol. Evol. 2015, 81, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Mallikaratchy, P. Evolution of complex target SELEX to identify aptamers against mammalian cell-surface antigens. Molecules 2017, 22, 215. [Google Scholar] [CrossRef] [PubMed]
- Ruscito, A.; DeRosa, M.C. Small-molecule binding aptamers: Selection strategies, characterization, and applications. Front. Chem. 2016, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lakhin, A.V.; Tarantul, V.Z.; Gening, L.V. Aptamers: Problems, solutions and prospects. Acta Nat. 2013, 5, 34–43. [Google Scholar]
- Šmuc, T.; Ahn, I.Y.; Ulrich, H. Nucleic acid aptamers as high affinity ligands in biotechnology and biosensorics. J. Pharm. Biomed. Anal. 2013, 81–82, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.; Zheng, C.; Zhou, S.F.; Qiao, S.; Tran, P.H.; Pu, C.; Li, Y.; Kong, L.; Kouzani, A.Z.; Lin, J.; Liu, K.; Li, L.; Shigdar, S.; Duan, W. Superior performance of aptamer in tumor penetration over antibody: implication of aptamer-based theranostics in solid tumors. Theranostics 2015, 5, 1083–1097. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.W.M.; Shima, D.T.; Calias, P.; Cunningham, E.T., Jr.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef] [PubMed]
- N.I.H., U.S. National Library of Medicine. ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/home (accessed on 10 October 2018).
- Gold, L. SELEX: How it happened and where it will go. J. Mol. Evol. 2015, 81, 140–143. [Google Scholar] [CrossRef] [PubMed]
- World Malaria Report 2017; World Health Organization: Geneva, Switzerland, 2017. Licence: CC BY-NC-SA 3.0 IGO. Available online: https://www.who.int/malaria/publications/world-malaria-report-2017/ report/en/ (accessed on October 2018).
- Teixeira, S.M.; de Paiva, R.M.; Kangussu-Marcolino, M.M.; Darocha, W.D. Trypanosomatid comparative genomics: Contributions to the study of parasite biology and different parasitic diseases. Genet. Mol. Biol. 2012, 35, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Jackson, T.F. Epidemiology. In Amebiasis; Ravdin, J.I., Ed.; Imperial College Press: London, UK, 2000; pp. 47–63. ISBN 1-86094-133-8. [Google Scholar]
- Mor, S.M.; Tzipori, S. Cryptosporidiosis in children in sub-Saharan Africa: A lingering challenge. Clin. Infect. Dis. 2008, 47, 915–921. [Google Scholar] [CrossRef] [PubMed]
- Manley, J.L. SELEX to identify protein-binding sites on RNA. Cold Spring Harb. Protoc. 2013, 2013, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.N.; Chatterjee, S.; Adhya, S. Mitochondrial RNA import in Leishmania tropica: Aptamers homologous to multiple tRNA domains that interact cooperatively or antagonistically at the inner membrane. Mol. Cell. Biol. 2002, 22, 4372–4382. [Google Scholar] [CrossRef] [PubMed]
- Ramos, E.; Piñeiro, D.; Soto, M.; Abanades, D.R.; Martín, M.E.; Salinas, M.; González, V.M. A DNA aptamer population specifically detects Leishmania infantum H2A antigen. Lab. Investig. 2007, 87, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Ramos, E.; Moreno, M.; Martín, M.E.; Soto, M.; Gonzalez, V.M. In vitro selection of Leishmania infantum H3-binding ssDNA aptamers. Oligonucleotides 2010, 20, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Pérez, N.; Ramos, E.; García-Hernández, M.; Pinto, C.; Soto, M.; Martín, M.E.; González, V.M. Molecular and functional characterization of ssDNA aptamers that Specifically bind Leishmania infantum PABP. PLoS ONE 2015, 10, e0140048. [Google Scholar] [CrossRef] [PubMed]
- Barfod, A.; Persson, T.; Lindh, J. In vitro selection of RNA aptamers against a conserved region of the Plasmodium falciparum erythrocyte membrane protein 1. Parasitol. Res. 2009, 105, 1557–1566. [Google Scholar] [CrossRef] [PubMed]
- Cheung, Y.W.; Kwok, J.; Law, A.W.; Watt, R.M.; Kotaka, M.; Tanner, J.A. Structural basis for discriminatory recognition of Plasmodium lactate dehydrogenase by a DNA aptamer. Proc. Natl. Acad. Sci. USA 2013, 110, 15967–15972. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Song, K.M.; Jeon, W.; Jo, H.; Shim, Y.B.; Ban, C. A highly sensitive aptasensor towards Plasmodium lactate dehydrogenase for the diagnosis of malaria. Biosens. Bioelectron. 2012, 35, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.; Chakma, B.; Singh, N.K.; Patra, S.; Goswami, P. Aromatic surfactant as aggregating agent for aptamer-gold nanoparticle-based detection of Plasmodium lactate dehydrogenase. Mol. Biotechnol. 2016, 58, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Ospina-Villa, J.D.; Dufour, A.; Weber, C.; Ramirez-Moreno, E.; Zamorano-Carrillo, A.; Guillén, N.; Lopez-Camarillo, C.; Marchat, L.A. Targeting the polyadenylation EhCFIm25 with RNA aptamers controls survival in Entamoeba histolytica. Sci. Rep. 2018, 8, 5720. [Google Scholar] [CrossRef] [PubMed]
- Song, K.M.; Lee, S.; Ban, C. Aptamers and their biological applications. Sensors 2012, 12, 612–631. [Google Scholar] [CrossRef] [PubMed]
- Almasi, F.; Gargari, S.L.M.; Bitaraf, F.; Rasoulinejad, S. Development of a single stranded DNA aptamer as a molecular probe for LNCap cells using cell-SELEX. Avicenna J. Med. Biotechnol. 2016, 8, 104–111. [Google Scholar] [PubMed]
- Kong, H.Y.; Byun, J. Nucleic acid aptamers: New methods for selection, stabilization, and application in biomedical science. Biomol. Ther. 2013, 21, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Homann, M.; Göringer, H.U. Combinatorial selection of high affinity RNA ligands to live African trypanosomes. Nucl. Acids Res. 1999, 27, 2006–2014. [Google Scholar] [CrossRef] [PubMed]
- Lorger, M.; Engstler, M.; Homann, M.; Göringer, H.U. Targeting the variable surface of African trypanosomes with variant surface glycoprotein-specific, serum-stable RNA aptamers. Eukaryot. Cell 2003, 2, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, H.; Magdesian, M.H.; Alves, M.J.; Colli, W. In vitro selection of RNA aptamers that bind to cell adhesion receptors of Trypanosoma cruzi and inhibit cell invasion. J. Biol. Chem. 2002, 277, 20756–20762. [Google Scholar] [CrossRef] [PubMed]
- Nagarkatti, R.; Bist, V.; Sun, S.; Fortes de Araujo, F.; Nakhasi, H.L.; Debrabant, A. Development of an aptamer-based concentration method for the detection of Trypanosoma cruzi in blood. PLoS ONE 2012, 7, e43533. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, A.; Labib, M.; Muharemagic, D.; Sattar, S.; Dixon, B.R.; Berezovski, M.V. Detection of Cryptosporidium parvum oocysts on fresh produce using DNA aptamers. PLoS ONE 2015, 10, e0137455. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Connell, G.J. An electrochemiluminescent aptamer switch for a high-throughput assay of an RNA editing reaction. RNA 2009, 15, 1929–1938. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.; Rincón, E.; Piñeiro, D.; Fernández, G.; Domingo, A.; Jiménez-Ruíz, A.; Salinas, M.; González, V.M. Selection of aptamers against KMP-11 using colloidal gold during the SELEX process. Biochem. Biophys. Res. Commun. 2003, 308, 214–218. [Google Scholar] [CrossRef]
- Birch, C.M.; Hou, H.W.; Han, J.; Niles, J.C. Identification of malaria parasite-infected red blood cell surface aptamers by inertial microfluidic SELEX (I-SELEX). Sci. Rep. 2015, 5, 11347. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Yao, H.; Wang, L.; Lu, J.; Jiang, F.; Lu, A.; Zhang, G. Chemical modifications of nucleic acid aptamers for therapeutic purposes. Int. J. Mol. Sci. 2017, 18, 1683. [Google Scholar] [CrossRef] [PubMed]
- Fooladi, A.A.I.; Ch, M.H.; Amin, M.; Amani, J. Applications and modifications of aptamers: Potential tool for medical microbiology. Rev. Med. Microbiol. 2016, 27, 107–120. [Google Scholar] [CrossRef]
- Homann, M.; Lorger, M.; Engstler, M.; Zacharias, M.; Göringer, H.U. Serum-stable RNA aptamers to an invariant surface domain of live African trypanosomes. Comb. Chem. High Throughput Screen. 2006, 9, 491–499. [Google Scholar] [PubMed]
- Adler, A.; Forster, N.; Homann, M.; Göringer, H.U. Post-SELEX chemical optimization of a trypanosome-specific RNA aptamer. Comb. Chem. High Throughput Screen. 2008, 11, 16–23. [Google Scholar] [PubMed]
- Nagarkatti, R.; de Araujo, F.F.; Gupta, C.; Debrabant, A. Aptamer based, non-PCR, non-serological detection of Chagas disease biomarkers in Trypanosoma cruzi infected mice. PLoS Negl. Trop. Dis. 2014, 8, e2650. [Google Scholar] [CrossRef] [PubMed]
- Crawford, M.; Woodman, R.; Ferrigno, P.K. Peptide aptamers: Tools for biology and drug discovery. Brief. Funct. Genomic Proteom. 2003, 2, 72–79. [Google Scholar] [CrossRef]
- Reverdatto, S.; Burz, D.; Shekhtman, A. Peptide Aptamers: Development and Applications. Curr. Top. Med. Chem. 2015, 15, 1082. [Google Scholar] [CrossRef] [PubMed]
- Mascini, M.; Palchetti, I.; Tombelli, S. Nucleic acid and peptide aptamers: Fundamentals and bioanalytical aspects. Angew. Chem. Int. Ed. Engl. 2012, 51, 1316–1332. [Google Scholar] [CrossRef] [PubMed]
- Seigneuric, R.; Gobbo, J.; Colas, P.; Garrido, C. Targeting cancer with peptide aptamers. Oncotarget 2011, 2, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Hall, M., 3rd; Misra, S.; Chaudhuri, M.; Chaudhuri, G. Peptide aptamer mimicking RAD51-binding domain of BRCA2 inhibits DNA damage repair and survival in Trypanosoma brucei. Microb. Pathog. 2011, 50, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Homann, M.; Göringer, H.U. Uptake and intracellular transport of RNA aptamers in African trypanosomes suggest therapeutic “piggy-back” approach. Bioorg. Med. Chem. 2001, 9, 2571–2580. [Google Scholar] [CrossRef]
- Zelada-Guillén, G.A.; Tweed-Kent, A.; Niemann, M.; Göringer, H.U.; Riu, J.; Rius, F.X. Ultrasensitive and real-time detection of proteins in blood using a potentiometric carbon-nanotube aptasensor. Biosens. Bioelectron. 2013, 41, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Martín, M.E.; García-Hernández, M.; García-Recio, E.M.; Gómez-Chacón, G.F.; Sánchez-López, M.; González, V.M. DNA aptamers selectively target Leishmania infantum H2A protein. PLoS ONE 2013, 8, e78886. [Google Scholar] [CrossRef] [PubMed]
- Dirkzwager, R.M.; Kinghorn, A.B.; Richards, J.S.; Tanner, J.A. APTEC: Aptamer-tethered enzyme capture as a novel rapid diagnostic test for malaria. Chem. Commun. Camb. 2015, 51, 4697–4700. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.J.; Ban, C. Crystal structure of a DNA aptamer bound to PvLDH elucidates novel single-stranded DNA structural elements for folding and recognition. Sci. Rep. 2016, 6, 34998. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Manjunatha, D.H.; Jeon, W.; Ban, C. Cationic surfactant-based colorimetric detection of Plasmodium lactate dehydrogenase, a biomarker for malaria, using the specific DNA aptamer. PLoS ONE 2014, 9, e100847. [Google Scholar] [CrossRef] [PubMed]
- Jeon, W.; Lee, S.; Manjunatha, D.H.; Ban, C. A colorimetric aptasensor for the diagnosis of malaria based on cationic polymers and gold nanoparticles. Anal. Biochem. 2013, 439, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.; Das, S.; Chakma, B.; Goswami, P. Aptamer-graphene oxide for highly sensitive dual electrochemical detection of Plasmodium lactate dehydrogenase Anal. Biochem. 2016, 514, 32–37. [Google Scholar]
- Godonoga, M.; Lin, T.Y.; Oshima, A.; Sumitomo, K.; Tang, M.S.; Cheung, Y.W.; Kinghorn, A.B.; Dirkzwager, R.M.; Zhou, C.; Kuzuya, A.; et al. A DNA aptamer recognising a malaria protein biomarker can function as part of a DNA origami assembly. Sci. Rep. 2016, 6, 21266. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.S.L.; Shiu, S.C.; Godonoga, M.; Cheung, Y.W.; Liang, S.; Dirkzwager, R.M.; Kinghorn, A.B.; Fraser, L.A.; Heddle, J.G.; Tanner, J.A. An aptamer-enabled DNA nanobox for protein sensing. Nanomedicine 2018, 14, 1161–1168. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.C.; Fraser, L.A.; Ding, Y.; Tanner, J.A. Aptamer display on diverse DNA polyhedron supports. Molecules 2018, 23, 1695. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Searson, P.C. Detection of Plasmodium lactate dehydrogenase antigen in buffer using aptamer-modified magnetic microparticles for capture, oligonucleotide-modified quantum dots for detection, and oligonucleotide-modified gold nanoparticles for signal amplification. Bioconjug. Chem. 2017, 28, 2230–2234. [Google Scholar] [CrossRef] [PubMed]
- Fraser, L.A.; Kinghorn, A.B.; Dirkzwager, R.M.; Liang, S.; Cheung, Y.W.; Lim, B.; Shiu, S.C.; Tang, M.S.L.; Andrew, D.; Manitta, J.; et al. A portable microfluidic aptamer-tethered enzyme capture (APTEC) biosensor for malaria diagnosis. Biosens. Bioelectron. 2018, 100, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Geldert, A.; Zhang, X.; Zhang, H.; Lim, C.T. Highly sensitive and selective aptamer-based fluorescence detection of a malarial biomarker using single-layer MoS2 nanosheets. ACS Sens. 2016, 1, 1315–1321. [Google Scholar] [CrossRef]
- Geldert, A.; Zhang, X.; Zhang, H.; Lim, C.T. Enhancing the sensing specificity of a MoS2 nanosheet-based FRET aptasensor using a surface blocking strategy. Analyst 2017, 142, 2570–2577. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.X.; Cheung, Y.W.; Dirkzwager, R.M.; Wong, W.C.; Tanner, J.A.; Li, H.W.; Wu, Y. Specific and sensitive detection of Plasmodium falciparum lactate dehydrogenase by DNA-scaffolded silver nanoclusters combined with an aptamer. Analyst 2017, 142, 800–807. [Google Scholar] [CrossRef] [PubMed]
- Cheung, Y.W.; Dirkzwager, R.M.; Wong, W.C.; Cardoso, J.; D’Arc Neves Costa, J.; Tanner, J.A. Aptamer-mediated Plasmodium-specific diagnosis of malaria. Biochimie 2018, 145, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Frith, K.A.; Fogel, R.; Goldring, J.P.D.; Krause, R.G.E.; Khati, M.; Hoppe, H.; Cromhout, M.E.; Jiwaji, M.; Limson, J.L.; Malar, J. Towards development of aptamers that specifically bind to lactate dehydrogenase of Plasmodium falciparum through epitopic targeting. Malar. J. 2018, 17, 191. [Google Scholar] [CrossRef] [PubMed]
- Niles, J.C.; Derisi, J.L.; Marletta, M.A. Inhibiting Plasmodium falciparum growth and heme detoxification pathway using heme-binding DNA aptamers. Proc. Natl. Acad. Sci. USA 2009, 106, 13266–13271. [Google Scholar] [CrossRef] [PubMed]
- Pezet-Valdez, M.; Fernández-Retana, J.; Ospina-Villa, J.D.; Ramírez-Moreno, M.E.; Orozco, E.; Charcas-López, S.; Soto-Sánchez, J.; Mendoza-Hernández, G.; López-Casamicha, M.; López-Camarillo, C.; et al. The 25 kDa subunit of cleavage factor Im is a RNA- binding proteina that interacts with the poly(A) polymerase in Entamoeba histolytica. PLoS ONE 2013, 8, e67977. [Google Scholar] [CrossRef] [PubMed]
- Ospina-Villa, J.D.; Zamorano-Carrillo, A.; Lopez-Camarillo, C.; Castañon-Sanchez, C.A.; Soto-Sanchez, J.; Ramirez-Moreno, E.; Marchat, L.A. Amino acid residues Leu135 and Tyr236 are required for RNA binding activity of CFIm25 in Entamoeba histolytica. Biochimie 2015, 115, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Ospina-Villa, J.D.; García-Contreras, J.; Rosas-Trigueros, J.L.; Ramírez-Moreno, E.; López-Camarillo, C.; Zamora-López, B.; Marchat, L.A.; Zamorano-Carrillo, A. Importance of amino acids Leu135 and Tyr236 for the interaction between EhCFIm25 and RNA: A molecular dynamics simulation study. J. Mol. Model. 2018, 4, 202. [Google Scholar] [CrossRef] [PubMed]
- Ospina-Villa, J.D.; Guillén, N.; Lopez-Camarillo, C.; Soto-Sanchez, J.; Ramirez-Moreno, E.; Garcia-Vazquez, R.; Castañon-Sanchez, C.A.; Betanzos, A.; Marchat, L.A. Silencing the cleavage factor CFIm25 as a new strategy to control Entamoeba histolytica parasite. J. Microbiol. 2017, 55, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Gilmartin, G.; Doublie, S. Structural basis of UGUA recognition by the Nudix protein CFIm25 and implications for a regulatory role in mRNA 3′ processing. Proc. Natl. Acad. Sci. USA 2010, 107, 10062–10067. [Google Scholar] [CrossRef] [PubMed]
| Key Points | Aptamers | Monoclonal Antibodies |
|---|---|---|
| Synthesis | Production process is purely chemical | Requires an immune response in an animal model |
| Target | Almost any type of molecule | Immunological molecule |
| Modification | Chemical modification to improve resistance to nucleases and bioavailability | Antibodies are typically conjugated with one type of signaling or binding molecule |
| Stability | Aptamers are fairly stable at ambient temperature and are easily refolded if denatured | Antibodies are susceptible to high temperatures and pH changes; denatured antibodies cannot be repaired |
| Long-term availability | Once the nucleotide sequence is known, the aptamers can be produced chemically when necessary | Frozen cell stocks must be maintained for monoclonal antibody production |
| Size | 12–30 kDa (~30–80 nucleotides) | ~150–170 kDa (IgG) |
| Production time | ~1–3 months | ~4–6 months |
| Modification | Position | Effect | Parasite/Application | Reference |
|---|---|---|---|---|
| Sugar modification | 2′-OH to 2′-fluorine, 2′-O-methyl | Increases half-life | T. brucei/treatment | [40,41, 42] |
| Conjugation with biotin | - | Blocks activity of 3′ exonucleases | T. cruzi/diagnosis | [32,43] |
| PEGylation | 5′conjugation | Increases half-life and solubility, offers protection from reticuloendothelial cells and proteolytic enzymes | T. brucei/treatment | [41,42] |
| Proposed Application | Nature of Aptamer | Protozoan | Parasite Target | Reference |
|---|---|---|---|---|
| Diagnosis | RNA | T. brucei | VSG proteins | [50] |
| Diagnosis | RNA | T. cruzi | live parasites in blood | [34] |
| Diagnosis | RNA | T. cruzi | TESA | [43] |
| Diagnosis | DNA | L. infantum | KMP-11 | [37] |
| Diagnosis | DNA | L. infantum | H2A | [20,51] |
| Diagnosis | DNA | L. infantum | H3 | [21] |
| Diagnosis | DNA | L. infantum | PABP | [22] |
| Diagnosis | DNA | P. falciparum | LDH | [24,52,65] |
| Diagnosis | DNA | P. vivax | LDH | [25] |
| Diagnosis | DNA | P. falciparum | LDH | [66] |
| Diagnosis | DNA | C. parvuum | oocyst | [35] |
| New drug | RNA | T. cruzi | Receptor of host ECM molecules | [33] |
| New drug | DNA | L. infantum | KMP-11 | [37] |
| New drug | DNA | L. infantum | PABP | [22] |
| New drug | RNA | P. falciparum | EMP1 | [23] |
| New drug | DNA | P. falciparum | hemozoin | [67] |
| New drug | DNA | P. falciparum | var2CSA | [38] |
| New drug | RNA | E. histolytica | CFIm25 | [27] |
| Drug delivery | RNA | T. brucei | 42 kDa protein | [49] |
| Drug delivery | RNA | T. brucei | VSG proteins | [32] |
| Molecular mechanisms | RNA | L. tropica | mitochondria | [19] |
| Molecular mechanisms | RNA | L. tarentolae | mitochondrial extracts | [34] |
| Protein purification | DNA | L. infantum | LiH2A | [20,51] |
| Protein purification | DNA | L. infantum | PABP | [22] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ospina-Villa, J.D.; López-Camarillo, C.; Castañón-Sánchez, C.A.; Soto-Sánchez, J.; Ramírez-Moreno, E.; Marchat, L.A. Advances on Aptamers against Protozoan Parasites. Genes 2018, 9, 584. https://doi.org/10.3390/genes9120584
Ospina-Villa JD, López-Camarillo C, Castañón-Sánchez CA, Soto-Sánchez J, Ramírez-Moreno E, Marchat LA. Advances on Aptamers against Protozoan Parasites. Genes. 2018; 9(12):584. https://doi.org/10.3390/genes9120584
Chicago/Turabian StyleOspina-Villa, Juan David, César López-Camarillo, Carlos A. Castañón-Sánchez, Jacqueline Soto-Sánchez, Esther Ramírez-Moreno, and Laurence A. Marchat. 2018. "Advances on Aptamers against Protozoan Parasites" Genes 9, no. 12: 584. https://doi.org/10.3390/genes9120584
APA StyleOspina-Villa, J. D., López-Camarillo, C., Castañón-Sánchez, C. A., Soto-Sánchez, J., Ramírez-Moreno, E., & Marchat, L. A. (2018). Advances on Aptamers against Protozoan Parasites. Genes, 9(12), 584. https://doi.org/10.3390/genes9120584






