Physiological Analysis and Transcriptome Profiling of Inverted Cuttings of Populus yunnanensis Reveal That Cell Wall Metabolism Plays a Crucial Role in Responding to Inversion
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Growth Comparison
2.3. Extraction of Endogenous Hormones and Quantification
2.4. Extraction of IAAO, PPO, and POD
2.5. RNA Isolation and Library Construction
2.6. De Novo Assembly and Functional Annotation
2.7. Sample Correlation and Differentially Expressed Gene Detection
2.8. Real-Time Quantitative PCR Analysis
3. Results
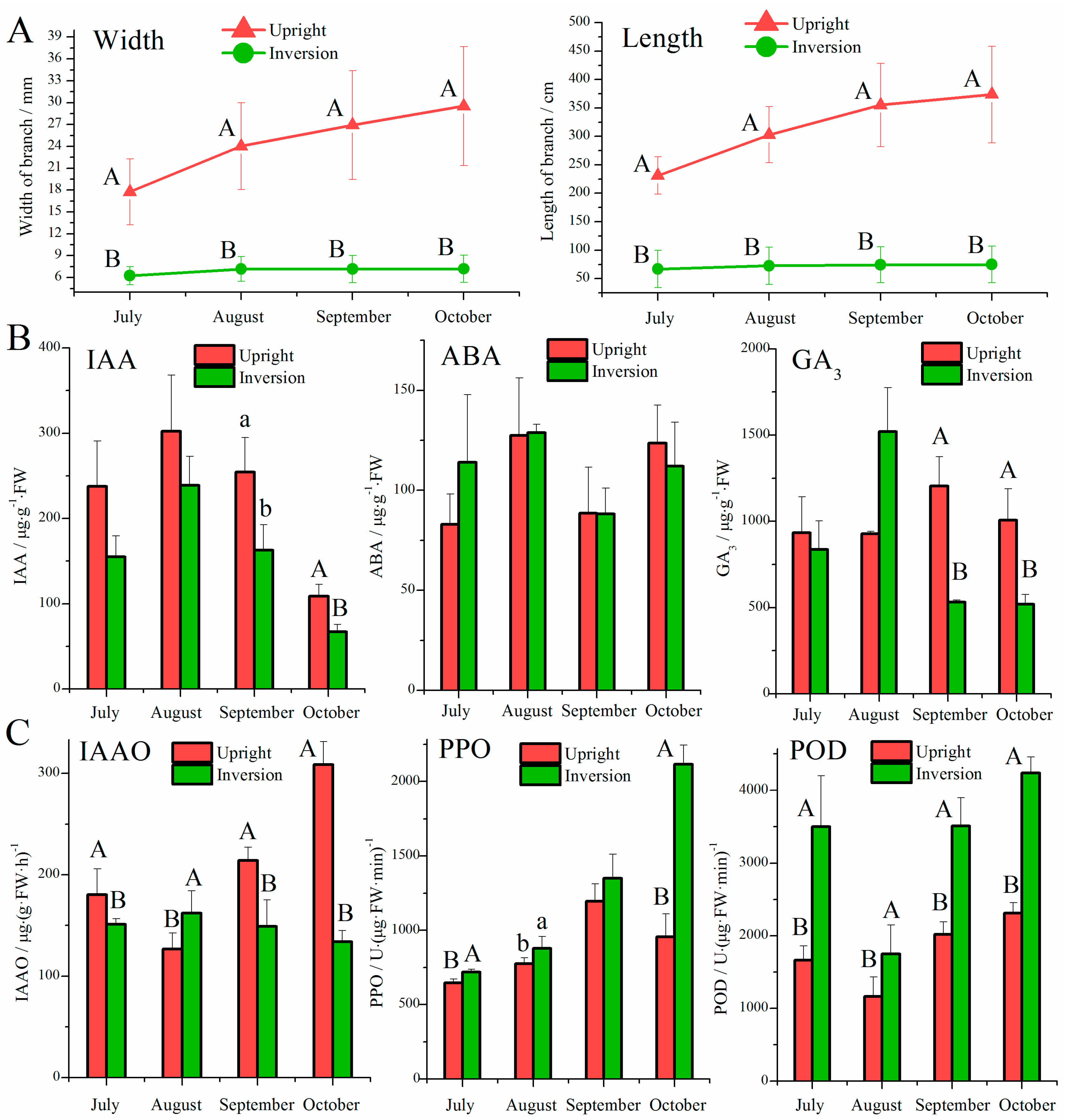
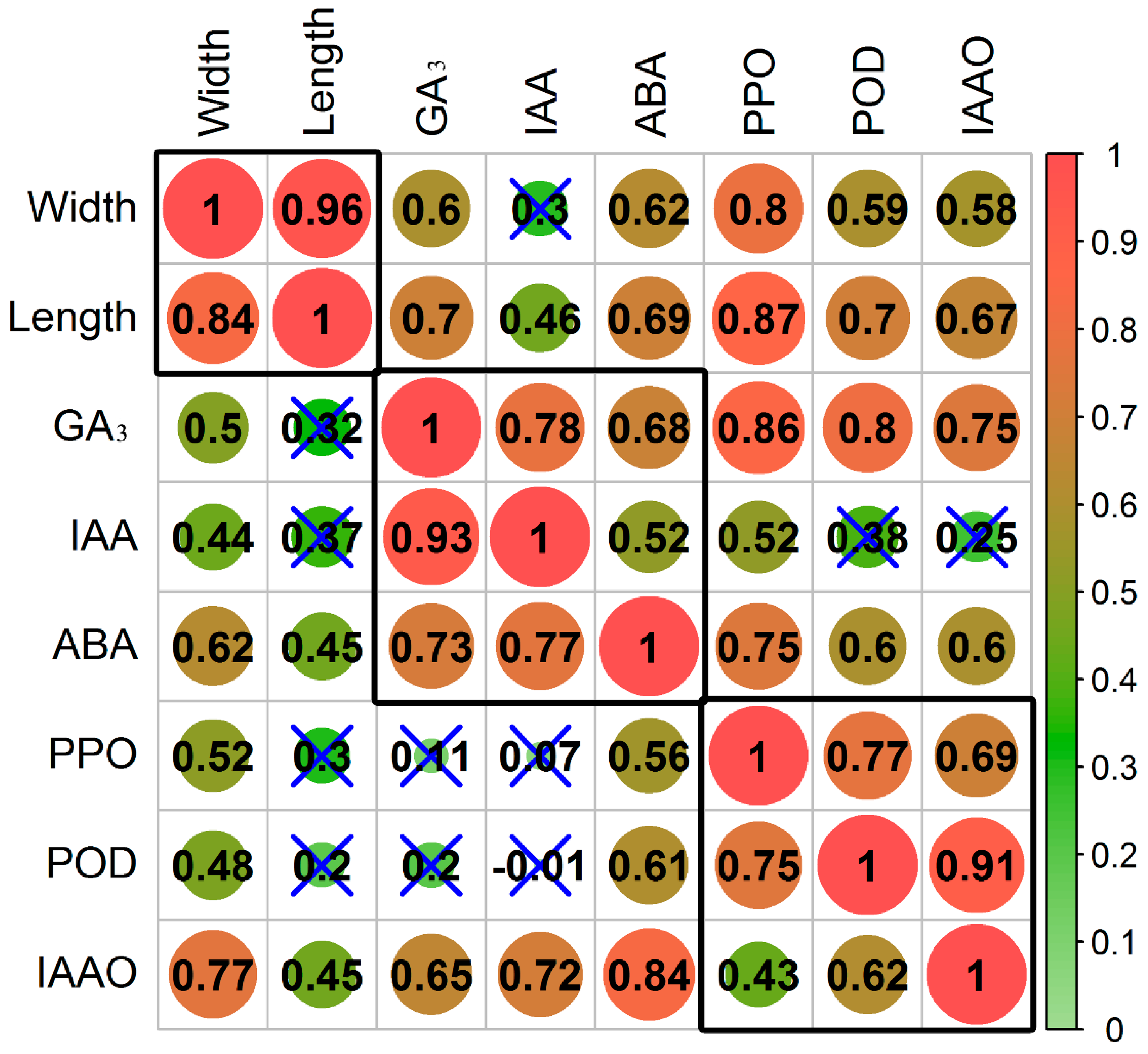
3.1. Effect of Inversion on Growth, Hormones, and Oxidases in Cuttings
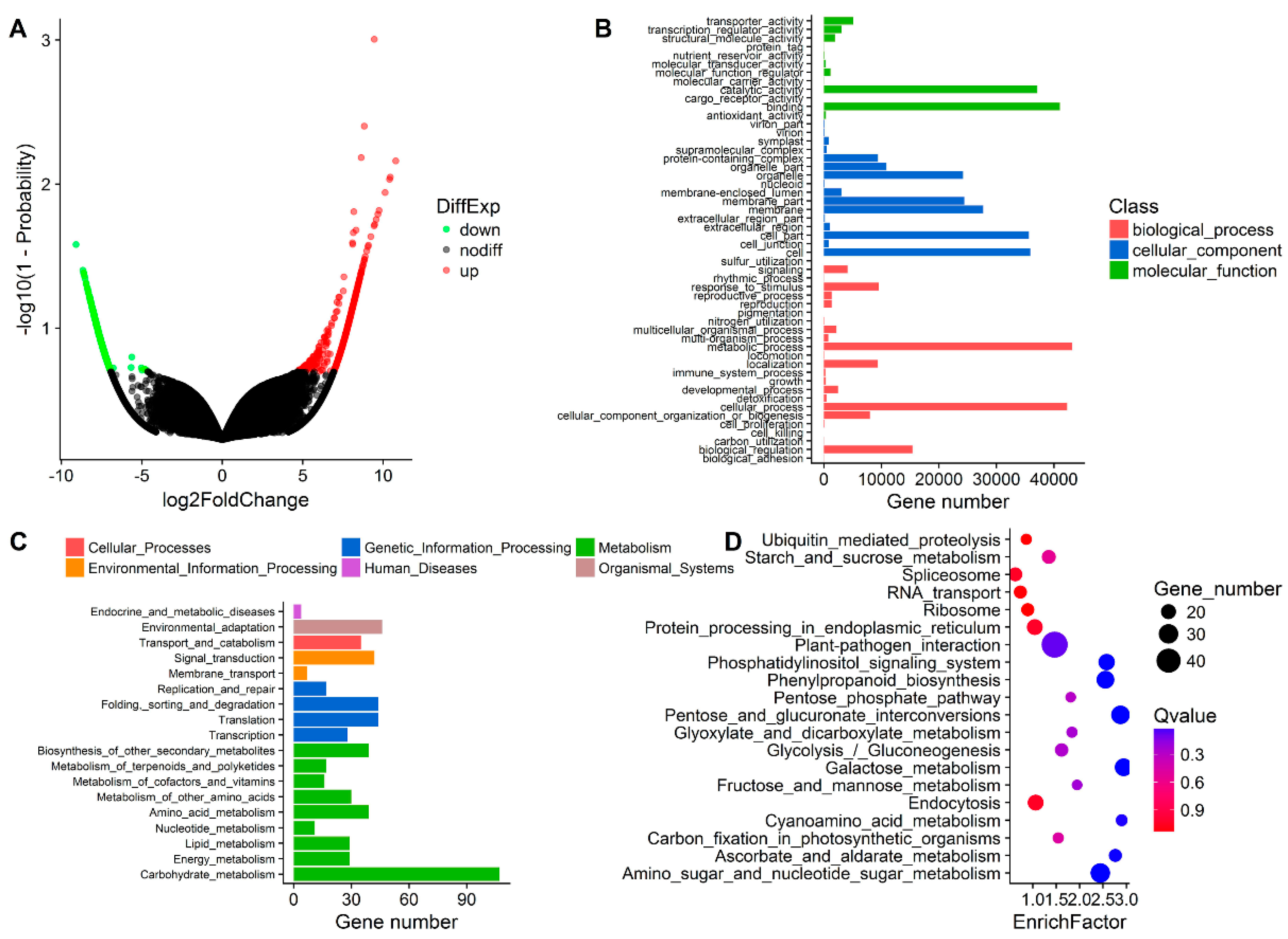
3.2. Transcriptome Sequencing, De Novo Assembly, and Functional Annotation
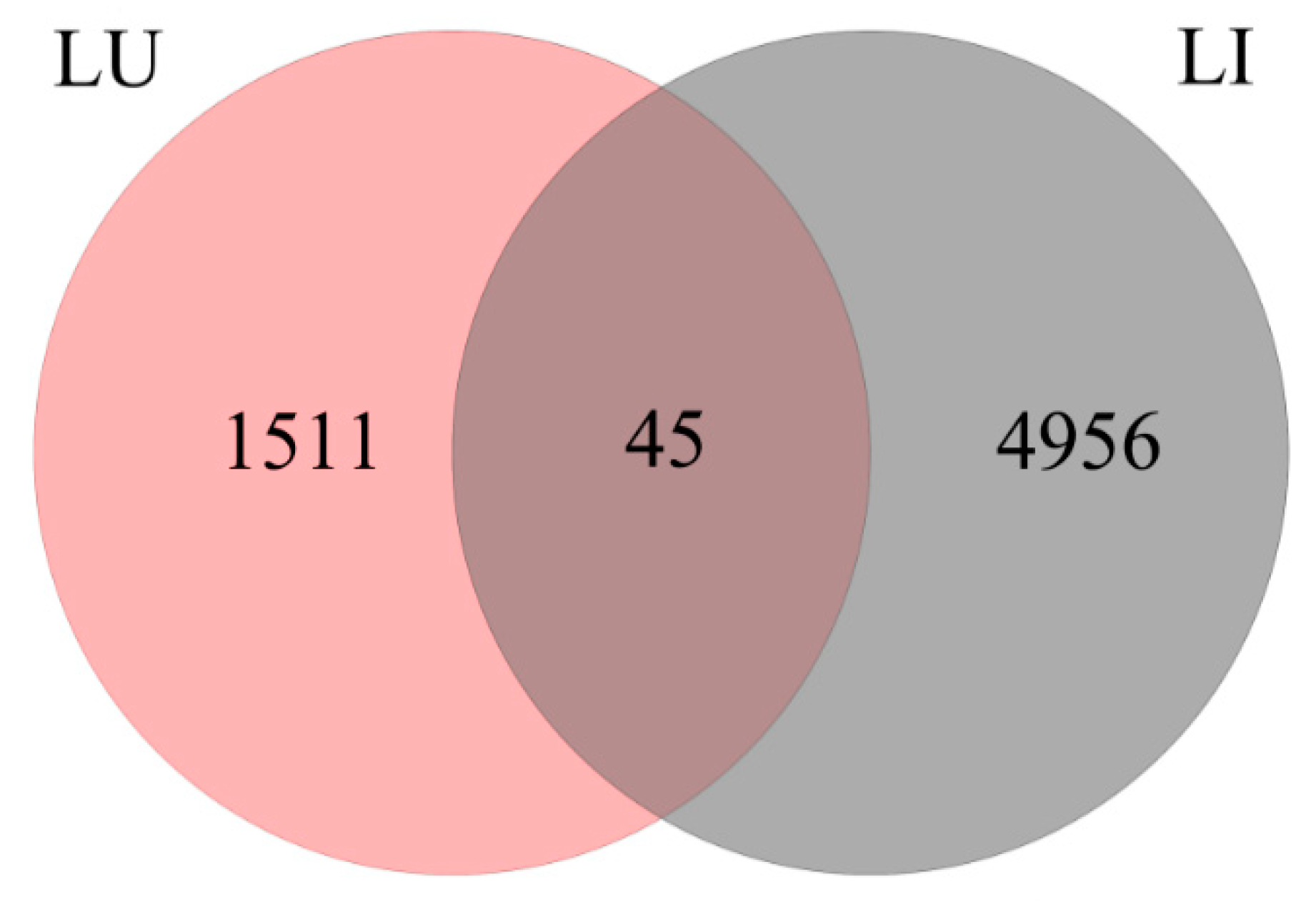
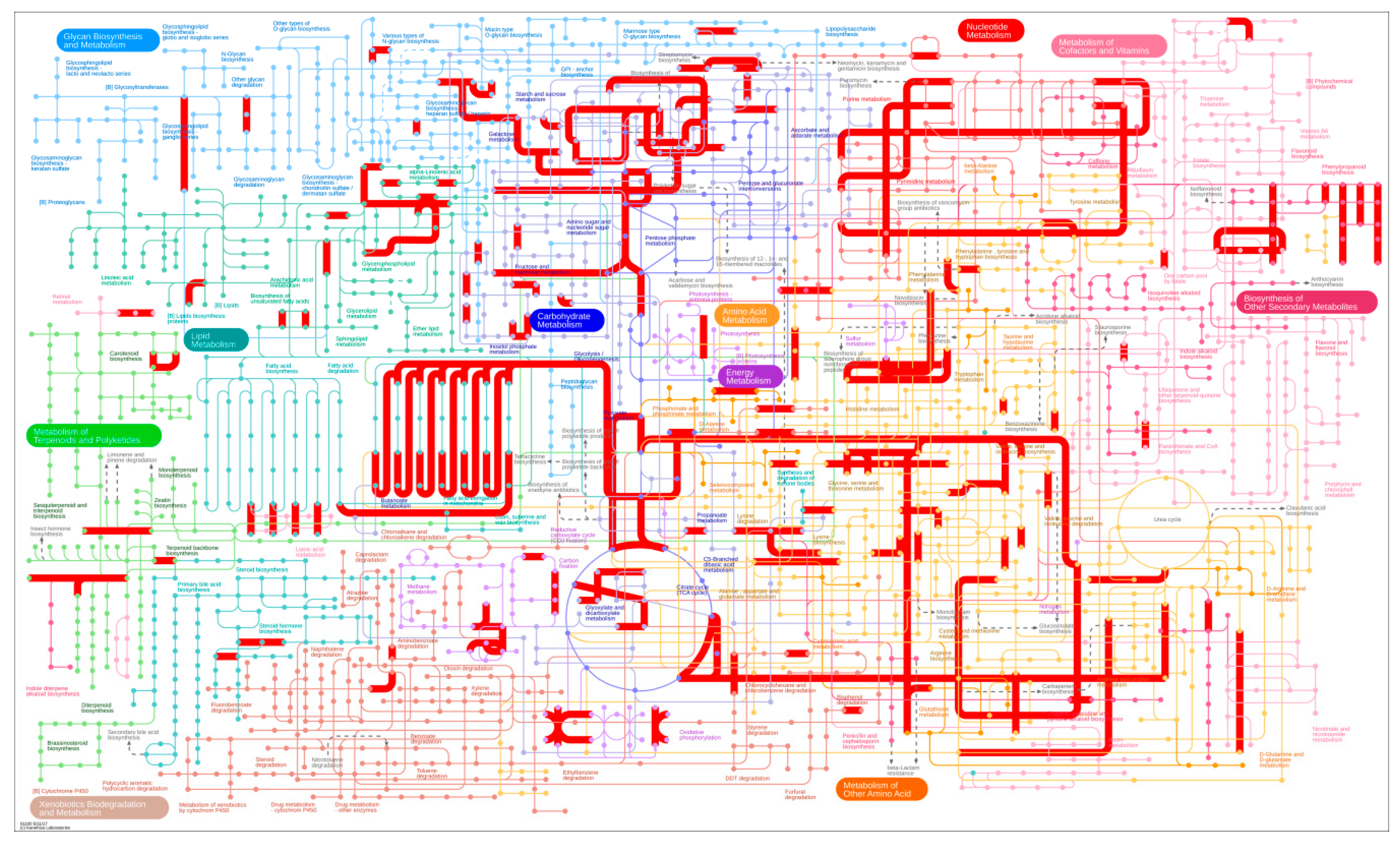
3.3. DEG Detection between Upright and Inverted Samples
3.4. Real-Time Quantitative PCR Analysis
4. Discussion
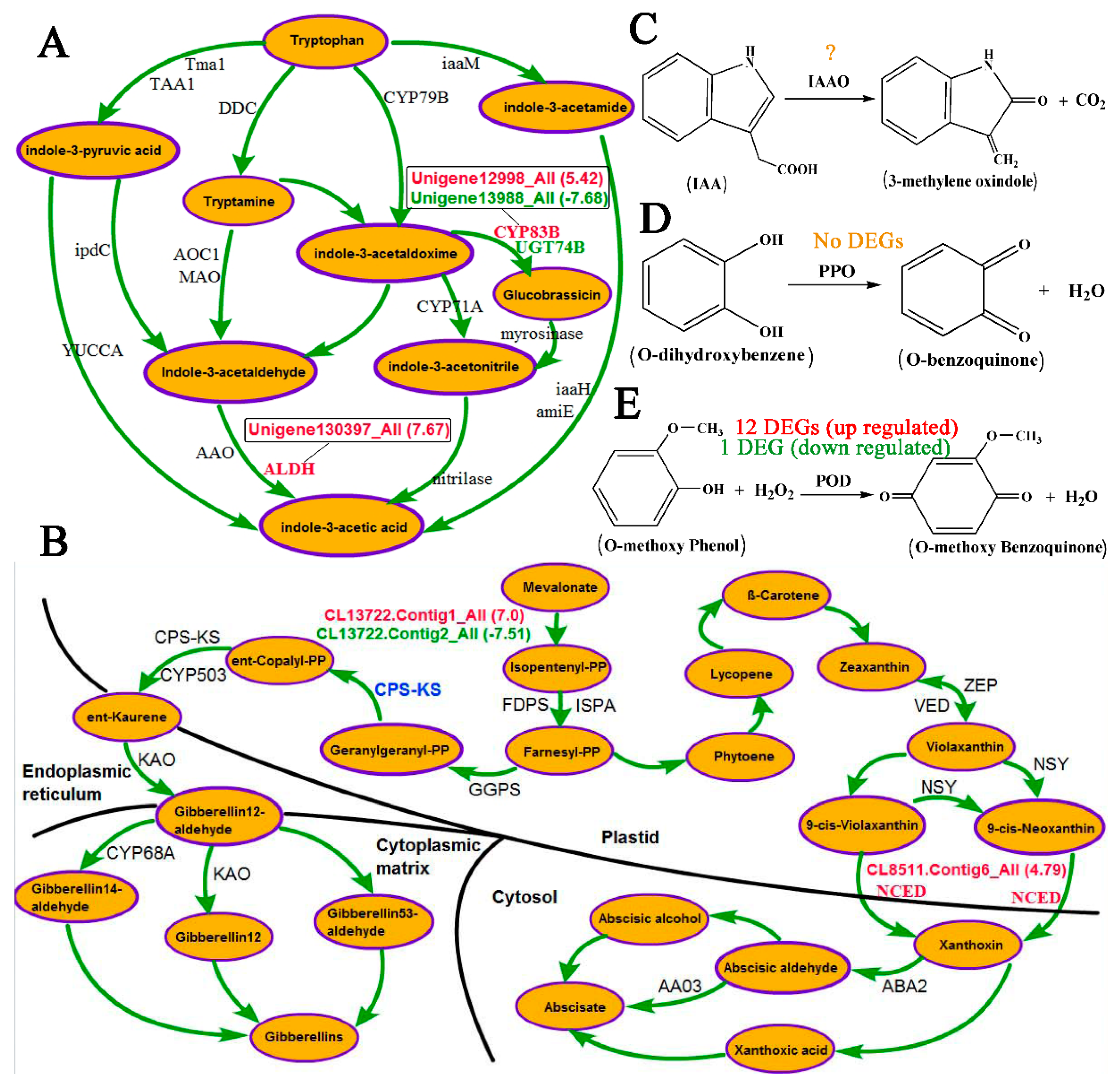
4.1. Role of Hormones in Response to the Inversion of Plants
4.2. Role of Oxidases in Response to Inversion of Plants
4.3. A Crucial Role of Cell Wall Metabolism in Response to Inversion of Plants is Revealed via RNA-Seq
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jiang, H.; Korpelainen, H.; Li, C. Populus yunnanensis males adopt more efficient protective strategies than females to cope with excess zinc and acid rain. Chemosphere 2013, 91, 1213–1220. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, Y.Q.; Sun, X.D.; Lin, H.M.; Chen, J.H.; Ren, J.; Hu, X.Y.; Yang, Y.P. Comparative physiological and proteomic analyses of poplar (Populus yunnanensis) plantlets exposed to high temperature and drought. PLoS ONE 2014, 9, e107605. [Google Scholar] [CrossRef] [PubMed]
- Belanger, K.D.; Quatrano, R.S. Polarity: The role of localized secretion. Curr. Opin. Plant Biol. 2000, 3, 67–72. [Google Scholar] [CrossRef]
- Qi, J.Y.; Wu, B.B.; Feng, S.L.; Lü, S.Q.; Guan, C.M.; Zhang, X.; Qiu, D.L.; Hu, Y.C.; Zhou, Y.H.; Li, C.Y.; et al. Mechanical regulation of organ asymmetry in leaves. Nat. Plants 2017, 3, 724–733. [Google Scholar] [CrossRef] [PubMed]
- Nick, P.; Furuya, M. Induction and fixation of polarity–early steps in plant morphogenesis. Dev. Growth Differ. 1992, 34, 115–125. [Google Scholar] [CrossRef]
- Bloch, R. Polarity in plants. Bot. Rev. 1943, 9, 261–310. [Google Scholar] [CrossRef]
- Niedergang-Kamien, E.; Skoog, F. Studies on polarity and auxin transport in plant. I. Modification of polarity and auxin transport by triiodobenzoic acid. Phys. Plant. 1956, 9, 60–73. [Google Scholar] [CrossRef]
- Bünning, E. Morphogenesis in plants. Surv. Biol. Prog. 1952, 2, 105–140. [Google Scholar] [CrossRef]
- Sachs, T. Polarity and the induction of organized vascular tissues. Ann. Bot. 1969, 33, 263–275. [Google Scholar] [CrossRef]
- Weisenseel, M.H. Induction of Polarity. In Encyclopedia of Plant Physiology; Haupt, W., Fenleib, M.E., Eds.; Springer: Berlin, Gramany, 1979; Volume 7, pp. 485–505. [Google Scholar]
- Robinson, K.R.; Cone, R. Polarization of Fucoid eggs by a calcium ionophore gradient. Science 1980, 270, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Nick, P.; Schäfer, E. Induction of transverse polarity by blue light: An all-or-none response. Planta 1991, 185, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Went, F.W. Polarity of auxin transport in inverted Tagetes cuttings. Bot. Gaz. 1941, 103, 386–390. [Google Scholar] [CrossRef]
- Sheldrake, A.R. The polarity of auxin transport in inverted cuttings. New Phytol. 1974, 73, 637–642. [Google Scholar] [CrossRef]
- Warren, W.J.; Warren, W.P.M. Mechanism of auxin regulation of structural and physiological polarity in plants, tissues, cells and embryos. Aust. J. Plant Physiol. 1993, 20, 555–571. [Google Scholar] [CrossRef]
- Souter, M.; Lindsey, K. Polarity and signaling in plant embryogenesis. J. Exp. Bot. 2000, 51, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Friml, J.; Benfey, P.; Benkova, E.; Bennett, M.; Berleth, T.; Geldner, N.; Grebe, M.; Heisler, M.; Hejatko, J.; Jurgens, G.; et al. Apical-basal polarity: Why plant cells don’t stand on their heads. Trends Plant Sci. 2006, 11, 12–14. [Google Scholar] [CrossRef] [PubMed]
- Hable, W.E.; Hart, P.E. Signaling mechanisms in the establishment of plant and fucoid algal polarity. Mol. Reprod. Dev. 2010, 77, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Medvedev, S.S. Mechanisms and physiological role of polarity in plants. Russ. J. Plant Physl. 2012, 59, 502–514. [Google Scholar] [CrossRef]
- Strzyz, P. Forcing cell polarity. Nat. Rev. Mol. Cell Biol. 2017, 18, 278. [Google Scholar] [CrossRef] [PubMed]
- Bornens, M. Cell polarity: Having and making sense of direction—On the evolutionary significance of the primary cilium/centrosome organ in Metazoa. Open Biol. 2018, 8, 180052. [Google Scholar] [CrossRef] [PubMed]
- Domagalska, M.A.; Leyser, O. Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 2011, 12, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.Y.; Lv, B.S.; Ding, T.T.; Bai, M.Y.; Ding, Z.J. Auxin-BR interaction regulates plant growth and development. Front. Plant Sci. 2018, 8, 2256. [Google Scholar] [CrossRef] [PubMed]
- Santner, A.; Calderon-Villalobos, L.I.; Estell, M. Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 2009, 5, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Santner, A.; Estelle, M. Recent advances and emerging trends in plant hormone signaling. Nature 2009, 459, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Wolters, H.; Jurgens, G. Survival of the flexible: Hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 2009, 10, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Depuydt, S.; Hardtke, C.S. Hormone signaling crosstalk in plant growth regulation. Curr. Biol. 2011, 21, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Band, L.R.; Ubeda-Tomas, S.; Dyson, R.J.; Middleton, A.M.; Hodgman, T.C.; Owen, M.R.; Jensen, O.E.; Bennett, M.J.; King, J.R. Growth-induced hormone dilution can explain the dynamics of plant root cell elongation. Proc. Natl. Acad. Sci. USA 2012, 109, 7577–7582. [Google Scholar] [CrossRef] [PubMed]
- Pacifici, E.; Polverari, L.; Sabatini, S. Plant hormone cross-talk: The pivot of root growth. J. Exp. Bot. 2015, 66, 1113–1121. [Google Scholar] [CrossRef] [PubMed]
- Gantait, S.; Sinniah, U.R.; Ali, M.N.; Sahu, N.C. Gibberellins—A multifaceted hormone in plant growth regulatory network. Curr. Protein Pept. Sci. 2015, 16, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Mroue, S.; Simeunovic, A.; Robert, H.S. Auxin production as an intergrator of environmental cues for developmental growth regulation. J. Exp. Bot. 2018, 69, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Jones, J.D. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Richards, D.E.; King, K.E.; Ait-ali, T.; Harberd, N.P. How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 67–88. [Google Scholar] [CrossRef] [PubMed]
- Nemhauser, J.L.; Mockler, T.C.; Chory, J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004, 2, e258. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.D. Auxin biaosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Choi, J.; An, G.; Kim, S.R. Ectopic expression of OsSta2 enhances salt stress tolerance in rice. Front. Plant Sci. 2017, 8, 316. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Gho, Y.S.; Jung, K.H.; Kim, S.R. Genome-wide identification and analysis of genes, conserved between japonica and indica rice cultivars, that respond to low-temperature stress at the vegetative growth stage. Front. Plant Sci. 2017, 8, 1120. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, C.; Sheng, H.; Wang, Y.; Zeng, J.; Kang, H.; Fan, X.; Sha, L.; Zhang, H.; Zhou, Y. Transcriptome-wide identification and expression analyses of ABC transporters in dwarf polish wheat under metal stresses. Biol. Plant. 2017, 61, 293–304. [Google Scholar] [CrossRef]
- Meloni, D.A.; Oliva, M.A.; Martinez, C.A.; Cambraia, J. Photosynthesis and activity of superoxide dismutase, peroxidase and glutathione reductase in cotton under salt stress. Environ. Exp. Bot. 2003, 49, 69–76. [Google Scholar] [CrossRef]
- Sirhindi, G.; Kumar, S.; Bhardwaj, R.; Kumar, M. Effects of 24-epibrassinolide and 28-homobrassinolide on the growth and antioxidant enzyme activities in the seedlings of Brassica juncea L. Physiol. Mol. Biol. Plants 2009, 15, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Smith, J.A.C.; Harberd, N.P.; Jiang, C.F. The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Mol. Biol. 2016, 91, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Hegedüs, A.; Erdei, S.; Horváth, G. Comparative studies of H2O2 detoxifying enzymes in green and greening barley seedlings under cadmium stress. Plant Sci. 2001, 160, 1085–1093. [Google Scholar] [CrossRef]
- Jain, P.; Bhatla, S.C. Tyrosine nitration of cytosolic peroxidase is probably triggered as a long distance signaling response in sunflower seedling cotyledons subjected to salt stress. PLoS ONE 2018, 13, e0197132. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.M. Polyphenol oxidases in plants and fungi: Going places? A review. Phytochemistry 2006, 67, 2318–2331. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, W.; Mahmoo, T. Response of rice polyphenol oxidase promoter to drought and salt stress. Pak. J. Bot. 2017, 49, 21–23. [Google Scholar]
- Li, S.W.; Leng, Y.; Feng, L.; Zeng, X.Y. Involvement of abscisic acid in regulating antioxidative defense systems and IAA-oxidase activity and improving adventitious rooting in mung bean [Vigna radiata (L.) Wilczek] seedlings under cadmium stress. Environ. Sci. Pollut. R. 2014, 21, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; Pandey, M.; et al. Abscisic acid signaling and abiotic stress tolerance in plants: A review on current knowledge and future prospects. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef] [PubMed]
- Di, F.F.; Jian, H.J.; Wang, T.Y.; Chen, X.P.; Ding, Y.R.; Du, H.; Lu, K.; Li, J.N.; Liu, L.Z. Genome-wide analysis of the PYL gene family and identification of PYL genes that respond to abiotic stress in Brassica napus. Genes 2018, 9, 156. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.X.; Wang, M.M.; Yin, Y.L.; Onac, E.; Zhou, G.F.; Peng, S.; Xia, X.J.; Shi, K.; Yu, J.Q.; Zhou, Y.H. RNA-seq analysis reveals the role of red light in resistance against Pseudomonas syringae pv. tomato DC3000 in tomato plants. BMC Genom. 2015, 16, 120. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.N.; Guo, X.J.; Cui, Y.Z.; Zheng, X.G.; Yang, H.Q. Comparative transcriptome analysis reveals hormone signaling genes involved in the launch of culm-shape differentiation in Dendrocalamus sinicus. Genes 2018, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Vashisth, D.; Kumar, R.; Rastogi, S.; Patel, V.K.; Kalra, A.; Gupta, M.M.; Gupta, A.K.; Shasany, A.K. Transcriptome changes induced by abiotic stresses in Artemisia annua. Sci. Rep. 2018, 8, 3423. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.W.; Hou, K.; Qin, P.; Liu, H.C.; Yi, B.; Yang, W.T.; Wu, W. RNA-seq for gene identification and transcript profiling of three Stevia rebaudiana genotypes. BMC Genom. 2014, 15, 571. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.J.; Chen, M.Q.; Dong, B.; Wang, N.H.; Yu, Q.; Wang, X.L.; Xuan, L.J.; Wang, Y.L.; Zhang, S.Z.; Shen, Y.M. Transcriptomic analysis of flower bud differentiation in Magnolia sinostellata. Genes 2018, 9, 212. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2005; Available online: www.R-project.org (accessed on 14 November 2018).
- Kar, M.; Mishra, D. Catalase, peroxidase and polyphenol oxidase activities during rice leaf senescence. Plant Physiol. 1976, 57, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Beffa, R.; Martin, H.V.; Pilet, P.E. In vitro oxidation of indoleacetic acid by soluble auxin-oxidases and peroxidases from maize roots. Plant Physiol. 1990, 94, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Rout, G.R. Effect of auxins on adventitious root development from single node cuttings of Camellia sinensis (L.) Kuntze and associated biochemical changes. Plant Growth Regul. 2006, 48, 111–117. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Tarazona, S.; García-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011, 21, 4436. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Xia, R.; Chen, H.; He, Y.H. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. BioRxiv 2018. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2—Elegant Graphics of Data Analysis; Springer: Berlin, Germany, 2016. [Google Scholar]
- Lalitha, S. Primer Premier 5. Biotech Softw. Internet Rep. 2000, 1, 270. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hauser, F.; Waadt, R.; Schroeder, J. Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 2011, 21, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Zhu, J.K. Regulation of abscisic acid biosynthesis. Plant Physiol. 2003, 133, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N. Abscisic acid and abiotic stress signaling. Plant Signal. Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.C.; Schwartz, S.H.; Zeevaart, J.A.D.; McCarty, D.R. Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA 1997, 94, 12235–12240. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Tan, B.C.; Gage, D.A.; Zeevaart, J.A.; McCarty, D.R. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 1997, 276, 1872–1874. [Google Scholar] [CrossRef] [PubMed]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Sah, S.K.; Reddy, K.R.; Li, J.X. Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.; Cho, Y.G. Plant hormones in salt stress tolerance. J. Plant Biol. 2015, 58, 147–155. [Google Scholar] [CrossRef]
- Suzuki, N.; Bassil, E.; Hamilton, J.S.; Inupakutika, M.A.; Zandalinas, I.; Tripathy, D.; Luo, Y.T.; Dion, E.; Fukui, G.; Kumazaki, A.; et al. ABA is required for plant acclimation to a combination of salt and heat stress. PLoS ONE 2016, 11, e0147625. [Google Scholar] [CrossRef] [PubMed]
- Verslues, P.E. ABA and cytokinins: Challenge and opportunity for plant stress research. Plant Mol. Biol. 2016, 91, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Wang, S.P. Insights into auxin signaling in plant-pathogen interactions. Front. Plant Sci. 2011, 2, 74. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.D. Auxin Biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 2012, 5, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Mano, Y.; Nemoto, K. The pathway of auxin biosynthesis in plants. J. Exp. Bot. 2012, 63, 2853–2872. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xie, W.F.; Zhang, L.; Valpuesta, V.; Ye, Z.W.; Gao, Q.H.; Duan, K. Auxin biosynthesis by the YUCCA6 flavin monooxygenase gene in woodland strawberry. J. Integr. Plant Biol. 2014, 56, 350–363. [Google Scholar] [CrossRef] [PubMed]
- Boeckx, T.; Winters, A.; Webb, K.J.; Kingston-Smith, A.H. Detection of potential chloroplastic substrates for polyphenol oxidase suggest a role in undamaged leaves. Front. Plant Sci. 2017, 8, 237. [Google Scholar] [CrossRef] [PubMed]
- Boeckx, T.; Webster, R.; Winter, A.L.; Webb, K.J.; Gay, A.; Kingston-Smith, A.H. Polyphenol oxidase-mediated protection against oxidative stress is not associated with enhanced photosynthetic efficiency. Ann. Bot. 2015, 116, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Kaur, L.; Zhawar, V.K. Phenolic parameters under exogenous ABA, water stress, salt stress in two wheat cultivars varying in drought tolerance. Ind. J. Plant Physiol. 2015, 20, 151–156. [Google Scholar] [CrossRef]
- Chibbar, R.N.; van Huystee, R.B. Characterization of peroxidase in plant cell. Plant Physiol. 1984, 75, 956–958. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.Q.; Wang, Y.C.; Liu, G.F.; Wang, C.; Jiang, J.; Yang, C.P. Cloning of ten peroxidase genes from Tamarix hispida and characterization of their responses to abiotic stress. Plant Mol. Biol. Rep. 2010, 28, 77–89. [Google Scholar] [CrossRef]
- Zhang, J.X.; Kirkham, M.B. Drought-stress-induced changes in activities of superoxide dismutase, catalase, and peroxidase in wheat species. Plant Cell Physiol. 1994, 35, 785–791. [Google Scholar] [CrossRef]
- Moura, J.C.M.S.; Bonine, C.A.V.; Viana, J.D.O.F.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.Q.; Luo, L.; Zheng, L.Q. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef] [PubMed]
- Kleczkowski, L.A.; Kunz, S.; Wilczynska, M. Mechanisms of UDP-glucose synthesis in plants. Crit. Rev. Plant Sci. 2010, 29, 191–203. [Google Scholar] [CrossRef]
- Wagstaff, B.A.; Rejzek, M.; Pesnot, T.; Tedaldi, L.M.; Caputi, L.; O’Neill, E.C.; Benini, S.; Wagner, G.K.; Field, R.A. Enzymatic synthesis of nucleobase-modified UDP-sugars: Scope and limitations. Carbohyd. Res. 2015, 404, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Wahl, C.; Spiertz, M.; Elling, L. Characterization of a new UDP-sugar pyrophosphorylase from Hordeum vulgare (barley). J. Biotechnol. 2017, 258, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Klezkowski, L.A.; Decker, D.; Wilczynska, M. UDP-sugar pyrophosphorylase—A new old mechanism for sugar activation. Plant Physiol. 2011, 156, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Geserick, C.; Tenhaken, R. UDP-sugar pyrophosphorylase is essential for arabinose and xylose recycling, and is required during vegetative and reproductive growth in Arabidopsis. Plant J. 2013, 74, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Schnurr, J.A.; Storey, K.K.; Jung, H.G.; Somers, D.A.; Gronwald, J.W. UDP-sugar pyrophosphorylase is essential for pollen development in Arabidopsis. Planta 2006, 224, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Coleman, H.D.; Canam, T.; Kang, K.; Ellis, D.D.; Mansfield, S.D. Over-expression of UDP-glucose pyrophosphorylase in hybrid poplar affects carbon allocation. J. Exp. Bot. 2007, 58, 4257–4268. [Google Scholar] [CrossRef] [PubMed]
- Micheli, F. Pectin methylesterases: Cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 2001, 6, 414–419. [Google Scholar] [CrossRef]
- Fruk, G.; Cmelik, Z.; Jemric, T.; Hribar, J.; Vidrih, R. Pectin role in woolliness development in peaches and nectarines: A review. Sci. Hortic. 2014, 180, 1–5. [Google Scholar] [CrossRef]
- Kyriakidis, N.B. Use of pectinesterase for detection of hydrocolloids addition in natural orange juice. Food Hydrocoll. 1999, 19, 497–500. [Google Scholar] [CrossRef]
- Daher, F.B.; Braybrook, S.A. How to let go: Pectin and plant cell adhesion. Front. Plant Sci. 2015, 6, 523. [Google Scholar] [CrossRef] [PubMed]
- Pelloux, J.; Rustérucci, C.; Mellerowicz, E.J. New insights into pectin methylesterase structure and function. Trends Plant Sci. 2007, 12, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Grebnev, G.; Ntefidou, M.; Kost, B. Secretion and endocytosis in pollen tubes: Models of tip growth in the spot light. Front. Plant Sci. 2017, 8, 154. [Google Scholar] [CrossRef] [PubMed]
- Leśniewska, J.; Öhman, D.; Krzesłowska, M.; Kushwah, S.; Barciszewska-Pacak, M.; Kleczkowski, L.; Sundberg, B.; Moritz, T.; Mellerowicz, E.J. Defense responses in aspen with altered pectin methylesterase activity reveal the hormonal inducers of tyloses. Plant Physiol. 2017, 173, 1409–1419. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.N.; Sun, Y.; Pei, Y.K.; Zhang, X.Y.; Wang, P.; Li, X.C.; Li, F.G.; Hou, Y.X. A pectin methylesterase inhibitor enhances resistance to verticillium wilt. Plant Physiol. 2018, 176, 2202–2220. [Google Scholar] [CrossRef] [PubMed]
- Hasunuma, T.; Fukusaki, E.; Kobayashi, A. Expression of fungal pectin methylesterase in transgenic tobacco leads to alteration in cell wall metabolism and a dwarf phenotype. J. Biotechnol. 2004, 111, 241–251. [Google Scholar] [CrossRef] [PubMed]
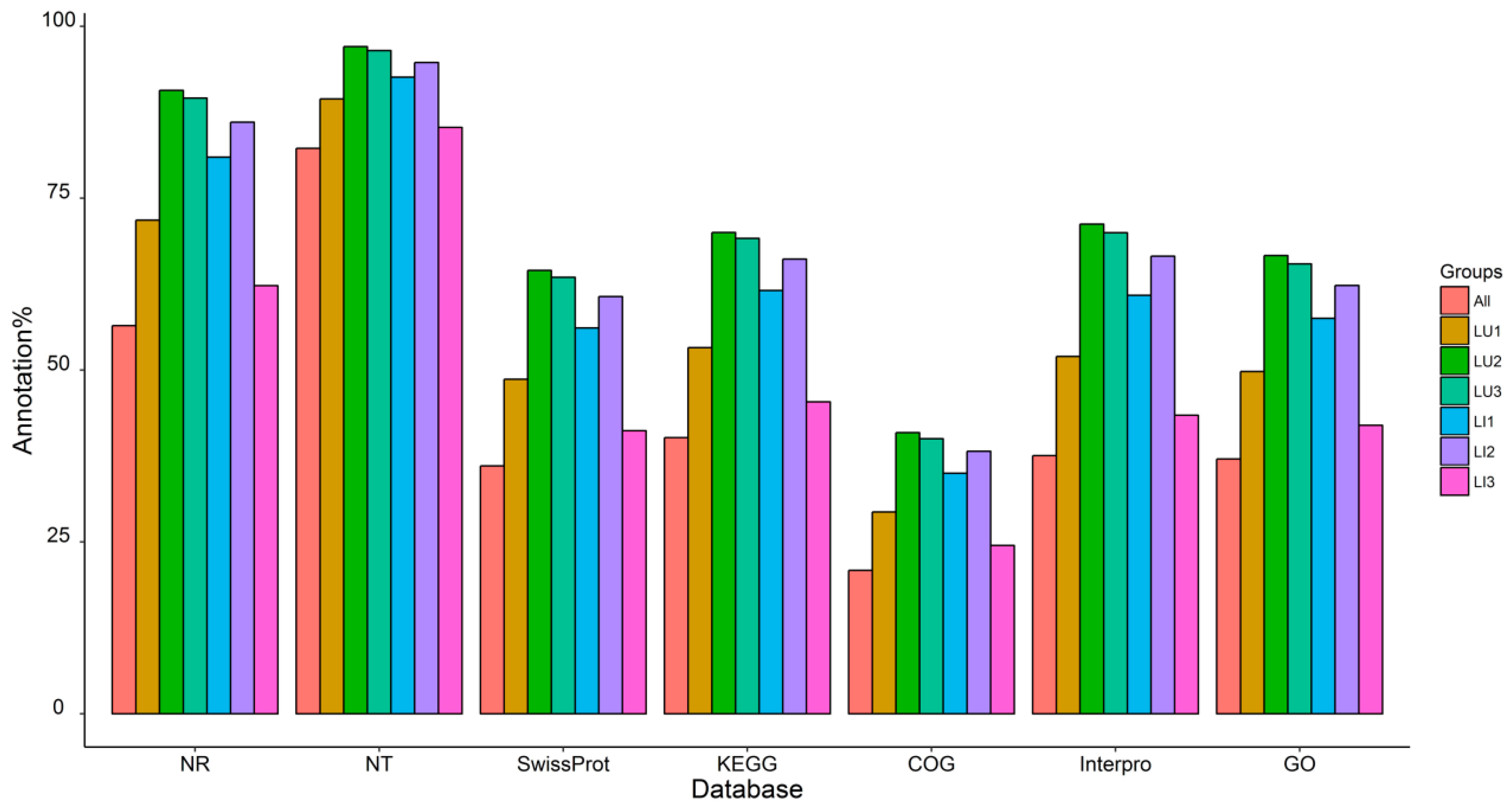
| Samples | Reads | Unigenes | ||||||
|---|---|---|---|---|---|---|---|---|
| Total Raw Reads (Mb) | Total Clean Reads (Mb) | Clean Reads Q20 (%) | Clean Reads Q30 (%) | Total Number | Mean Length (nt) | N50 | N90 | |
| LU1 | 64.78 | 44.75 | 98.09 | 94.47 | 127876 | 543 | 826 | 238 |
| LU2 | 64.78 | 44.39 | 98 | 94.19 | 68747 | 805 | 1336 | 310 |
| LU3 | 63.16 | 44.8 | 98.05 | 94.32 | 73913 | 857 | 1471 | 323 |
| LI1 | 63.16 | 44.86 | 98.03 | 94.27 | 100268 | 741 | 1354 | 272 |
| LI2 | 64.78 | 45.17 | 97.94 | 94.03 | 86133 | 784 | 1386 | 285 |
| LI3 | 64.78 | 44.31 | 97.86 | 93.9 | 210767 | 560 | 801 | 246 |
| All | - | - | - | - | 296815 | 693 | 1017 | 294 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, A.-P.; Zong, D.; Gan, P.-H.; Zou, X.-L.; Fei, X.; Zhong, Y.-Y.; He, C.-Z. Physiological Analysis and Transcriptome Profiling of Inverted Cuttings of Populus yunnanensis Reveal That Cell Wall Metabolism Plays a Crucial Role in Responding to Inversion. Genes 2018, 9, 572. https://doi.org/10.3390/genes9120572
Zhou A-P, Zong D, Gan P-H, Zou X-L, Fei X, Zhong Y-Y, He C-Z. Physiological Analysis and Transcriptome Profiling of Inverted Cuttings of Populus yunnanensis Reveal That Cell Wall Metabolism Plays a Crucial Role in Responding to Inversion. Genes. 2018; 9(12):572. https://doi.org/10.3390/genes9120572
Chicago/Turabian StyleZhou, An-Pei, Dan Zong, Pei-Hua Gan, Xin-Lian Zou, Xuan Fei, Yuan-Yuan Zhong, and Cheng-Zhong He. 2018. "Physiological Analysis and Transcriptome Profiling of Inverted Cuttings of Populus yunnanensis Reveal That Cell Wall Metabolism Plays a Crucial Role in Responding to Inversion" Genes 9, no. 12: 572. https://doi.org/10.3390/genes9120572
APA StyleZhou, A.-P., Zong, D., Gan, P.-H., Zou, X.-L., Fei, X., Zhong, Y.-Y., & He, C.-Z. (2018). Physiological Analysis and Transcriptome Profiling of Inverted Cuttings of Populus yunnanensis Reveal That Cell Wall Metabolism Plays a Crucial Role in Responding to Inversion. Genes, 9(12), 572. https://doi.org/10.3390/genes9120572





