Terpenoid Metabolic Engineering in Photosynthetic Microorganisms
Abstract
1. Introduction
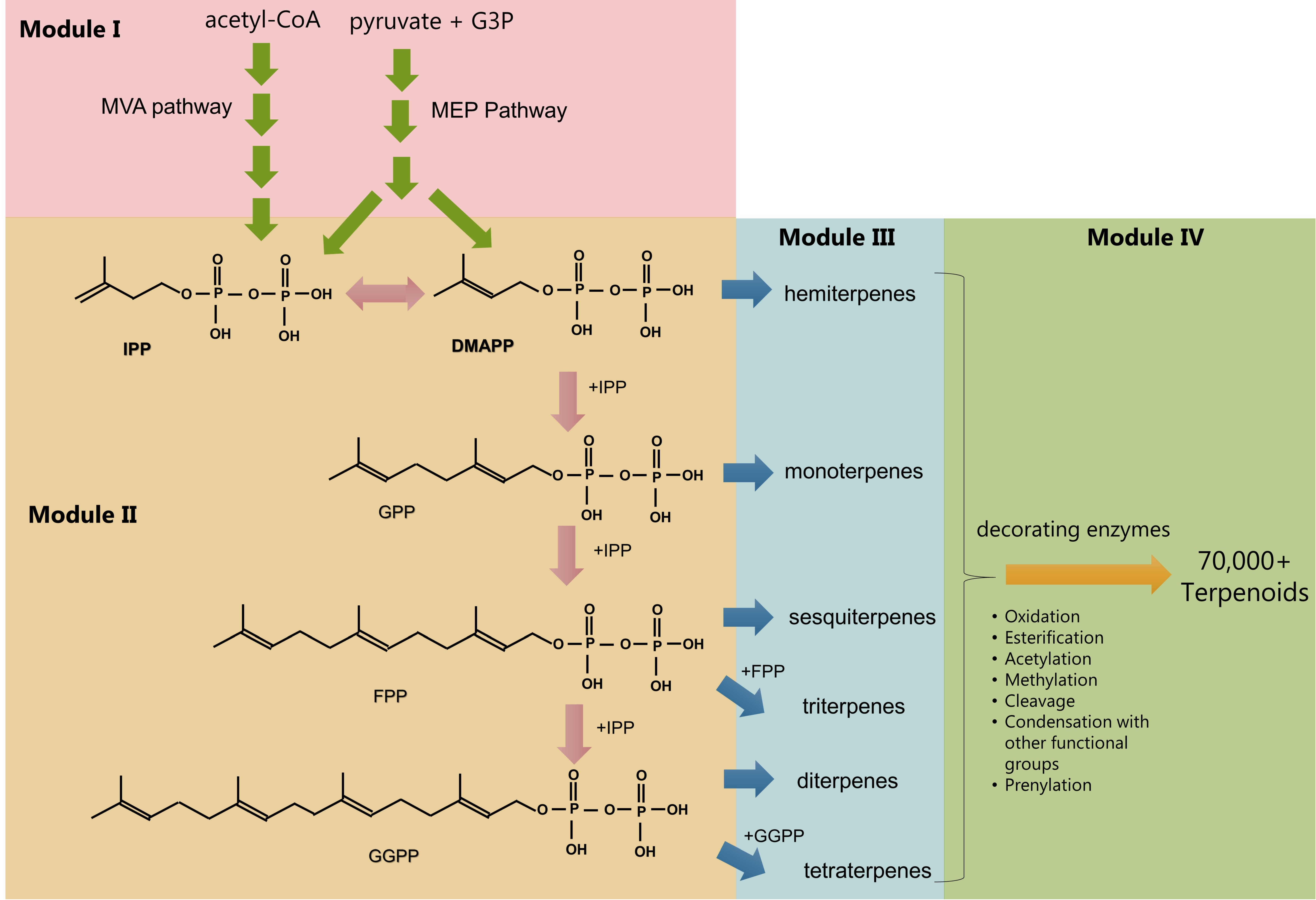
2. Pathway Modules in Terpenoid Biosynthesis
2.1. Module I: Redirection of Resources Towards Isopentenyl Pyrophosphate and Dimethylallyl Pyrophosphate
2.2. Module II: Prenyl Phosphate Metabolism
2.3. Modules III and IV: Terpene Synthases, Skeleton Decorations, and Further Modifications
2.4. General Considerations Spanning Across Nodes
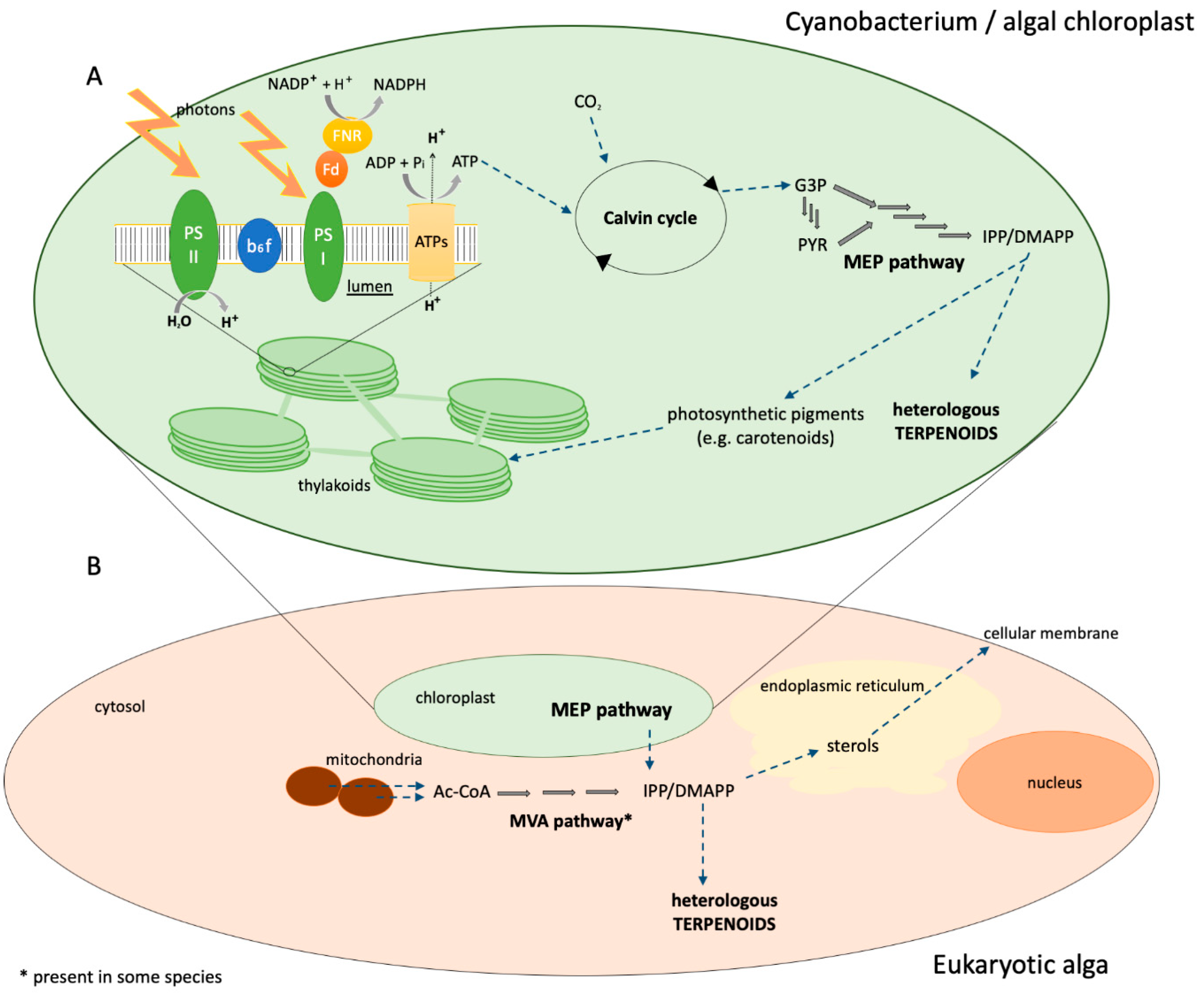
3. Photosynthetic Microorganisms as Terpenoid Production Hosts
3.1. Cyanobacteria
3.2. Eukaryotic Algae
3.2.1. Chlamydomonas reinhardtii
3.2.2. Diatoms
4. Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Pichersky, E.; Raguso, R.A. Why do plants produce so many terpenoid compounds? New Phytol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Gil, J.; Rodríguez-Concepción, M.; Vickers, C.E. Formation of Isoprenoids. In Biogenesis of Fatty Acids, Lipids and Membranes; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–29. ISBN 978-3-319-43676-0. [Google Scholar]
- Tholl, D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol. 2006, 9, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Tholl, D. Biosynthesis and Biological Functions of Terpenoids in Plants. In Advances in Biochemical Engineering/Biotechnology; Springer: Cham, Switzerland, 2015; Volume 148, pp. 63–106. [Google Scholar]
- Pateraki, I.; Heskes, A.M.; Hamberger, B. Cytochromes P450 for terpene functionalisation and metabolic engineering. Adv. Biochem. Eng. Biotechnol. 2015, 148, 107–139. [Google Scholar] [CrossRef] [PubMed]
- Lassen, L.M.; Nielsen, A.Z.; Ziersen, B.; Gnanasekaran, T.; Møller, B.L.; Jensen, P.E. Redirecting Photosynthetic Electron Flow into Light-Driven Synthesis of Alternative Products Including High-Value Bioactive Natural Compounds. ACS Synth. Biol. 2014, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, S.; Chu, H.; Felding, J.; Baran, P.S. Nineteen-step total synthesis of (+)-phorbol. Nature 2016, 532, 90–93. [Google Scholar] [CrossRef] [PubMed]
- Arendt, P.; Pollier, J.; Callewaert, N.; Goossens, A. Synthetic biology for production of natural and new-to-nature terpenoids in photosynthetic organisms. Plant J. 2016, 87, 16–37. [Google Scholar] [CrossRef] [PubMed]
- Ignea, C.; Athanasakoglou, A.; Ioannou, E.; Georgantea, P.; Trikka, F.A.; Loupassaki, S.; Roussis, V.; Makris, A.M.; Kampranis, S.C. Carnosic acid biosynthesis elucidated by a synthetic biology platform. Proc. Natl. Acad. Sci. USA 2016, 113, 3681–3686. [Google Scholar] [CrossRef] [PubMed]
- Pateraki, I.; Andersen-Ranberg, J.; Jensen, N.B.; Wubshet, S.G.; Heskes, A.M.; Forman, V.; Hallström, B.; Hamberger, B.; Motawia, M.S.; Olsen, C.E.; et al. Total biosynthesis of the cyclic AMP booster forskolin from Coleus forskohlii. eLife 2017, 6, e23001. [Google Scholar] [CrossRef] [PubMed]
- Ignea, C.; Ioannou, E.; Georgantea, P.; Loupassaki, S.; Trikka, F.A.; Kanellis, A.K.; Makris, A.M.; Roussis, V.; Kampranis, S.C. Reconstructing the chemical diversity of labdane-type diterpene biosynthesis in yeast. Metab. Eng. 2015, 28, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Zi, J.; Mafu, S.; Peters, R.J. To Gibberellins and Beyond! Surveying the Evolution of (Di)Terpenoid Metabolism. Annu. Rev. Plant Biol. 2014, 65, 259–286. [Google Scholar] [CrossRef] [PubMed]
- Zerbe, P.; Hamberger, B.B.; Yuen, M.M.S.; Chiang, A.; Sandhu, H.K.; Madilao, L.L.; Nguyen, A.; Hamberger, B.B.; Bach, S.S.; Bohlmann, J. Gene discovery of modular diterpene metabolism in nonmodel systems. Plant Physiol. 2013, 162, 1073–1091. [Google Scholar] [CrossRef] [PubMed]
- Paddon, C.J.; Westfall, P.J.; Pitera, D.J.; Benjamin, K.; Fisher, K.; McPhee, D.; Leavell, M.D.; Tai, A.; Main, A.; Eng, D.; et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 2013, 496, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Ajikumar, P.K.; Xiao, W.-H.; Tyo, K.E.J.; Wang, Y.; Simeon, F.; Leonard, E.; Mucha, O.; Phon, T.H.; Pfeifer, B.; Stephanopoulos, G. Isoprenoid Pathway Optimization for Taxol Precursor Overproduction in Escherichia coli. Science 2010, 330, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Vickers, C.E.; Bongers, M.; Liu, Q.; Delatte, T.; Bouwmeester, H. Metabolic engineering of volatile isoprenoids in plants and microbes. Plant Cell Environ. 2014, 37, 1753–1775. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.-D.; MacNevin, G.; Ro, D.-K. De novo synthesis of high-value plant sesquiterpenoids in yeast. Methods Enzymol. 2012, 517, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Bian, G.; Deng, Z.; Liu, T. Strategies for terpenoid overproduction and new terpenoid discovery. Curr. Opin. Biotechnol. 2017, 48, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Davies, F.K.; Jinkerson, R.E.; Posewitz, M.C. Toward a photosynthetic microbial platform for terpenoid engineering. Photosynth. Res. 2015, 123, 265–284. [Google Scholar] [CrossRef] [PubMed]
- Ward, V.C.A.; Chatzivasileiou, A.O.; Stephanopoulos, G. Metabolic engineering of Escherichia coli for the production of isoprenoids. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef] [PubMed]
- Bian, G.; Han, Y.; Hou, A.; Yuan, Y.; Liu, X.; Deng, Z.; Liu, T. Releasing the potential power of terpene synthases by a robust precursor supply platform. Metab. Eng. 2017, 42, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Andersen-Ranberg, J.; Kongstad, K.T.; Nielsen, M.T.; Jensen, N.B.; Pateraki, I.; Bach, S.S.; Hamberger, B.; Zerbe, P.; Staerk, D.; Bohlmann, J.; et al. Expanding the Landscape of Diterpene Structural Diversity through Stereochemically Controlled Combinatorial Biosynthesis. Angew. Chem. Int. Ed. Engl. 2016, 128, 2182–2186. [Google Scholar] [CrossRef]
- Zurbriggen, A.; Kirst, H.; Melis, A. Isoprene Production via the Mevalonic Acid Pathway in Escherichia coli (Bacteria). BioEnergy Res. 2012, 5, 814–828. [Google Scholar] [CrossRef]
- Bentley, F.K.; Zurbriggen, A.; Melis, A. Heterologous expression of the mevalonic acid pathway in cyanobacteria enhances endogenous carbon partitioning to isoprene. Mol. Plant 2014, 7, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Maury, J.; Asadollahi, M.A.; Møller, K.; Schalk, M.; Clark, A.; Formenti, L.R.; Nielsen, J. Reconstruction of a bacterial isoprenoid biosynthetic pathway in Saccharomyces cerevisiae. FEBS Lett. 2008, 582, 4032–4038. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Guo, L. Biosynthesis of β-carotene in engineered E. coli using the MEP and MVA pathways. Microb. Cell Fact. 2014, 13, 160. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Gao, X.; Jiang, Y.; Sun, B.; Gao, F.; Yang, S. Synergy between methylerythritol phosphate pathway and mevalonate pathway for isoprene production in Escherichia coli. Metab. Eng. 2016, 37, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Gutierrez, J.; Chan, R.; Batth, T.S.; Adams, P.D.; Keasling, J.D.; Petzold, C.J.; Lee, T.S. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metab. Eng. 2013, 19, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Calabria, A.R.; Cervin, M.A.; Chotani, G.K.; La Duca, R.; McAuliffe, J.C.; Miller, M.C.; Sabo, T.A.; Sanford, K.J.; Spring, E.L.; Whited, G.M. Compositions and Methods for Producing Isoprene Free of c5 Hydrocarbons under Decoupling Conditions and/or Safe Operating Ranges. Patent WO 2010/003007 A3, 7 January 2010. [Google Scholar]
- Kim, S.-W.; Keasling, J.D. Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol. Bioeng. 2001, 72, 408–415. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, J.; Qin, B.; Li, Y.; Sun, Y.; Su, S.; Xian, M. Biosynthesis of isoprene in Escherichia coli via methylerythritol phosphate (MEP) pathway. Appl. Microbiol. Biotechnol. 2011, 90, 1915–1922. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, J.L.; Brown, M.S. Regulation of the mevalonate pathway. Nature 1990, 343, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xian, M.; Su, S.; Zhao, G.; Nie, Q.; Jiang, X.; Zheng, Y.; Liu, W. Enhancing Production of Bio-Isoprene Using Hybrid MVA Pathway and Isoprene Synthase in E. coli. PLoS ONE 2012, 7, e33509. [Google Scholar] [CrossRef] [PubMed]
- George, K.W.; Thompson, M.G.; Kim, J.; Baidoo, E.E.K.; Wang, G.; Benites, V.T.; Petzold, C.J.; Chan, L.J.G.; Yilmaz, S.; Turhanen, P.; et al. Integrated analysis of isopentenyl pyrophosphate (IPP) toxicity in isoprenoid-producing Escherichia coli. Metab. Eng. 2018, 47, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Meadows, A.L.; Hawkins, K.M.; Tsegaye, Y.; Antipov, E.; Kim, Y.; Raetz, L.; Dahl, R.H.; Tai, A.; Mahatdejkul-Meadows, T.; Xu, L.; et al. Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nature 2016, 537, 694–697. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Plan, M.R.; Chrysanthopoulos, P.; Hodson, M.P.; Nielsen, L.K.; Vickers, C.E. A squalene synthase protein degradation method for improved sesquiterpene production in Saccharomyces cerevisiae. Metab. Eng. 2017, 39, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Jensen, E.D.; Ferreira, R.; Jakočiūnas, T.; Arsovska, D.; Zhang, J.; Ding, L.; Smith, J.D.; David, F.; Nielsen, J.; Jensen, M.K.; et al. Transcriptional reprogramming in yeast using dCas9 and combinatorial gRNA strategies. Microb. Cell Fact. 2017, 16, 46. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Ye, L.; Lv, X.; Xu, H.; Yu, H. Sequential control of biosynthetic pathways for balanced utilization of metabolic intermediates in Saccharomyces cerevisiae. Metab. Eng. 2015, 28, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Tarshis, L.C.; Yan, M.; Poulter, C.D.; Sacchettini, J.C. Crystal Structure of Recombinant Farnesyl Diphosphate Synthase at 2.6-.ANG. Resolution. Biochemistry 1994, 33, 10871–10877. [Google Scholar] [CrossRef] [PubMed]
- Ignea, C.; Pontini, M.; Maffei, M.E.; Makris, A.M.; Kampranis, S.C. Engineering monoterpene production in yeast using a synthetic dominant negative geranyl diphosphate synthase. ACS Synth. Biol. 2014, 3, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Nielsen, L.K.; Kampranis, S.C.; Vickers, C.E. Engineered protein degradation of farnesyl pyrophosphate synthase is an effective regulatory mechanism to increase monoterpene production in Saccharomyces cerevisiae. Metab. Eng. 2018. [Google Scholar] [CrossRef] [PubMed]
- Brückner, K.; Tissier, A. High-level diterpene production by transient expression in Nicotiana benthamiana. Plant Methods 2013, 9, 46. [Google Scholar] [CrossRef] [PubMed]
- Gnanasekaran, T.; Vavitsas, K.; Andersen-Ranberg, J.; Nielsen, A.Z.; Olsen, C.E.; Hamberger, B.; Jensen, P.E. Heterologous expression of the isopimaric acid pathway in Nicotiana benthamiana and the effect of N-terminal modifications of the involved cytochrome P450 enzyme. J. Biol. Eng. 2015, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Ignea, C.; Trikka, F.A.; Nikolaidis, A.K.; Georgantea, P.; Ioannou, E.; Loupassaki, S.; Kefalas, P.; Kanellis, A.K.; Roussis, V.; Makris, A.M.; et al. Efficient diterpene production in yeast by engineering Erg20p into a geranylgeranyl diphosphate synthase. Metab. Eng. 2015, 27, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Christianson, D.W. Structural and Chemical Biology of Terpenoid Cyclases. Chem. Rev. 2017, 117, 11570–11648. [Google Scholar] [CrossRef] [PubMed]
- Formighieri, C.; Melis, A. Regulation of β-phellandrene synthase gene expression, recombinant protein accumulation, and monoterpene hydrocarbons production in Synechocystis transformants. Planta 2014, 240, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Denby, C.M.; Li, R.A.; Vu, V.T.; Costello, Z.; Lin, W.; Chan, L.J.G.; Williams, J.; Donaldson, B.; Bamforth, C.W.; Petzold, C.J.; et al. Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer. Nat. Commun. 2018, 9, 965. [Google Scholar] [CrossRef] [PubMed]
- Wichmann, J.; Baier, T.; Wentnagel, E.; Lauersen, K.J.; Kruse, O. Tailored carbon partitioning for phototrophic production of (E)-α-bisabolene from the green microalga Chlamydomonas reinhardtii. Metab. Eng. 2018, 45, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef] [PubMed]
- Vattekkatte, A.; Garms, S.; Brandt, W.; Boland, W. Enhanced structural diversity in terpenoid biosynthesis: Enzymes, substrates and cofactors. Org. Biomol. Chem. 2018, 16, 348–362. [Google Scholar] [CrossRef] [PubMed]
- Moses, T.; Pollier, J.; Almagro, L.; Buyst, D.; Van Montagu, M.; Pedreño, M.A.; Martins, J.C.; Thevelein, J.M.; Goossens, A. Combinatorial biosynthesis of sapogenins and saponins in Saccharomyces cerevisiae using a C-16α hydroxylase from Bupleurum falcatum. Proc. Natl. Acad. Sci. USA 2014, 111, 1634–1639. [Google Scholar] [CrossRef] [PubMed]
- Bak, S.; Beisson, F.; Bishop, G.; Hamberger, B.; Höfer, R.; Paquette, S.; Werck-Reichhart, D. Cytochromes P450. Arabidopsis Book 2011, 9, e0144. [Google Scholar] [CrossRef] [PubMed]
- Munro, A.W.; Girvan, H.M.; Mason, A.E.; Dunford, A.J.; McLean, K.J. What makes a P450 tick? Trends Biochem. Sci. 2013, 38, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Mellor, S.B.; Vavitsas, K.; Nielsen, A.Z.; Jensen, P.E. Photosynthetic fuel for heterologous enzymes: The role of electron carrier proteins. Photosynth. Res. 2017, 134, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Munro, A.W.; Girvan, H.M.; McLean, K.J. Variations on a (t)heme—Novel mechanisms, redox partners and catalytic functions in the cytochrome P450 superfamily. Nat. Prod. Rep. 2007, 24, 585–609. [Google Scholar] [CrossRef] [PubMed]
- Barnes, H.J.; Arlotto, M.P.; Waterman, M.R. Expression and enzymatic activity of recombinant cytochrome P450 17 alpha-hydroxylase in Escherichia coli. Proc. Natl. Acad. Sci. USA 1991, 88, 5597–5601. [Google Scholar] [CrossRef] [PubMed]
- Paddon, C.J.; Keasling, J.D. Semi-synthetic artemisinin: A model for the use of synthetic biology in pharmaceutical development. Nat. Rev. Microbiol. 2014, 12, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.Y.; Farasat, I.; Maranas, C.D.; Salis, H.M. Rational design of a synthetic Entner–Doudoroff pathway for improved and controllable NADPH regeneration. Metab. Eng. 2015, 29, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Chou, H.H.; Keasling, J.D. Programming adaptive control to evolve increased metabolite production. Nat. Commun. 2013, 4, 2595. [Google Scholar] [CrossRef] [PubMed]
- Özaydın, B.; Burd, H.; Lee, T.S.; Keasling, J.D. Carotenoid-based phenotypic screen of the yeast deletion collection reveals new genes with roles in isoprenoid production. Metab. Eng. 2013, 15, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Trikka, F.A.; Nikolaidis, A.; Athanasakoglou, A.; Andreadelli, A.; Ignea, C.; Kotta, K.; Argiriou, A.; Kampranis, S.C.; Makris, A.M. Iterative carotenogenic screens identify combinations of yeast gene deletions that enhance sclareol production. Microb. Cell Fact. 2015, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Bongers, M.; Chrysanthopoulos, P.K.; Behrendorff, J.B.Y.H.; Hodson, M.P.; Vickers, C.E.; Nielsen, L.K. Systems analysis of methylerythritol-phosphate pathway flux in E. coli: Insights into the role of oxidative stress and the validity of lycopene as an isoprenoid reporter metabolite. Microb. Cell Fact. 2015, 14, 193. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, Y.J.; Siewers, V.; Nielsen, J. Enabling Technologies to Advance Microbial Isoprenoid Production. Adv. Biochem. Eng. Biotechnol. 2015, 148, 143–160. [Google Scholar] [PubMed]
- Ma, S.M.; Garcia, D.E.; Redding-Johanson, A.M.; Friedland, G.D.; Chan, R.; Batth, T.S.; Haliburton, J.R.; Chivian, D.; Keasling, J.D.; Petzold, C.J.; et al. Optimization of a heterologous mevalonate pathway through the use of variant HMG-CoA reductases. Metab. Eng. 2011, 13, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Kozak, B.U.; van Rossum, H.M.; Luttik, M.A.H.; Akeroyd, M.; Benjamin, K.R.; Wu, L.; de Vries, S.; Daran, J.-M.; Pronk, J.T.; van Maris, A.J.A. Engineering acetyl coenzyme A supply: Functional expression of a bacterial pyruvate dehydrogenase complex in the cytosol of Saccharomyces cerevisiae. MBio 2014, 5, e01696-14. [Google Scholar] [CrossRef] [PubMed]
- Van Rossum, H.M.; Kozak, B.U.; Pronk, J.T.; van Maris, A.J.A. Engineering cytosolic acetyl-coenzyme A supply in Saccharomyces cerevisiae: Pathway stoichiometry, free-energy conservation and redox-cofactor balancing. Metab. Eng. 2016, 36, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Bogorad, I.W.; Lin, T.-S.; Liao, J.C. Synthetic non-oxidative glycolysis enables complete carbon conservation. Nature 2013, 502, 693–697. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Qiao, K.; Edgar, S.; Stephanopoulos, G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat. Biotechnol. 2015, 33, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Vickers, C.E.; Williams, T.C.; Peng, B.; Cherry, J. Recent advances in synthetic biology for engineering isoprenoid production in yeast. Curr. Opin. Chem. Biol. 2017, 40, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Vickers, C.E.; Gershenzon, J.; Lerdau, M.T.; Loreto, F. A unified mechanism of action for volatile isoprenoids in plant abiotic stress. Nat. Chem. Biol. 2009, 5, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.Z.; Mellor, S.B.; Vavitsas, K.; Wlodarczyk, A.J.; Gnanasekaran, T.; Perestrello Ramos H de Jesus, M.; King, B.C.; Bakowski, K.; Jensen, P.E. Extending the biosynthetic repertoires of cyanobacteria and chloroplasts. Plant J. 2016, 87, 87–102. [Google Scholar] [CrossRef] [PubMed]
- Chaves, J.E.; Melis, A. Biotechnology of cyanobacterial isoprene production. Appl. Microbiol. Biotechnol. 2018, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Gao, F.; Liu, D.; Zhang, H.; Nie, X.; Yang, C. Engineering the methylerythritol phosphate pathway in cyanobacteria for photosynthetic isoprene production from CO2. Energy Environ. Sci. 2016, 9, 1400–1411. [Google Scholar] [CrossRef]
- Formighieri, C.; Melis, A. Sustainable heterologous production of terpene hydrocarbons in cyanobacteria. Photosynth. Res. 2016, 130, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Davies, F.K.; Work, V.H.; Beliaev, A.S.; Posewitz, M.C. Engineering Limonene and Bisabolene Production in Wild Type and a Glycogen-Deficient Mutant of Synechococcus sp. PCC 7002. Front. Bioeng. Biotechnol. 2014, 2, 21. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.-C.; Saha, R.; Zhang, F.; Pakrasi, H.B. Metabolic engineering of the pentose phosphate pathway for enhanced limonene production in the cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 2017, 7, 17503. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, C.; Gu, L.; Gibbons, W.; Zhou, R. Genetically engineering cyanobacteria to convert CO2, water, and light into the long-chain hydrocarbon farnesene. Appl. Microbiol. Biotechnol. 2014, 98, 9869–9877. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.Y.; Lee, H.J.; Choi, J.; Kim, J.; Sim, S.J.; Um, Y.; Kim, Y.; Lee, T.S.; Keasling, J.D.; Woo, H.M. Photosynthetic conversion of CO2 to farnesyl diphosphate-derived phytochemicals (amorpha-4,11-diene and squalene) by engineered cyanobacteria. Biotechnol. Biofuels 2016, 9, 202. [Google Scholar] [CrossRef] [PubMed]
- Englund, E.; Pattanaik, B.; Ubhayasekera, S.J.K.; Stensjö, K.; Bergquist, J.; Lindberg, P. Production of Squalene in Synechocystis sp. PCC 6803. PLoS ONE 2014, 9, e90270. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Lee, J.; Lee, S.-M.; Um, Y.; Kim, Y.; Sim, S.J.; Choi, J.; Woo, H.M. Direct Conversion of CO2 to α-Farnesene Using Metabolically Engineered Synechococcus elongatus PCC 7942. J. Agric. Food Chem. 2017, 65, 10424–10428. [Google Scholar] [CrossRef] [PubMed]
- Formighieri, C.; Melis, A. Heterologous synthesis of geranyllinalool, a diterpenol plant product, in the cyanobacterium Synechocystis. Appl. Microbiol. Biotechnol. 2017, 101, 2791–2800. [Google Scholar] [CrossRef] [PubMed]
- Vavitsas, K.; Rue, E.Ø.; Stefánsdóttir, L.K.; Gnanasekaran, T.; Blennow, A.; Crocoll, C.; Gudmundsson, S.; Jensen, P.E. Responses of Synechocystis sp. PCC 6803 to heterologous biosynthetic pathways. Microb. Cell Fact. 2017, 16, 140. [Google Scholar] [CrossRef] [PubMed]
- Lauersen, K.J.; Baier, T.; Wichmann, J.; Wördenweber, R.; Mussgnug, J.H.; Hübner, W.; Huser, T.; Kruse, O. Efficient phototrophic production of a high-value sesquiterpenoid from the eukaryotic microalga Chlamydomonas reinhardtii. Metab. Eng. 2016, 38, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Lauersen, K.J.; Wichmann, J.; Baier, T.; Kampranis, S.C.; Pateraki, I.; Møller, B.L.; Kruse, O. Phototrophic production of heterologous diterpenoids and a hydroxy-functionalized derivative from Chlamydomonas reinhardtii. Metab. Eng. 2018, 49, 116–127. [Google Scholar] [CrossRef] [PubMed]
- D’Adamo, S.; Schiano di Visconte, G.; Lowe, G.; Szaub-Newton, J.; Beacham, T.; Landels, A.; Allen, M.J.; Spicer, A.; Matthijs, M. Engineering the unicellular alga Phaeodactylum tricornutum for high-value plant triterpenoid production. Plant Biotechnol. J. 2018. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Cano, M.; Wang, B.; Douchi, D.; Yu, J. The plasticity of cyanobacterial carbon metabolism. Curr. Opin. Chem. Biol. 2017, 41, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Ducat, D.C.; Way, J.C.; Silver, P.A. Engineering cyanobacteria to generate high-value products. Trends Biotechnol. 2011, 29, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Matson, M.M.; Atsumi, S. Photomixotrophic chemical production in cyanobacteria. Curr. Opin. Biotechnol. 2018, 50, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Gudmundsson, S.; Nogales, J. Cyanobacteria as photosynthetic biocatalysts: A systems biology perspective. Mol. Biosyst. 2015, 11, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Broddrick, J.T.; Rubin, B.E.; Welkie, D.G.; Du, N.; Mih, N.; Diamond, S.; Lee, J.J.; Golden, S.S.; Palsson, B.O. Unique attributes of cyanobacterial metabolism revealed by improved genome-scale metabolic modeling and essential gene analysis. Proc. Natl. Acad. Sci. USA 2016, 113, 8344–8353. [Google Scholar] [CrossRef] [PubMed]
- Rubin, B.E.; Wetmore, K.M.; Price, M.N.; Diamond, S.; Shultzaberger, R.K.; Lowe, L.C.; Curtin, G.; Arkin, A.P.; Deutschbauer, A.; Golden, S.S. The essential gene set of a photosynthetic organism. Proc. Natl. Acad. Sci. USA 2015, 112, 6634–6643. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Cengic, I.; Anfelt, J.; Hudson, E.P. Multiple Gene Repression in Cyanobacteria Using CRISPRi. ACS Synth. Biol. 2016, 5, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Shen, C.R.; Huang, C.-H.; Sung, L.-Y.; Wu, M.-Y.; Hu, Y.-C. CRISPR-Cas9 for the genome engineering of cyanobacteria and succinate production. Metab. Eng. 2016, 38, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Wang, S.; Rommelfanger, S.; Balassy, A.; Barba-Ostria, C.; Gu, P.; Galazka, J.M.; Zhang, F. Developing a Cas9-based tool to engineer native plasmids in Synechocystis sp. PCC 6803. Biotechnol. Bioeng. 2018. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Li, S.; Song, X.; Diao, J.; Chen, L.; Zhang, W. Toolboxes for cyanobacteria: Recent advances and future direction. Biotechnol. Adv. 2018. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, R.; Gale, G.A.; Schiavon, A.A.; Puzorjov, A.; Malin, J.; Gillespie, M.D.; Vavitsas, K.; Zulkower, V.; Wang, B.; Howe, C.J.; et al. CyanoGate: A Golden Gate modular cloning suite for engineering cyanobacteria based on the plant MoClo syntax. bioRxiv 2018, 426700. [Google Scholar] [CrossRef]
- Lindberg, P.; Park, S.; Melis, A. Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab. Eng. 2010, 12, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Chaves, J.E.; Rueda-Romero, P.; Kirst, H.; Melis, A. Engineering Isoprene Synthase Expression and Activity in Cyanobacteria. ACS Synth. Biol. 2017, 6, 2281–2292. [Google Scholar] [CrossRef] [PubMed]
- Englund, E.; Shabestary, K.; Hudson, E.P.; Lindberg, P. Systematic overexpression study to find target enzymes enhancing production of terpenes in Synechocystis PCC 6803, using isoprene as a model compound. Metab. Eng. 2018. [Google Scholar] [CrossRef] [PubMed]
- Bentley, F.K.; García-Cerdán, J.G.; Chen, H.-C.; Melis, A. Paradigm of Monoterpene (β-phellandrene) Hydrocarbons Production via Photosynthesis in Cyanobacteria. BioEnergy Res. 2013, 6, 917–929. [Google Scholar] [CrossRef]
- Englund, E.; Andersen-Ranberg, J.; Miao, R.; Hamberger, B.; Lindberg, P. Metabolic Engineering of Synechocystis sp. PCC 6803 for Production of the Plant Diterpenoid Manoyl Oxide. ACS Synth. Biol. 2015, 4, 1270–1278. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.C.; Lee, H.J.; Choi, S.Y.; Choi, J.; Woo, H.M. Bio-solar cell factories for photosynthetic isoprenoids production. Planta 2018, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ducat, D.C.; Avelar-Rivas, J.A.; Way, J.C.; Silver, P.A. Rerouting Carbon Flux To Enhance Photosynthetic Productivity. Appl. Environ. Microbiol. 2012, 78, 2660–2668. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, W.; Xin, C.; Zheng, Y.; Cheng, Y.; Sun, S.; Li, R.; Zhu, X.-G.; Dai, S.Y.; Rentzepis, P.M.; et al. Enhanced limonene production in cyanobacteria reveals photosynthesis limitations. Proc. Natl. Acad. Sci. USA 2016, 113, 14225–14230. [Google Scholar] [CrossRef] [PubMed]
- Wlodarczyk, A.; Gnanasekaran, T.; Nielsen, A.Z.; Zulu, N.N.; Mellor, S.B.; Luckner, M.; Thøfner, J.F.B.; Olsen, C.E.; Mottawie, M.S.; Burow, M.; et al. Metabolic engineering of light-driven cytochrome P450 dependent pathways into Synechocystis sp. PCC 6803. Metab. Eng. 2016, 33, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Berepiki, A.; Gittins, J.R.; Moore, C.M.; Bibby, T.S. Rational engineering of photosynthetic electron flux enhances light-powered cytochrome P450 activity. Synth. Biol. 2018. [Google Scholar] [CrossRef]
- Berepiki, A.; Hitchcock, A.; Moore, C.M.; Bibby, T.S. Tapping the Unused Potential of Photosynthesis with a Heterologous Electron Sink. ACS Synth. Biol. 2016, 5, 1369–1375. [Google Scholar] [CrossRef] [PubMed]
- Tomitani, A.; Knoll, A.H.; Cavanaugh, C.M.; Ohno, T. The evolutionary diversification of cyanobacteria: Molecular-phylogenetic and paleontological perspectives. Proc. Natl. Acad. Sci. USA 2006, 103, 5442–5447. [Google Scholar] [CrossRef] [PubMed]
- Sasso, S.; Pohnert, G.; Lohr, M.; Mittag, M.; Hertweck, C. Microalgae in the postgenomic era: A blooming reservoir for new natural products. FEMS Microbiol. Rev. 2012, 36, 761–785. [Google Scholar] [CrossRef] [PubMed]
- Slattery, S.S.; Diamond, A.; Wang, H.; Therrien, J.A.; Lant, J.T.; Jazey, T.; Lee, K.; Klassen, Z.; Desgagné-Penix, I.; Karas, B.J.; et al. An Expanded Plasmid-Based Genetic Toolbox Enables Cas9 Genome Editing and Stable Maintenance of Synthetic Pathways in Phaeodactylum tricornutum. ACS Synth. Biol. 2018, 7, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Karas, B.J.; Diner, R.E.; Lefebvre, S.C.; Mcquaid, J.; Phillips, A.P.R.; Noddings, C.M.; Brunson, J.K.; Valas, R.E.; Deerinck, T.J.; Jablanovic, J.; et al. Designer diatom episomes delivered by bacterial conjugation—Supplementary Information. Nat. Commun. 2015. [Google Scholar] [CrossRef] [PubMed]
- Crozet, P.; Navarro, F.J.; Willmund, F.; Mehrshahi, P.; Bakowski, K.; Lauersen, K.J.; Pérez-Pérez, M.-E.; Auroy, P.; Gorchs Rovira, A.; Sauret-Gueto, S.; et al. Birth of a photosynthetic chassis: A MoClo toolkit enabling synthetic biology in the microalga Chlamydomonas reinhardtii. ACS Synth. Biol. 2018, 7, 2074–2086. [Google Scholar] [CrossRef] [PubMed]
- Fabris, M.; Matthijs, M.; Rombauts, S.; Vyverman, W.; Goossens, A.; Baart, G.J.E. The metabolic blueprint of Phaeodactylum tricornutum reveals a eukaryotic Entner-Doudoroff glycolytic pathway. Plant J. 2012, 70, 1004–1014. [Google Scholar] [CrossRef] [PubMed]
- May, P.; Jochristianmpimp-golmmpgde, J.O.C.; Kempa, S.; Walthermpimp-golmmpgde, D.W.; Christian, J.-O.; Walther, D. ChlamyCyc: An integrative systems biology database and web-portal for Chlamydomonas reinhardtii. BMC Genom. 2009, 10, 209. [Google Scholar] [CrossRef] [PubMed]
- Kroth, P.G.; Chiovitti, A.; Gruber, A.; Martin-Jezequel, V.; Mock, T.; Parker, M.S.; Stanley, M.S.; Kaplan, A.; Caron, L.; Weber, T.; et al. A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS ONE 2008, 3, e1426. [Google Scholar] [CrossRef] [PubMed]
- Kliphuis, A.M.J.; Klok, A.J.; Martens, D.E.; Lamers, P.P.; Janssen, M.; Wijffels, R.H. Metabolic modeling of Chlamydomonas reinhardtii: Energy requirements for photoautotrophic growth and maintenance. J. Appl. Phycol. 2012, 24, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Fabris, M.; Baart, G.; Kim, M.K.; Goossens, A.; Vyverman, W.; Falkowski, P.G.; Lun, D.S. Flux balance analysis of primary metabolism in the diatom Phaeodactylum tricornutum. Plant J. 2016, 85, 161–176. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Quinn, A.H.; Sriram, G. Experimental evidence and isotopomer analysis of mixotrophic glucose metabolism in the marine diatom Phaeodactylum tricornutum. Microb. Cell Fact. 2013, 12, 109. [Google Scholar] [CrossRef] [PubMed]
- Boyle, N.R.; Morgan, J.A. Flux balance analysis of primary metabolism in Chlamydomonas reinhardtii. BMC Syst. Biol. 2009, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.L.; Ghamsari, L.; Manichaikul, A.; Hom, E.F.Y.; Balaji, S.; Fu, W.; Shen, Y.; Hao, T.; Palsson, B.Ø.; Salehi-Ashtiani, K.; et al. Metabolic network reconstruction of Chlamydomonas offers insight into light-driven algal metabolism. Mol. Syst. Biol. 2011, 7, 518. [Google Scholar] [CrossRef] [PubMed]
- Levering, J.; Broddrick, J.; Dupont, C.L.; Peers, G.; Beeri, K.; Mayers, J.; Gallina, A.A.; Allen, A.E.; Palsson, B.O.; Zengler, K. Genome-scale model reveals metabolic basis of biomass partitioning in a model diatom. PLoS ONE 2016, 11, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Neupert, J.; Karcher, D.; Bock, R. Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 2009, 57, 1140–1150. [Google Scholar] [CrossRef] [PubMed]
- Peralta-Yahya, P.P.; Zhang, F.; del Cardayre, S.B.; Keasling, J.D. Microbial engineering for the production of advanced biofuels. Nature 2012, 488, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Srikanta Dani, K.G.; Silva Benavides, A.M.; Michelozzi, M.; Peluso, G.; Torzillo, G.; Loreto, F. Relationship between isoprene emission and photosynthesis in diatoms, and its implications for global marine isoprene estimates. Mar. Chem. 2017, 189, 17–24. [Google Scholar] [CrossRef]
- Fabris, M.; Matthijs, M.; Carbonelle, S.; Moses, T.; Pollier, J.; Dasseville, R.; Baart, G.J.E.; Vyverman, W.; Goossens, A. Tracking the sterol biosynthesis pathway of the diatom Phaeodactylum tricornutum. New Phytol. 2014, 204, 521–535. [Google Scholar] [CrossRef] [PubMed]
- De Riso, V.; Raniello, R.; Maumus, F.; Rogato, A.; Bowler, C.; Falciatore, A. Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res. 2009, 37, e96. [Google Scholar] [CrossRef] [PubMed]
- Hopes, A.; Nekrasov, V.; Kamoun, S.; Mock, T. Editing of the urease gene by CRISPR-Cas in the diatom Thalassiosira pseudonana. Plant Methods 2016, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nymark, M.; Sharma, A.K.; Sparstad, T.; Bones, A.M.; Winge, P. A CRISPR/Cas9 system adapted for gene editing in marine algae. Sci. Rep. 2016, 6, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Eilers, U.; Bikoulis, A.; Breitenbach, J.; Büchel, C.; Sandmann, G. Limitations in the biosynthesis of fucoxanthin as targets for genetic engineering in Phaeodactylum tricornutum. J. Appl. Phycol. 2016, 123–129. [Google Scholar] [CrossRef]
- Vickers, C.E.; Blank, L.M.; Krömer, J.O. Grand Challenge Commentary: Chassis cells for industrial biochemical production. Nat. Chem. Biol. 2010, 6, 875–877. [Google Scholar] [CrossRef] [PubMed]
| Terpenoid | Production Organism | Titer | Reference |
|---|---|---|---|
| isoprene | Synechocystis sp. PCC 6803 | 12.30 mg g−1 | [72] |
| isoprene | Synechococcus elongatus PCC 7942 | 1.26 g L−1 | [73] |
| beta-phellandrene | Synechocystis sp. PCC 6803 | 3.20 mg g−1 | [74] |
| limonene | Synechococcus sp. PCC 7002 | 4 mg L−1 | [75] |
| limonene | Synechocystis sp. PCC 6803 | 6.70 mg L−1 | [76] |
| bisabolene | Synechococcus sp. PCC 7002 | 0.60 mg L−1 | [75] |
| farnesene | Anabaena sp. PCC 7120 | 0.08 mg g−1 | [77] |
| amorphadiene | Synechococcus elongatus PCC 7942 | 19.80 mg L−1 | [78] |
| squalene | Synechocystis sp. PCC 6803 | 1.20 mg g−1 | [79] |
| farnesene | Synechococcus elongatus PCC 7942 | 4.60 mg L−1 | [80] |
| geranyllinalool | Synechocystis sp. PCC 6803 | 0.36 mg g−1 | [81] |
| manoyl oxide | Synechocystis sp. PCC 6803 | 2 mg L−1 | [82] |
| (E)-alpha-bisabolene | Chlamydomonas reinhardtii | 11 mg L−1 | [48] |
| patchoulol | Chlamydomonas reinhardtii | 0.47 mg L−1 | [83] |
| manoyl oxide | Chlamydomonas reinhardtii | 50 mg L−1 | [84] |
| lupeol | Phaeodactylum tricornutum | 0.1 mg L−1 | [85] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vavitsas, K.; Fabris, M.; Vickers, C.E. Terpenoid Metabolic Engineering in Photosynthetic Microorganisms. Genes 2018, 9, 520. https://doi.org/10.3390/genes9110520
Vavitsas K, Fabris M, Vickers CE. Terpenoid Metabolic Engineering in Photosynthetic Microorganisms. Genes. 2018; 9(11):520. https://doi.org/10.3390/genes9110520
Chicago/Turabian StyleVavitsas, Konstantinos, Michele Fabris, and Claudia E. Vickers. 2018. "Terpenoid Metabolic Engineering in Photosynthetic Microorganisms" Genes 9, no. 11: 520. https://doi.org/10.3390/genes9110520
APA StyleVavitsas, K., Fabris, M., & Vickers, C. E. (2018). Terpenoid Metabolic Engineering in Photosynthetic Microorganisms. Genes, 9(11), 520. https://doi.org/10.3390/genes9110520







