Unusual Diversity of Sex Chromosomes in African Cichlid Fishes
Abstract
1. Introduction
1.1. The Canonical Model of Sex Chromosome Evolution
1.2. Ancient Sex Chromosomes of Mammals and Birds
2. Diversity of Sex Chromosomes in Fishes
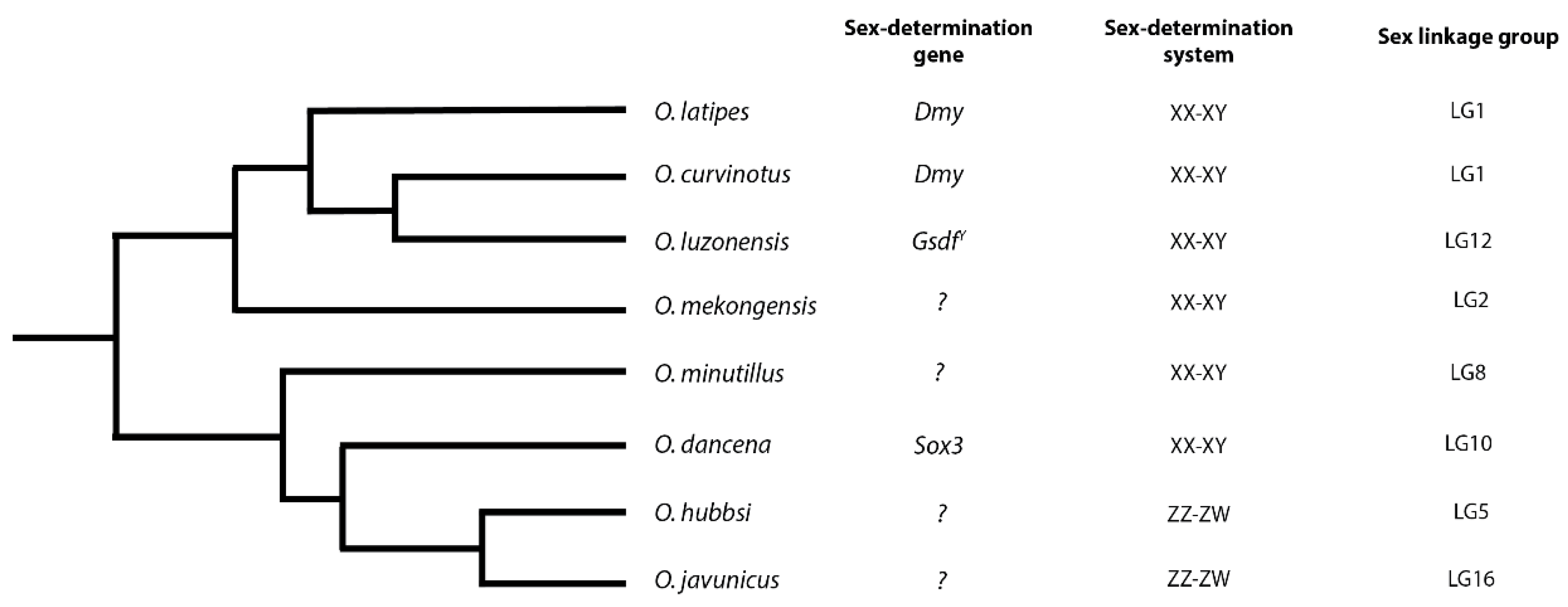
2.1. Molecular Basis of Sex-Determination in Ricefish
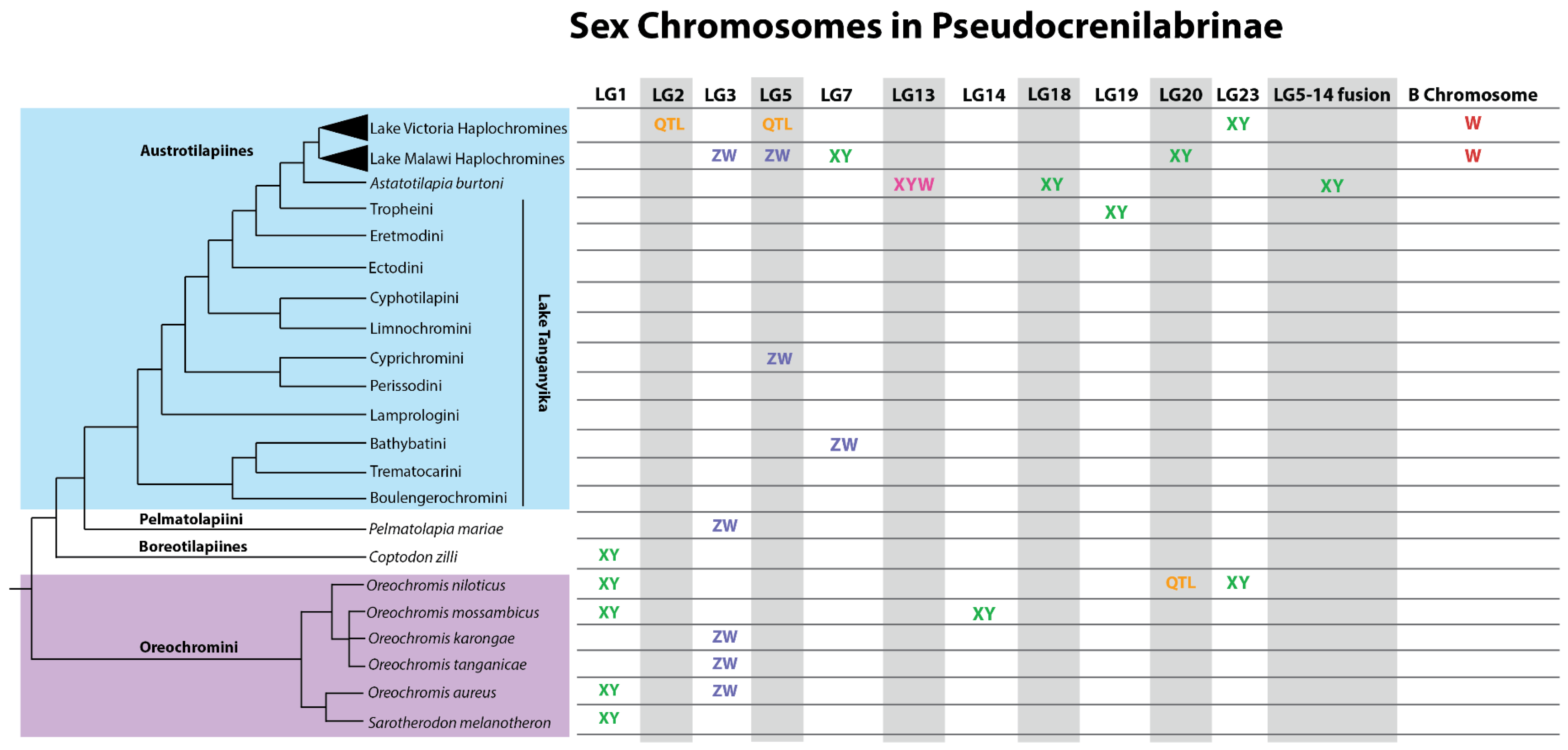
2.2. Cichlid Sex Chromosomes
2.2.1. Oreochromini
2.2.2. Boreotilapiines
2.2.3. Pelmatolapini
2.2.4. Austrotilapiines
2.2.5. Haplochromines
Lake Tanganyika Haplochromines
Lake Malawi Haplochromines
Lake Victoria Haplochromines
2.2.6. Remarkable Diversity of Sex Chromosomes
2.2.7. Stages in The Evolution of Sex Chromosomes
3. Why Do Cichlids Have so Many Sex Chromosome Transitions?
3.1. Number of Genes in the Sex-Determination Network
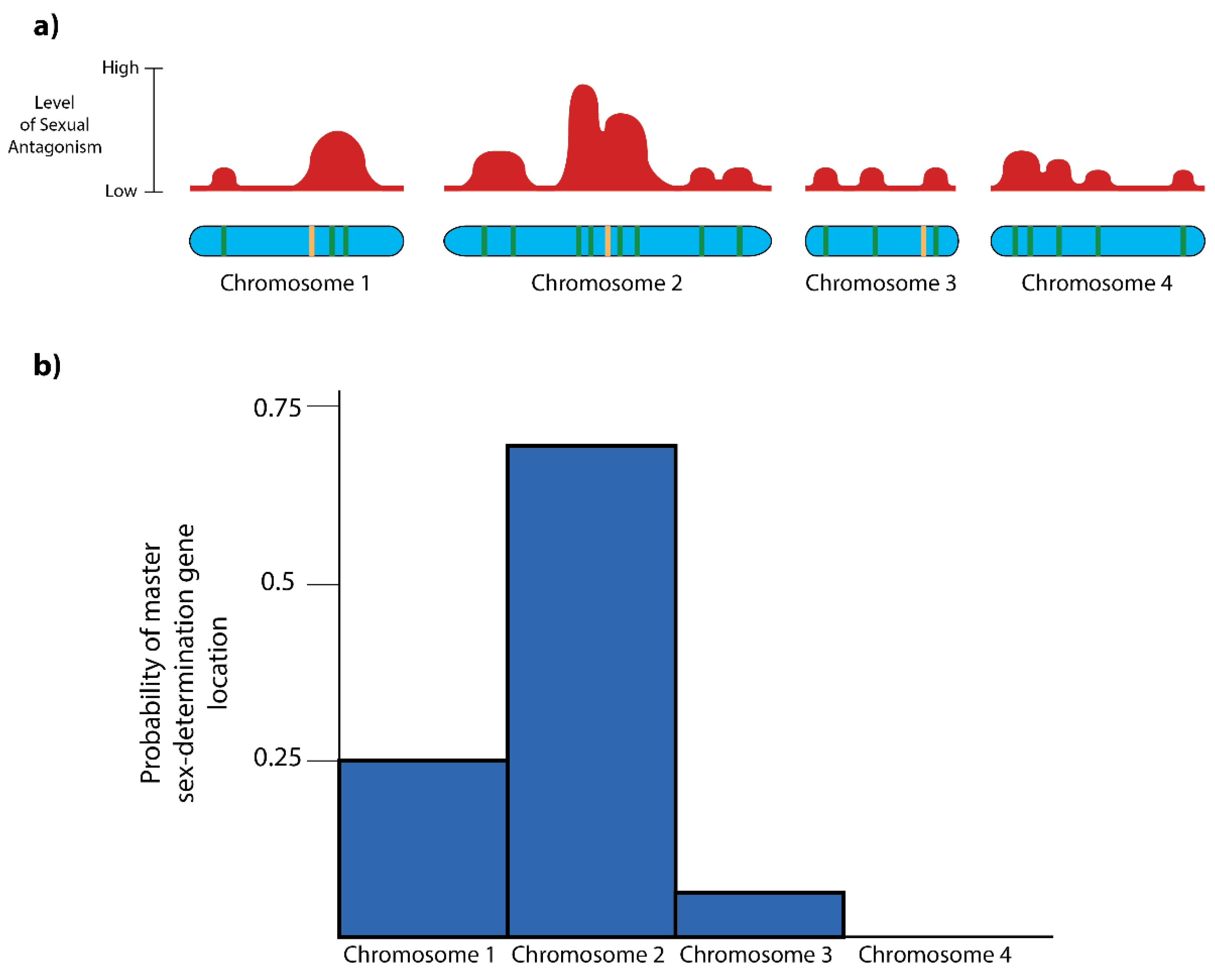
3.2. The Landscape of Sexual Antagonism
3.3. Predicting the Appearance of New Sex Chromosomes
4. Sex Chromosomes and Cichlid Speciation
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lindholm, A.; Breden, F. Sex chromosomes and sexual selection in Poeciliid fishes. Am. Nat. 2002, 160, S214–S224. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.B.; Ser, J.R.; Kocher, T.D. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science 2009, 326, 998–1001. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, B.; Coyne, J.A.; Barton, N.H. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 1987, 130, 113–146. [Google Scholar] [CrossRef]
- Presgraves, D.C. Sex chromosomes and speciation in Drosophila. Trends Genet. 2008, 24, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Haldane, J.B.S. Sex ratio and unisexual sterility in hybrid animals. J. Genet. 1922, 12, 101–109. [Google Scholar] [CrossRef]
- Bachtrog, D. Y-chromosome evolution: Emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 2013, 14, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Abbott, J.K.; Nordén, A.K.; Hansson, B. Sex chromosome evolution: Historical insights and future perspectives. Proc. R. Soc. B Biol. Sci. 2017, 284, 20162806. [Google Scholar] [CrossRef] [PubMed]
- Rice, W.R. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution 1987, 41, 911–914. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.A. The evolution of dominance. Biol. Rev. 1931, 6, 345–368. [Google Scholar] [CrossRef]
- van Doorn, G.S.; Kirkpatrick, M. Transitions between male and female heterogamety caused by sex-antagonistic selection. Genetics 2010, 186, 629–645. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.E.; Darolti, I.; Bloch, N.I.; Oostra, V.; Sandkam, B.; Buechel, S.D.; Kolm, N.; Breden, F.; Vicoso, B.; Mank, J.E. Convergent recombination suppression suggests role of sexual selection in guppy sex chromosome formation. Nat. Commun. 2017, 8, 14251. [Google Scholar] [CrossRef] [PubMed]
- Muller, H. The relation of recombination to mutational advance. Mutat. Res. 1964, 1, 2–9. [Google Scholar] [CrossRef]
- Bachtrog, D.; Kirkpatrick, M.; Mank, J.E.; McDaniel, S.F.; Pires, J.C.; Rice, W.; Valenzuela, N. Are all sex chromosomes created equal? Trends Genet. 2011, 27, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Koopman, P.; Gubbay, J.; Vivian, N.; Goodfellow, P.; Lovell-Badge, R. Male development of chromosomally female mice transgenic for Sry. Nature 1991, 351, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Cortez, D.; Marin, R.; Toledo-Flores, D.; Froidevaux, L.; Liechti, A.; Waters, P.D.; Grützner, F.; Kaessmann, H. Origins and functional evolution of Y chromosomes across mammals. Nature 2014, 508, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Suzuki, H.; Itoh, M. An unusual sex chromosome constitution found in the Amami spinous country-rat, Tokudaia osimensis osimensis. Jpn. J. Genet. 1977, 52, 247–249. [Google Scholar] [CrossRef]
- Honda, T.; Suzuki, H.; Itoh, M.; Hayashi, K. Karyotypical differences of the Amami spinous country-rats, Tokudaia osimensis osimensis obtained from two neighboring islands. Jpn. J. Genet. 1978, 53, 297–299. [Google Scholar] [CrossRef]
- Sutou, S.; Mitsui, Y.; Tsuchiya, K. Sex determination without the Y chromosome in two Japanese rodents Tokudaia osimensis osimensis and Tokudaia osimensis spp. Mamm. Genome 2001, 12, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Vorontsov, N.N.; Lyapunova, E.A.; Borissov, Y.M.; Dovgal, V.E. Variability of sex chromosomes in mammals. Genetica 1980, 52, 361–372. [Google Scholar] [CrossRef]
- Kolomiets, O.L.; Vorontsov, N.N.; Lyapunova, E.A.; Mazurova, T.F. Ultrastructure, meiotic behavior, and evolution of sex chromosomes of the genus Ellobius. Genetica 1991, 84, 179–189. [Google Scholar] [CrossRef]
- Just, W.; Baumstark, A.; Süß, A.; Graphodatsky, A.; Rens, W.; Schäfer, N.; Bakloushinskaya, I.; Hameister, H.; Vogel, W. Ellobius lutescens: Sex determination and sex chromosome. Sex. Dev. 2007, 1, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A.M. Avian sex, sex chromosomes, and dosage compensation in the age of genomics. Chromosom. Res. 2014, 22, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Stiglec, R.; Ezaz, T.; Graves, J.A.M. A new look at the evolution of avian sex chromosomes. Cytogenet. Genome Res. 2007, 117, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H. Evolution of the avian sex chromosomes and their role in sex determination. Trends Ecol. Evol. 2000, 15, 188–192. [Google Scholar] [CrossRef]
- Ogawa, A.; Murata, K.; Mizuno, S. The location of Z- and W-linked marker genes and sequence on the homomorphic sex chromosomes of the ostrich and the emu. Proc. Natl. Acad. Sci. USA 1998, 95, 4415–4418. [Google Scholar] [CrossRef] [PubMed]
- Gamble, T. A review of sex determining mechanisms in geckos (Gekkota: Squamata). Sex. Dev. 2010, 4, 88–103. [Google Scholar] [CrossRef] [PubMed]
- Bachtrog, D.; Mank, J.E.; Peichel, C.L.; Kirkpatrick, M.; Otto, S.P.; Ashman, T.-L.; Hahn, M.W.; Kitano, J.; Mayrose, I.; Ming, R.; et al. The tree of sex consortium sex determination: Why so many ways of doing it? PLoS Biol. 2014, 12, e1001899. [Google Scholar] [CrossRef] [PubMed]
- Vicoso, B.; Bachtrog, D. Numerous transitions of sex chromosomes in diptera. PLoS Biol. 2015, 13, e1002078. [Google Scholar] [CrossRef] [PubMed]
- Devlin, R.H.; Nagahama, Y. Sex determination and sex differentiation in fish: An overview of genetic, physiological, and environmental influences. Aquaculture 2002, 208, 191–364. [Google Scholar] [CrossRef]
- Gomelsky, B. Chromosome set manipulation and sex control in common carp: A review. Aquat. Living Resour. 2003, 16, 408–415. [Google Scholar] [CrossRef]
- Collares-Pereira, M.J.; Próspero, M.I.; Biléu, R.I.; Rodrigues, E.M. Leuciscus (Pisces, Cyprinidae) karyotypes: Transect of Portuguese populations. Genet. Mol. Biol. 1998, 21. [Google Scholar] [CrossRef]
- Volff, J.-N.; Schartl, M. Variability of genetic sex determination in Poeciliid fishes. Genetica 2001, 111, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Artoni, R.F.; Venere, P.C.; Bertollo, L.A.C. A heteromorphic ZZ/ZW sex chromosome system in fish, genus Hypostomus (Loricariidae). Cytologia 1998, 63, 421–425. [Google Scholar] [CrossRef]
- Andreata, A.A.; de Almeida-Toledo, L.F.; Oliveira, C.; de Almeida Toledo Filho, S. Chromosome studies in Hypoptopomatinae (Pisces, Siluriformes, Loricariidade). Cytologia 1992, 57, 369–372. [Google Scholar] [CrossRef]
- Pennell, M.W.; Mank, J.E.; Peichel, C.L. Transitions in sex determination and sex chromosomes across vertebrate species. Mol. Ecol. 2018, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Uwa, H.; Ojima, Y. Detailed and banding karyotype analyses the medaka, Oryzias latipes in cultured cells. Proc. Jpn. Acad. Ser. B 1981, 57, 39–43. [Google Scholar] [CrossRef]
- Uwa, H.; Iwanatsu, T.; Ojima, Y. Karyotype and banding analyses of Oryzias celebensis (Oryziatidae, Pisces) in cultured cells. Proc. Jpn. Acad. Ser. B 1981, 57, 95–99. [Google Scholar] [CrossRef]
- Matsuda, M.; Nagahama, Y.; Shinomiya, A.; Sato, T.; Matsuda, C.; Kobayashi, T.; Morrey, C.E.; Shibata, N.; Asakawa, S.; Shimizu, N.; et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 2002, 417, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Nanda, I.; Kondo, M.; Hornung, U.; Asakawa, S.; Winkler, C.; Shimizu, A.; Shan, Z.; Haaf, T.; Shimizu, N.; Shima, A.; et al. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. USA 2002, 99, 11778–11783. [Google Scholar] [CrossRef] [PubMed]
- Nagai, T.; Takehana, Y.; Hamaguchi, S.; Sakaizumi, M. Identification of the sex-determining locus in the Thai medaka, Oryzias minutillus. Cytogenet. Genome Res. 2008, 121, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Takehana, Y.; Naruse, K.; Hamaguchi, S.; Sakaizumi, M. Evolution of ZZ/ZW and XX/XY sex-determination systems in the closely related medaka species, Oryzias hubbsi and O. dancena. Chromosoma 2007, 116, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Takehana, Y.; Hamaguchi, S.; Sakaizumi, M. Different origins of ZZ/ZW sex chromosomes in closely related medaka fishes, Oryzias javanicus and O. hubbsi. Chromosom. Res. 2008, 16, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Takehana, Y.; Matsuda, M.; Myosho, T.; Suster, M.L.; Kawakami, K.; Shin-I, T.; Kohara, Y.; Kuroki, Y.; Toyoda, A.; Fujiyama, A.; et al. Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat. Commun. 2014, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Takehana, Y.; Naruse, K.; Sakaizumi, M. Molecular phylogeny of the medaka fishes genus Oryzias (Beloniformes: Adrianichthyidae) based on nuclear and mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2005, 36, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Setiamarga, D.H.E.; Miya, M.; Yamanoue, Y.; Azuma, Y.; Inoue, J.G.; Ishiguro, N.B.; Mabuchi, K.; Nishida, M. Divergence time of the two regional medaka populations in Japan as a new time scale for comparative genomics of vertebrates. Biol. Lett. 2009, 5, 812–816. [Google Scholar] [CrossRef] [PubMed]
- Kirchmaier, S.; Naruse, K.; Wittbrodt, J.; Loosli, F. The genomic and genetic toolbox of the teleost medaka (Oryzias latipes). Genetics 2015, 199, 905–918. [Google Scholar] [CrossRef] [PubMed]
- Myosho, T.; Otake, H.; Masuyama, H.; Matsuda, M.; Kuroki, Y.; Fujiyama, A.; Naruse, K.; Hamaguchi, S.; Sakaizumi, M. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 2012, 191, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Hattori, R.S.; Murai, Y.; Oura, M.; Masuda, S.; Majhi, S.K.; Sakamoto, T.; Fernandino, J.I.; Somoza, G.M.; Yokota, M.; Strüssmann, C. A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc. Natl. Acad. Sci. USA 2012, 109, 2955–2959. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, T.; Kai, W.; Tasumi, S.; Oka, A.; Matsunaga, T.; Mizuno, N.; Fujita, M.; Suetake, H.; Suzuki, S.; Hosoya, S.; et al. A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet. 2012, 8, e1002798. [Google Scholar] [CrossRef] [PubMed]
- Reichwald, K.; Petzold, A.; Koch, P.; Downie, B.R.; Hartmann, N.; Pietsch, S.; Baumgart, M.; Chalopin, D.; Felder, M.; Bens, M.; et al. Insights into sex chromosome evolution and aging from the genome of a short-lived fish. Cell 2015, 163, 1527–1538. [Google Scholar] [CrossRef] [PubMed]
- Yano, A.; Guyomard, R.; Nicol, B.; Jouanno, E.; Quillet, E.; Klopp, C.; Cabau, C.; Bouchez, O.; Fostier, A.; Guiguen, Y. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr. Biol. 2012, 22, 1423–1428. [Google Scholar] [CrossRef] [PubMed]
- Faber-Hammond, J.J.; Phillips, R.B.; Brown, K.H. Comparative analysis of the shared sex-determination region (sdr) among salmonid fishes. Genome Biol. Evol. 2015, 7, 1972–1987. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.S.; Matschiner, M.; Salzburger, W. A tribal level phylogeny of Lake Tanganyika cichlid fishes based on a genomic multi-marker approach. Mol. Phylogenet. Evol. 2015, 83, 56–71. [Google Scholar] [CrossRef] [PubMed]
- Dunz, A.R.; Schliewen, U.K. Molecular phylogeny and revised classification of the haplotilapiine cichlid fishes formerly referred to as “Tilapia”. Mol. Phylogenet. Evol. 2013, 68, 64–80. [Google Scholar] [CrossRef] [PubMed]
- Nagl, S.; Tichy, H.; Mayer, W.E.; Samonte, I.E.; McAndrew, B.J.; Klein, J. Classification and phylogenetic relationships of African tilapiine fishes inferred from mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2001, 20, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Klett, V.; Meyer, A. What, if anything, is a Tilapia?—mitochondrial ND2 phylogeny of tilapiines and the evolution of parental care systems in the African cichlid fishes. Mol. Biol. Evol. 2002, 19, 865–883. [Google Scholar] [CrossRef] [PubMed]
- Farias, I.P.; Ortí, G.; Sampaio, I.; Schneider, H.; Meyer, A. Mitochondrial DNA phylogeny of the family Cichlidae: Monophyly and fast molecular evolution of the neotropical assemblage. J. Mol. Evol. 1999, 48, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Sparks, J.S.; Smith, W.L. Phylogeny and biogeography of cichlid fishes (Teleostei: Perciformes: Cichlidae). Cladistics 2004, 20, 501–517. [Google Scholar] [CrossRef]
- McMahan, C.D.; Chakrabarty, P.; Sparks, J.S.; Smith, W.M.L.; Davis, M.P. Temporal patterns of diversification across global cichlid biodiversity (Acanthomorpha: Cichlidae). PLoS ONE 2013, 8, e71162. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M.; Keck, B.P.; Dornburg, A.; Eytan, R.I.; Martin, C.H.; Darrin, C.; Wainwright, P.C.; Near, T.J.; Hulsey, C.D. Molecular and fossil evidence place the origin of cichlid fishes long after Gondwanan rifting. Proc. Biol. Sci. 2013, 280, 20131733. [Google Scholar] [CrossRef] [PubMed]
- Matschiner, M.; Musilová, Z.; Barth, J.M.I.; Starostová, Z.; Salzburger, W.; Steel, M.; Bouckaert, R. Bayesian phylogenetic estimation of clade ages supports trans-Atlantic dispersal of cichlid fishes. Syst. Biol. 2017, 66, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Irisarri, I.; Singh, P.; Koblmüller, S.; Torres-Dowdall, J.; Henning, F.; Franchini, P.; Fischer, C.; Lemmon, A.R.; Lemmon, E.M.; Thallinger, G.G.; et al. Phylogenomics uncovers early hybridization and adaptive loci shaping the radiation of Lake Tanganyika cichlid fishes. Nat. Commun. 2018, 9, 3159. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, J.; Misof, B.; Tautz, D.; Schliewen, U.K. The root of the East African cichlid radiations. BMC Evol. Biol. 2009, 9, 186. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-Y.; Penman, D.J.; Kocher, T.D. Identification of a sex-determining region in Nile tilapia (Oreochromis niloticus) using bulked segregant analysis. Anim. Genet. 2003, 34, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Ezaz, M.T.; Harvey, S.C.; Boonphakdee, C.; Teale, A.J.; McAndrew, B.J.; Penman, D.J. Isolation and physical mapping of sex-linked AFLP markers in nile tilapia (Oreochromis niloticus L.). Mar. Biotechnol. 2004, 6, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-Y.; Coutanceau, J.-P.; Ozouf-Costaz, C.; D’Cotta, H.; Baroiller, J.-F.; Kocher, T.D. Genetic and physical mapping of sex-linked AFLP markers in Nile tilapia (Oreochromis niloticus). Mar. Biotechnol. 2011, 13, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Cnaani, A.; Lee, B.-Y.; Zilberman, N.; Ozouf-Costaz, C.; Hulata, G.; Ron, M.; D’Hont, A.; Baroiller, J.-F.; D’Cotta, H.; Penman, D.J.; et al. Genetics of sex determination in tilapiine species. Sex. Dev. 2008, 2, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Palaiokostas, C.; Bekaert, M.; Khan, M.G.Q.; Taggart, J.B.; Gharbi, K.; McAndrew, B.J.; Penman, D.J. Mapping and validation of the major sex-determining region in Nile tilapia (Oreochromis niloticus L.) using RAD sequencing. PLoS ONE 2013, 8, e68389. [Google Scholar] [CrossRef]
- Palaiokostas, C.; Bekaert, M.; Khan, M.G.Q.; Taggart, J.B.; Gharbi, K.; McAndrew, B.J.; Penman, D.J. A novel sex-determining QTL in Nile tilapia (Oreochromis niloticus). BMC Genomics 2015, 16, 171. [Google Scholar] [CrossRef] [PubMed]
- Gammerdinger, W.J.; Conte, M.A.; Acquah, E.A.; Roberts, R.B.; Kocher, T.D. Structure and decay of a proto-Y region in tilapia, Oreochromis niloticus. BMC Genomics 2014, 15, 975. [Google Scholar] [CrossRef] [PubMed]
- Gammerdinger, W.J.; Conte, M.A.; Baroiller, J.-F.; D’Cotta, H.; Kocher, T.D. Comparative analysis of a sex chromosome from the blackchin tilapia, Sarotherodon melanotheron. BMC Genomics 2016, 17, 808. [Google Scholar] [CrossRef] [PubMed]
- Ocalewicz, K.; Mota-Velasco, J.C.; Campos-Ramos, R.; Penman, D.J. FISH and DAPI staining of the synaptonemal complex of the Nile tilapia (Oreochromis niloticus) allow orientation of the unpaired region of bivalent 1 observed during early pachytene. Chromosom. Res. 2009, 17, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Campos-Ramos, R.; Harvey, S.C.; Penman, D.J. Sex-specific differences in the synaptonemal complex in the genus Oreochromis (Cichlidae). Genetica 2009, 135, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Foresti, F.; Oliveira, C.; Galetti Junior, P.M.; Almeida-Toledo, L.F.D. Synaptonemal complex analysis in spermatocytes of tilapia, Oreochromis niloticus (Pisces, Cichlidae). Genome 1993, 36, 1124–1128. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, L.A.P.; Penman, D.J.; Bromage, N. Evidence for the presence of sex chromosomes in the Nile tilapia (Oreochromis niloticus) from synaptonemal complex analysis of XX, XY and YY genotypes. Aquaculture 1999, 173, 207–218. [Google Scholar] [CrossRef]
- Conte, M.A.; Gammerdinger, W.J.; Bartie, K.L.; Penman, D.J.; Kocher, T.D. A high quality assembly of the Nile tilapia (Oreochromis niloticus) genome reveals the structure of two sex determination regions. BMC Genomics 2017, 18, 341. [Google Scholar] [CrossRef] [PubMed]
- Harvey, S.C.; Powell, S.F.; Kennedy, D.D.; Mcandrew, B.J.; Penman, D.J. Karyotype analysis of Oreochromis mortimeri (Trewavas) and Sarotherodon melanotheron (Rüppell). Aquac. Res. 2002, 33, 339–342. [Google Scholar] [CrossRef]
- Majumdar, K.C.; McAndrew, B.J. Relative DNA content of somatic nuclei and chromosomal studies in three genera, Tilapia, Sarotherodon, and Oreochromis of the tribe Tilapiini (Pisces, Cichlidae). Genetica 1984, 68, 175–188. [Google Scholar] [CrossRef]
- Cnaani, A.; Zilberman, N.; Tinman, S.; Hulata, G.; Ron, M. Genome-scan analysis for quantitative trait loci in an F2 tilapia hybrid. Mol. Genet. Genomics 2004, 272, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-Y.; Hulata, G.; Kocher, T.D. Two unlinked loci controlling the sex of blue tilapia (Oreochromis aureus). Heredity 2004, 92, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Thompson, K.W. Karyotypes of six species of African Cichlidae (Pisces: Perciformes). Experientia 1981, 37, 351–352. [Google Scholar] [CrossRef] [PubMed]
- Campos-Ramos, R.; Harvey, S.C.; Masabanda, J.S.; Carrasco, L.A.P.; Griffin, D.K.; McAndrew, B.J.; Bromage, N.R.; Penman, D.J. Identification of putative sex chromosomes in the blue tilapia, Oreochromis aureus, through synaptonemal complex and FISH analysis. Genetica 2001, 111, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Cnaani, A.; Kocher, T.D. Sex-linked markers and microsatellite locus duplication in the cichlid species Oreochromis tanganicae. Biol. Lett. 2008, 4, 700–703. [Google Scholar] [CrossRef] [PubMed]
- Harvey, S.C.; Campos-Ramos, R.; Kennedy, D.D.; Ezaz, M.T.; Bromage, N.R.; Griffin, D.K.; Penman, D.J. Karyotype evolution in Tilapia: Mitotic and meiotic chromosome analysis of Oreochromis karongae and O. niloticus x O. karongae hybrids. Genetica 2002, 115, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Mota-Velasco, J.C.; Ferreira, I.A.; Cioffi, M.B.; Ocalewicz, K.; Campos-Ramos, R.; Shirak, A.; Lee, B.-Y.; Martins, C.; Penman, D.J. Characterisation of the chromosome fusions in Oreochromis karongae. Chromosom. Res. 2010, 18, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Shirak, A.; Palti, Y.; Cnaani, A.; Korol, A.; Hulata, G.; Ron, M.; Avtalion, R.R. Association between loci with deleterious alleles and distorted sex ratios in an inbred line of tilapia (Oreochromis aureus). J. Hered. 2002, 93, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Cnaani, A.; Hallerman, E.M.; Ron, M.; Weller, J.I.; Indelman, M.; Kashi, Y.; Gall, G.A.E.; Hulata, G. Detection of a chromosomal region with two quantitative trait loci, affecting cold tolerance and fish size, in an F2 tilapia hybrid. Aquaculture 2003, 223, 117–128. [Google Scholar] [CrossRef]
- Eshel, O.; Shirak, A.; Weller, J.I.; Slossman, T.; Hulata, G.; Cnaani, A.; Ron, M. Fine-mapping of a locus on linkage group 23 for sex determination in Nile tilapia (Oreochromis niloticus). Anim. Genet. 2011, 42, 222–224. [Google Scholar] [CrossRef] [PubMed]
- Eshel, O.; Shirak, A.; Weller, J.I.; Hulata, G.; Ron, M. Linkage and physical mapping of sex region on LG23 of Nile tilapia (Oreochromis niloticus). G3 2012, 2, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Eshel, O.; Shirak, A.; Dor, L.; Band, M.; Zak, T.; Markovich-Gordon, M.; Chalifa-Caspi, V.; Feldmesser, E.; Weller, J.I.; Seroussi, E.; et al. Identification of male-specific amh duplication, sexually differentially expressed genes and microRNAs at early embryonic development of Nile tilapia (Oreochromis niloticus). BMC Genomics 2014, 15, 774. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Sun, Y.; Zhao, J.; Shi, H.; Zeng, S.; Ye, K.; Jiang, D.; Zhou, L.; Sun, L.; Tao, W.; et al. A tandem duplicate of anti-müllerian hormone with a missense SNP on the Y Chromosome is essential for male sex determination in Nile Tilapia, Oreochromis niloticus. PLoS Genet. 2015, 11, e1005678. [Google Scholar] [CrossRef] [PubMed]
- Gammerdinger, W.J.; Conte, M.A.; Sandkam, B.A.; Penman, D.J.; Kocher, T.D. Characterization of sex chromosomes in three deeply diverged species of Pseudocrenilabrinae (Teleostei: Cichlidae). Hydrobiologia. in press. [CrossRef]
- Kide, N.G.; Dunz, A.; Agnèse, J.F.; Dilyte, J.; Pariselle, A.; Carneiro, C.; Correia, E.; Brito, J.C.; Yarba, L.O.; Kone, Y.; Durand, J.-D. Cichlids of the Banc d’Arguin National Park, Mauritania: Insight into the diversity of the genus Coptodon. J. Fish Biol. 2016, 88, 1369–1393. [Google Scholar] [CrossRef] [PubMed]
- Sofy, H.I.; Layla, A.M.; Iman, M.K.A. Karyotypic diversity of some tilapia species. Nat. Sci. 2008, 6, 19–27. [Google Scholar]
- Ferreira, I.A.; Poletto, A.B.; Kocher, T.D.; Mota-Velasco, J.C.; Penman, D.J.; Martins, C. Chromosome evolution in african cichlid fish: Contributions from the physical mapping of repeated DNAs. Cytogenet. Genome Res. 2010, 129, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Swanepoel, A. ’n Vergelykende studie van die kariotipes van Tilapia rendalli, Tilapia sparrmanii en Oreochromis mossambicus (Cichlidae). Master’s Thesis, University of Johannesburg, Johannesburg, South Africa, 1990. [Google Scholar]
- Michele, J.L.; Takahashi, C.S. Comparative cytology of Tilapia rendalli and Geophagus brasiliensis (Cichlidae, Pisces). Cytologia 1977, 42, 535–537. [Google Scholar] [CrossRef]
- Vervoort, A. The karyotypes of seven species of Tilapia (Teleostei: Cichlidae). Cytologia 1980, 45, 651–656. [Google Scholar] [CrossRef]
- Ferreira, I.A.; Martins, C. Physical chromosome mapping of repetitive DNA sequences in Nile tilapia Oreochromis niloticus: Evidences for a differential distribution of repetitive elements in the sex chromosomes. Micron 2008, 39, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Rizzon, C.; Marais, G.; Gouy, M.; Biémont, C. Recombination rate and the distribution of transposable elements in the Drosophila melanogaster genome. Genome Res. 2002, 12, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Jensen-Seaman, M.; Furey, T.S.; Payseur, B.A.; Lu, Y.; Roskin, K.M.; Chen, C.-F.; Thomas, M.A.; Haussler, D.; Jacob, H.J. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 2004, 14, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, B.; Sniegowski, P.; Stephan, W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 1994, 371, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.S.; Soreghan, M.J.; Scholz, C.A. Estimating the age of formation of lakes: An example from Lake Tanganyika, East African Rift system. Geology 1993, 21, 511–514. [Google Scholar] [CrossRef]
- Turner, G.F.; Seehausen, O.; Knight, M.E.; Allender, C.J.; Robinson, R.L. How many species of cichlid fishes are there in African lakes? Mol. Ecol. 2001, 10, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Terai, Y.; Nishida, M.; Okada, N. Phylogenetic relationships and ancient incomplete lineage sorting among cichlid fishes in Lake Tanganyika as revealed by analysis of the insertion of retroposons. Mol. Biol. Evol. 2001, 18, 2057–2066. [Google Scholar] [CrossRef] [PubMed]
- Gammerdinger, W.J.; Conte, M.A.; Sandkam, B.A.; Ziegelbecker, A.; Koblmüller, S.; Kocher, T.D. Novel sex chromosomes in three cichlid fishes from Lake Tanganyika. J. Hered. 2018, 1, 12. [Google Scholar]
- Salzburger, W.; Meyer, A.; Baric, S.; Verheyen, E.; Sturmbauer, C. Phylogeny of the Lake Tanganyika cichlid species flock and its relationship to the Central and East African haplochromine cichlid fish faunas. Syst. Biol. 2002, 51, 113–135. [Google Scholar] [CrossRef] [PubMed]
- Koblmüller, S.; Duftner, N.; Katongo, C.; Phiri, H.; Sturmbauer, C. Ancient divergence in bathypelagic Lake Tanganyika deepwater cichlids: Mitochondrial phylogeny of the tribe Bathybatini. J. Mol. Evol. 2005, 60, 297–314. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Sota, T. A robust phylogeny among major lineages of the East African cichlids. Mol. Phylogenet. Evol. 2016, 100, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Ozouf-Costaz, C.; Coutanceau, J.-P.; Bonillo, C.; Mercot, H.; Fermon, Y.; Guidi-Rontani, C. New insights into the chromosomal differentiation patterns among cichlids from Africa and Madagascar. Cybium Int. J. Ichthyol. 2017, 41, 35–43. [Google Scholar]
- Koblmüller, S.; Schliewen, U.K.; Duftner, N.; Sefc, K.M.; Katongo, C.; Sturmbauer, C. Age and spread of the haplochromine cichlid fishes in Africa. Mol. Phylogenet. Evol. 2008, 49, 153–169. [Google Scholar] [CrossRef] [PubMed]
- Genner, M.J.; Seehausen, O.; Lunt, D.H.; Joyce, D.A.; Shaw, P.W.; Carvalho, G.R.; Turner, G.F. Age of cichlids: New dates for ancient lake fish radiations. Mol. Biol. Evol. 2007, 24, 1269–1282. [Google Scholar] [CrossRef] [PubMed]
- Salzburger, W.; Mack, T.; Verheyen, E.; Meyer, A. Out of Tanganyika: Genesis, explosive speciation, key-innovations and phylogeography of the haplochromine cichlid fishes. BMC Evol. Biol. 2005, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Kocher, T.D. Adaptive evolution and explosive speciation: The cichlid fish model. Nat. Rev. Genet. 2004, 5, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Böhne, A.; Wilson, C.A.; Postlethwait, J.H.; Salzburger, W. Variations on a theme: Genomics of sex determination in the cichlid fish Astatotilapia burtoni. BMC Genomics 2016, 17, 883. [Google Scholar] [CrossRef] [PubMed]
- Roberts, N.B.; Juntti, S.A.; Coyle, K.P.; Dumont, B.L.; Stanley, M.K.; Ryan, A.Q.; Fernald, R.D.; Roberts, R.B. Polygenic sex determination in the cichlid fish, Astatotilapia burtoni. BMC Genomics 2016, 17, 835. [Google Scholar] [CrossRef] [PubMed]
- Sturmbauer, C.; Baric, S.; Salzburger, W.; Ruber, L.; Verheyen, E. Lake level fluctuations synchronized genetic divergence of cichlid fishes in African lakes. Mol. Biol. Evol. 2001, 18, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Kocher, T.D.; Conroy, J.A.; McKaye, K.R.; Stauffer, J.R.; Lockwood, S.F. Evolution of NADH Dehydrogenase Subunit 2 in East African cichlid fish. Mol. Phylogenet. Evol. 1995, 4, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.; Kocher, T.D.; Basasibwaki, P.; Wilson, A.C. Monophyletic orign of Lake Victoria cichlid fishes suggested by mitochondrial DNA sequences. Nature 1990, 347, 550–553. [Google Scholar] [CrossRef] [PubMed]
- Streelman, J.T.; Albertson, R.C.; Kocher, T.D. Genome mapping of the orange blotch colour pattern in cichlid fishes. Mol. Ecol. 2003, 12, 2465–2471. [Google Scholar] [CrossRef] [PubMed]
- Ser, J.R.; Roberts, R.B.; Kocher, T.D. Multiple interacting loci control sex determination in Lake Malawi cichlid fish. Evolution 2010, 64, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Parnell, N.F.; Streelman, J.T. Genetic interactions controlling sex and color establish the potential for sexual conflict in Lake Malawi cichlid fishes. Heredity 2013, 110, 239–246. [Google Scholar] [CrossRef] [PubMed]
- O’Quin, C.T. The Genetic Basis of Pigment Pattern Differentiation in Lake Malawi African Cichlids. Ph.D. Thesis, University of Maryland at College Park, College Park, MD, USA, 2014. [Google Scholar]
- Peterson, E.N.; Cline, M.E.; Moore, E.C.; Roberts, N.B.; Roberts, R.B. Genetic sex determination in Astatotilapia calliptera, a prototype species for the Lake Malawi cichlid radiation. Sci. Nat. 2017, 104, 41. [Google Scholar] [CrossRef] [PubMed]
- Clark, F.E.; Conte, M.A.; Ferreira-Bravo, I.A.; Poletto, A.B.; Martins, C.; Kocher, T.D. Dynamic sequence evolution of a sex-associated B chromosome in Lake Malawi cichlid fish. J. Hered. 2017, 108, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Martis, M.M.; Klemme, S.; Banaei-Moghaddam, A.M.; Blattner, F.R.; Macas, J.; Schmutzer, T.; Scholz, U.; Gundlach, H.; Wicker, T.; Simkova, H.; et al. Selfish supernumerary chromosome reveals its origin as a mosaic of host genome and organellar sequences. Proc. Natl. Acad. Sci. USA 2012, 109, 13343–13346. [Google Scholar] [CrossRef] [PubMed]
- Valente, G.T.; Conte, M.A.; Fantinatti, B.E.A.; Cabral-De-Mello, D.C.; Carvalho, R.F.; Vicari, M.R.; Kocher, T.D.; Martins, C. Origin and evolution of B chromosomes in the cichlid fish Astatotilapia latifasciata based on integrated genomic analyses. Mol. Biol. Evol. 2014, 31, 2061–2072. [Google Scholar] [CrossRef] [PubMed]
- Verheyen, E.; Salzburger, W.; Snoeks, J.; Meyer, A. Origin of the superflock of cichlid fishes from Lake Victoria, East Africa. Science 2003, 300, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Kudo, Y.; Nikaido, M.; Kondo, A.; Suzuki, H.; Yoshida, K.; Kikuchi, K.; Okada, N. A microsatellite-based genetic linkage map and putative sex-determining genomic regions in Lake Victoria cichlids. Gene 2015, 560, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Feulner, P.G.D.; Schwarzer, J.; Haesler, M.P.; Meier, J.I.; Seehausen, O. A dense linkage map of Lake Victoria cichlids improved the Pundamilia genome assembly and revealed a major QTL for sex-determination. G3 Genes Genomes Genet. 2018, 200207. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Terai, Y.; Mizoiri, S.; Aibara, M.; Nishihara, H.; Watanabe, M.; Kuroiwa, A.; Hirai, H.; Hirai, Y.; Matsuda, Y.; Okada, N. B chromosomes have a functional effect on female sex determination in Lake Victoria cichlid fishes. PLoS Genet. 2011, 7, e1002203. [Google Scholar] [CrossRef] [PubMed]
- Blaser, O.; Neuenschwander, S.; Perrin, N. Sex-chromosome turnovers: The hot-potato model. Am. Nat. 2014, 183, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Blaser, O.; Grossen, C.; Neuenschwander, S.; Perrin, N. Sex-chromsome turnovers induced by deleterious mutation load. Evolution 2012, 67, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Rice, W.R. On the instability of polygenic sex determination: The effect of sex-specific selection. Evolution 1986, 40, 633–639. [Google Scholar] [CrossRef] [PubMed]
- Bellott, D.W.; Hughes, J.F.; Skaletsky, H.; Brown, L.G.; Pyntikova, T.; Cho, T.-J.; Koutseva, N.; Zaghlul, S.; Graves, T.; Rock, S.; et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 2014, 508, 494–499. [Google Scholar] [CrossRef] [PubMed]
- White, M.A.; Kitano, J.; Peichel, C.L. Purifying selection maintains dosage-sensitive genes during degeneration of the threespine stickleback Y chromosome. Mol. Biol. Evol. 2015, 32, 1981–1995. [Google Scholar] [CrossRef] [PubMed]
- Eggers, S.; Ohnesorg, T.; Sinclair, A. Genetic regulation of mammalian gonad development. Nat. Rev. Endocrinol. 2014, 10, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Eggers, S.; Sinclair, A. Mammalian sex determination-insights from humans and mice. Chromosom. Res. 2012, 20, 215–238. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A.M.; Peichel, C.L. Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biol. 2010, 11, 205. [Google Scholar] [CrossRef] [PubMed]
- Matson, C.K.; Zarkower, D. Sex and the singular DM domain: Insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 2012, 13, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, N.B.; Balshine-Earn, S.; Reynolds, J.D. Evolutionary transitions in parental care in cichlid fish. Proc. Biol. Sci. 1998, 265, 2265–2272. [Google Scholar] [CrossRef]
- Mank, J.E. Sex chromosomes and the evolution of sexual dimorphism: Lessons from the genome. Am. Nat. 2009, 173, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, G.; Shao, C.; Huang, Q.; Liu, G.; Zhang, P.; Song, W.; An, N.; Chalopin, D.; Volff, J.-N.; et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 2014, 46, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Ranz, J.; Castillo-Davis, C.; Meiklejohn, C.; Hartl, D. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science 2003, 300, 1742–1745. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H.; Parsch, J. The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 2007, 8, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Innocenti, P.; Morrow, E.H. The sexually antagonistic genes of Drosophila melanogaster. PLoS Biol. 2010, 8, e1000335. [Google Scholar] [CrossRef] [PubMed]
- Crespi, B.; Nosil, P. Conflictual speciation: Species formation via genomic conflict. Trends Ecol. Evol. 2013, 28, 48–57. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gammerdinger, W.J.; Kocher, T.D. Unusual Diversity of Sex Chromosomes in African Cichlid Fishes. Genes 2018, 9, 480. https://doi.org/10.3390/genes9100480
Gammerdinger WJ, Kocher TD. Unusual Diversity of Sex Chromosomes in African Cichlid Fishes. Genes. 2018; 9(10):480. https://doi.org/10.3390/genes9100480
Chicago/Turabian StyleGammerdinger, William J., and Thomas D. Kocher. 2018. "Unusual Diversity of Sex Chromosomes in African Cichlid Fishes" Genes 9, no. 10: 480. https://doi.org/10.3390/genes9100480
APA StyleGammerdinger, W. J., & Kocher, T. D. (2018). Unusual Diversity of Sex Chromosomes in African Cichlid Fishes. Genes, 9(10), 480. https://doi.org/10.3390/genes9100480




