Salt-Stress Response Mechanisms Using de Novo Transcriptome Sequencing of Salt-Tolerant and Sensitive Corchorus spp. Genotypes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Salt-Stress Treatment
2.2. RNA Isolation
2.3. Transcriptome Sequencing
2.4. Transcriptome Data Analysis and Annotation
2.5. Identification and Biological Analysis of Differentially Expressed Genes
2.6. Real-Time Quantitative PCR Analysis
3. Results
3.1. Transcriptome Sequencing and Assembly
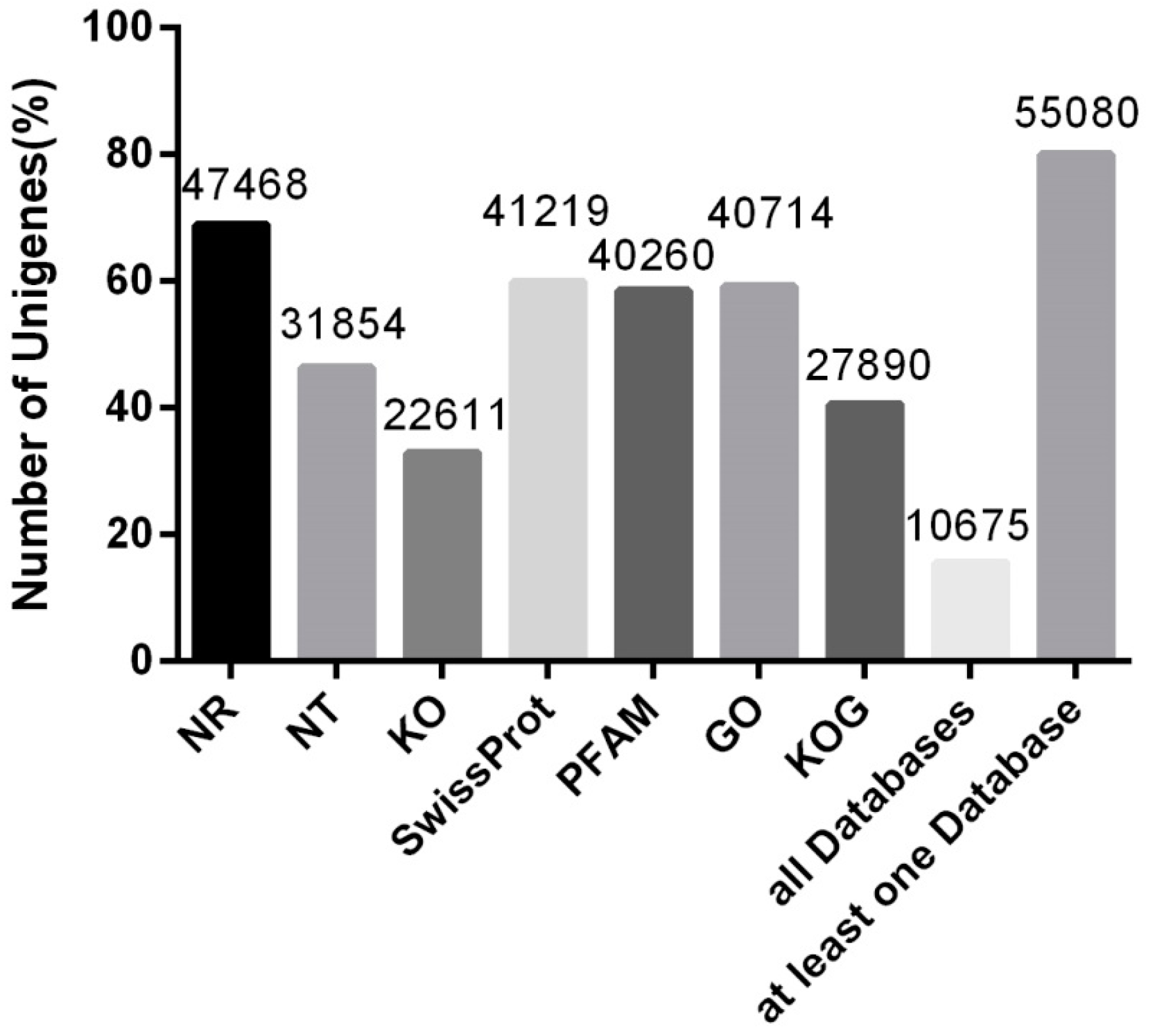
3.2. Unigene Functional Annotation
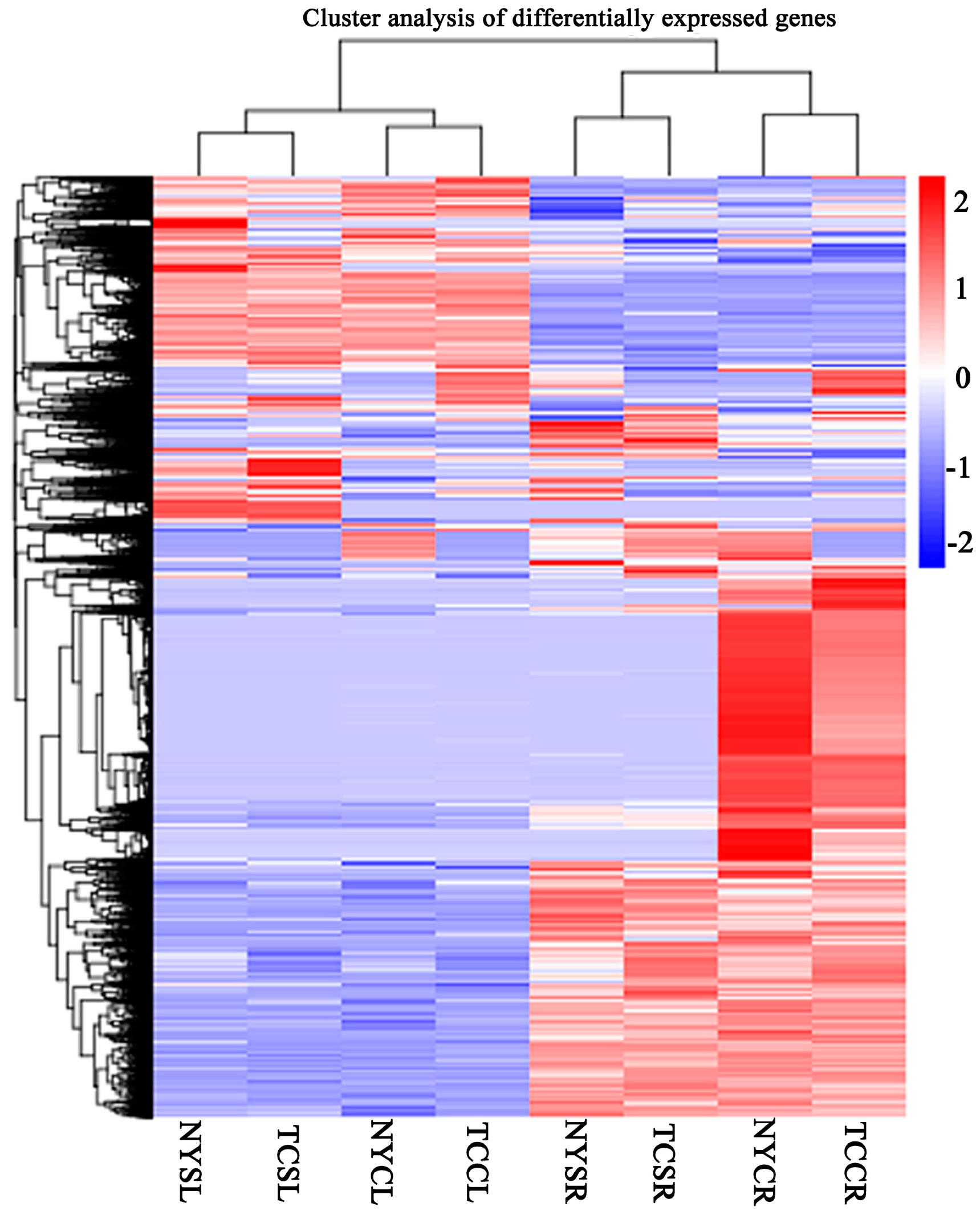
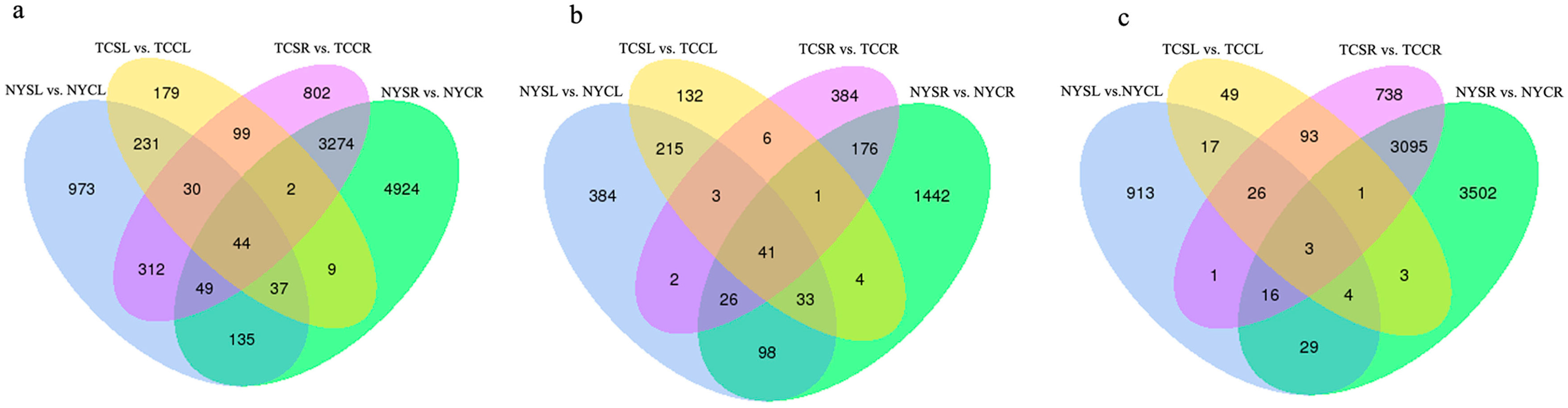
3.3. Differential Gene Expression in Response to Salt-Stress Treatments
3.4. Differential Expression of Transcription Factors
3.5. Functional Categorization of DEGs in Salt-Stressed Jute
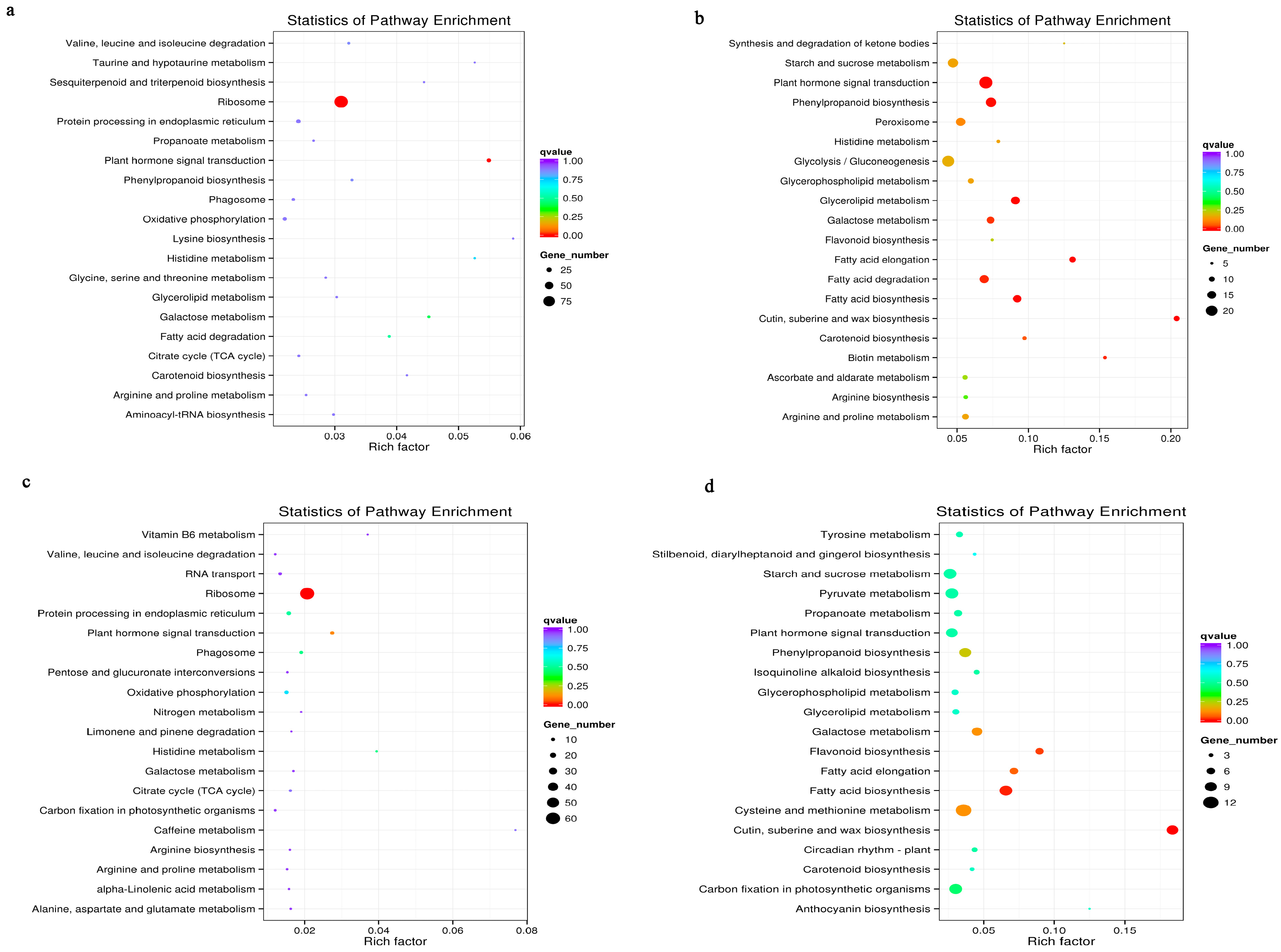
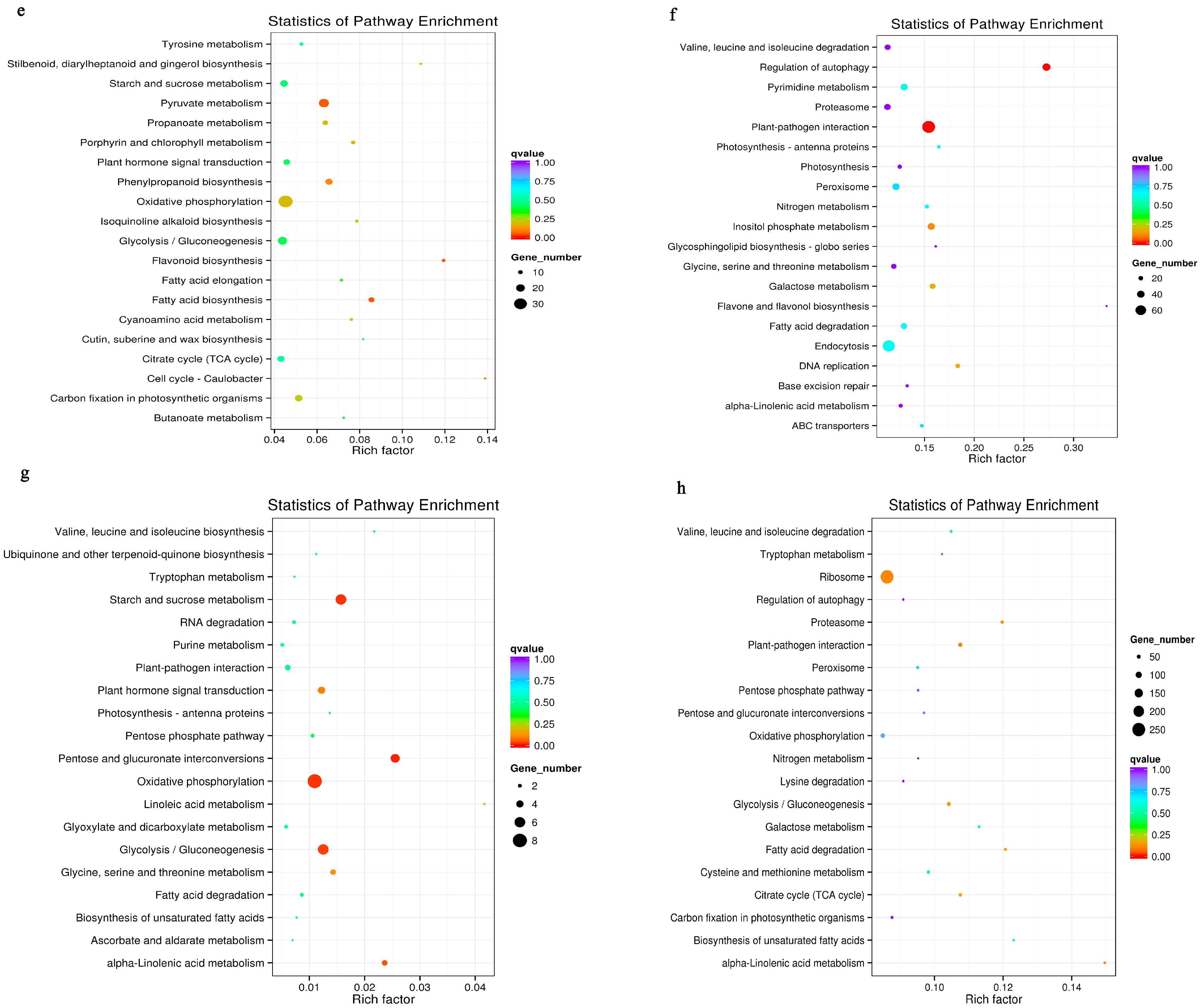
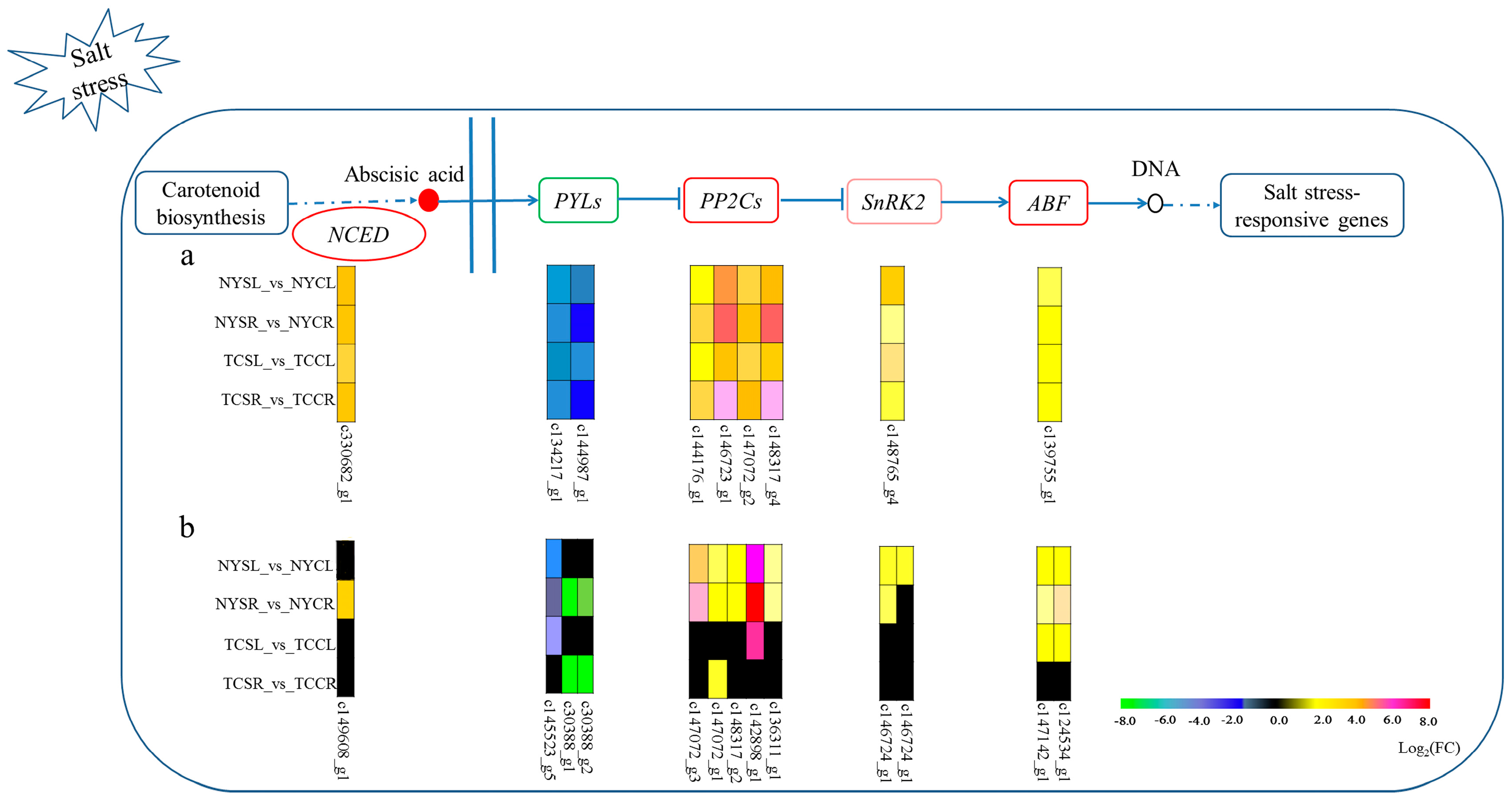
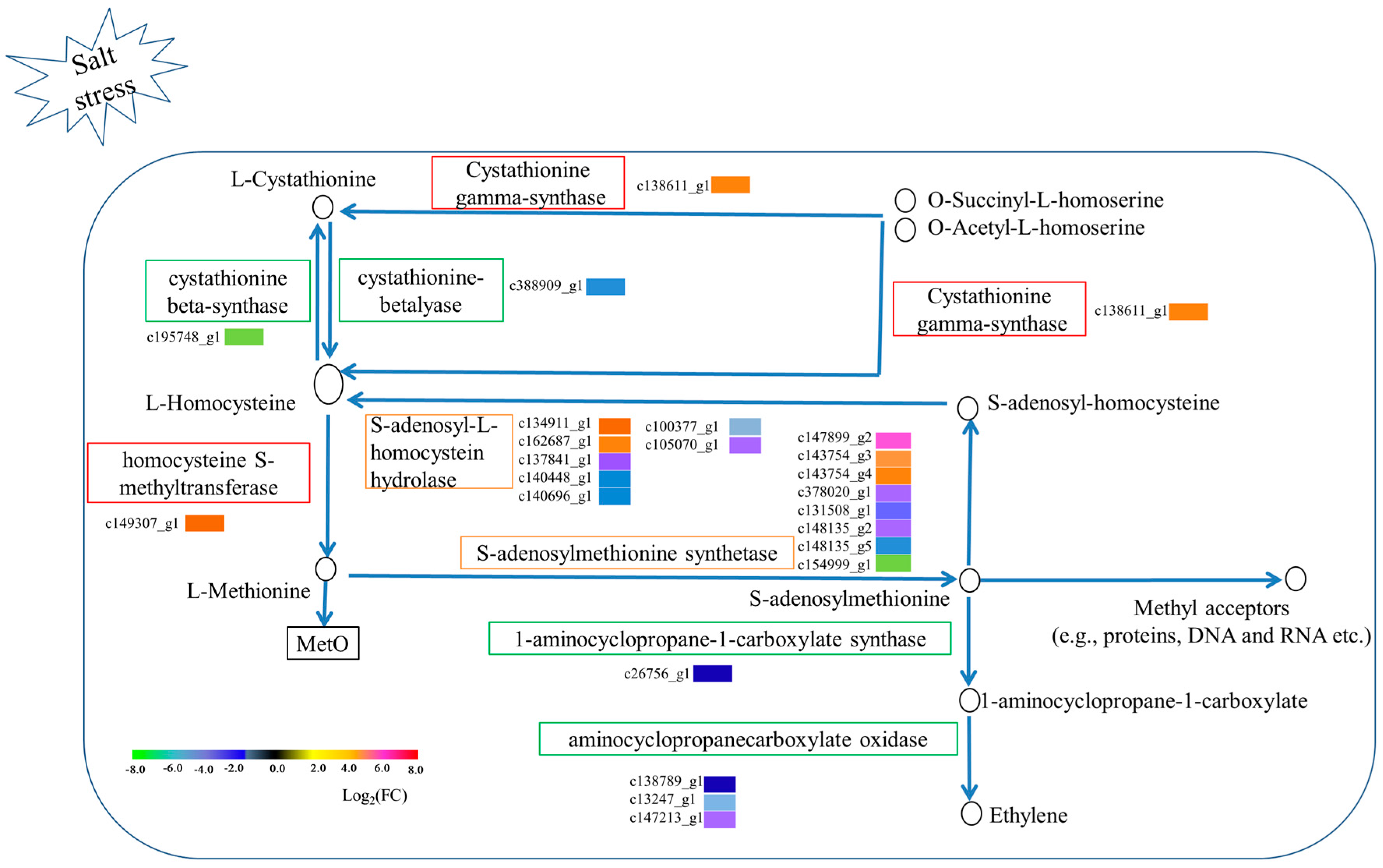
3.6. Metabolic Pathways Enhanced under Salt Stress Conditions
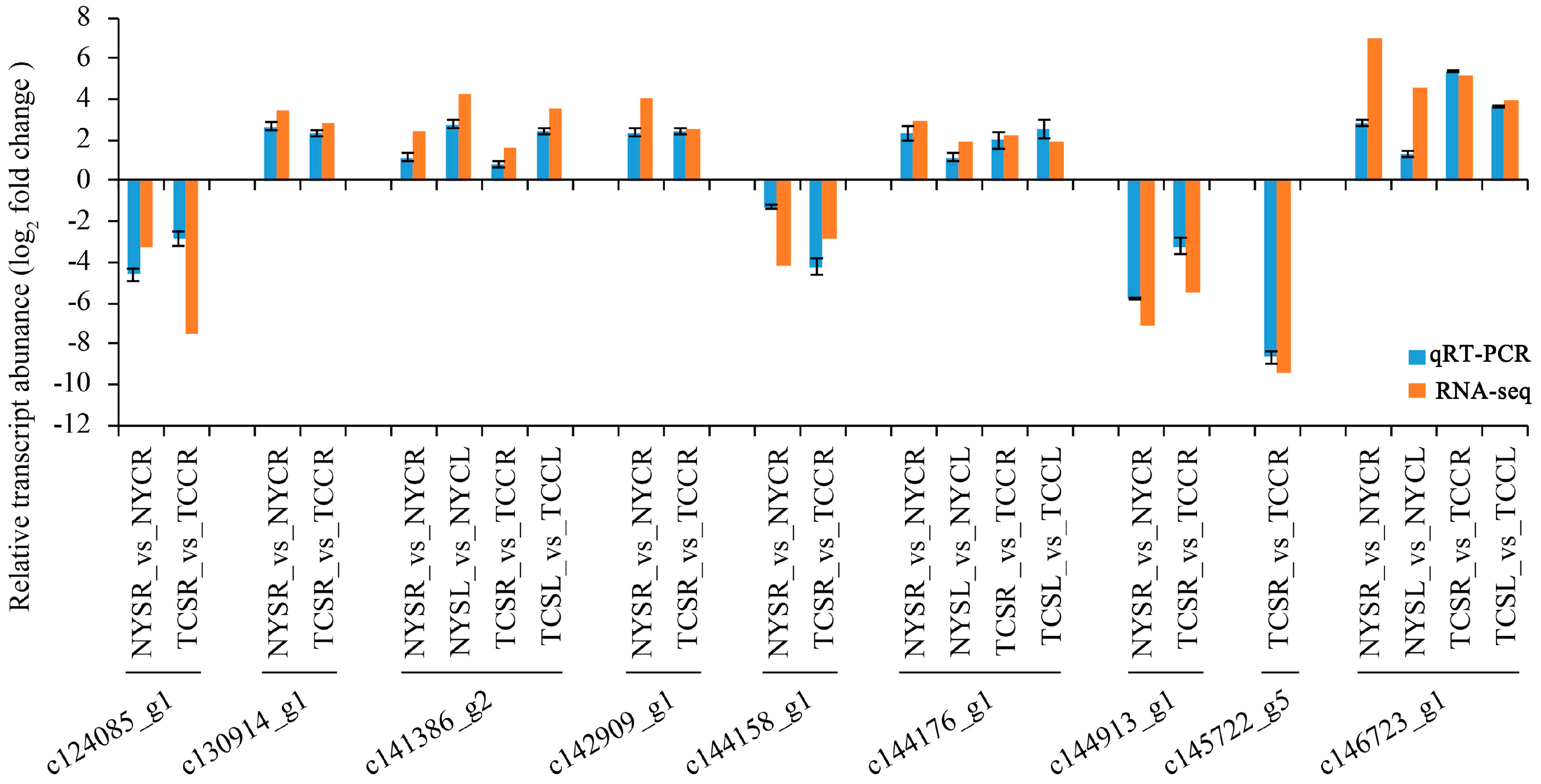
3.7. Validation of DEGs
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Yang, R.; Song, L.; Yang, Y.; Wang, Q.; Wang, Z.; Ren, C.; Ma, H.; Zafar, S.; Ashraf, M.Y. Differential proteomic analysis of salt stress response in jute (Corchorus Capsularis & olitorius L.) seedling roots. Pak. J. Bot. 2015, 47, 385–396. [Google Scholar]
- Li, S.; Fan, C.; Li, Y.; Zhang, J.; Sun, J.; Chen, Y.; Tian, C.; Su, X.; Lu, M.; Liang, C.; et al. Effects of drought and salt-stresses on gene expression in Caragana korshinskii seedlings revealed by RNA-Seq. BMC Genom. 2016, 17, 200. [Google Scholar] [CrossRef] [PubMed]
- De Zelicourt, A.; Colcombet, J.; Hirt, H. The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 2016, 21, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Zhu, J.-K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef] [PubMed]
- Furihata, T.; Maruyama, K.; Fujita, Y.; Umezawa, T.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 2006, 103, 1988–1993. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-J.; Kim, M.-D.; Deng, X.-P.; Kwak, S.-S.; Chen, W. Enhanced salt stress tolerance in transgenic potato plants expressing IBMYB1, a sweet potato transcription factor. J. Microbiol. Biotechnol. 2013, 23, 1737–1746. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhu, G.; Du, L.; Shang, X.; Cheng, C.; Yang, B.; Hu, Y.; Cai, C.; Guo, W. Genetic regulation of salt stress tolerance revealed by RNA-seq in cotton diploid wild species, Gossypium davidsonii. Sci. Rep. 2016, 6, 20582. [Google Scholar] [CrossRef] [PubMed]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2014, 5, 771. [Google Scholar] [CrossRef] [PubMed]
- Zagorchev, L.; Seal, C.E.; Kranner, I.; Odjakova, M. A central role for thiols in plant tolerance to abiotic stress. Int. J. Mol. Sci. 2013, 14, 7405–7432. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, S.B.; Aung, B.; Amyot, L.; Lalin, I.; Lachâal, M.; Karray-Bouraoui, N.; Hannoufa, A. Salt stress (NaCl) affects plant growth and branch pathways of carotenoid and flavonoid biosyntheses in Solanum nigrum. Acta Physiol. Plant. 2016, 38, 72. [Google Scholar] [CrossRef]
- Islam, M.M. Biochemistry, medicinal and food values of jute (Corchorus capsularis L. and C. olitorius L.) leaf: A review. Int. J. Enhanc. Res. Sci. Technol. Eng. 2013, 2, 135–144. [Google Scholar]
- Ogunkunle, C.O.; Ziyath, A.M.; Adewumi, F.E.; Fatoba, P.O. Bioaccumulation and associated dietary risks of Pb, Cd, and Zn in amaranth (Amaranthus cruentus) and jute mallow (Corchorus olitorius) grown on soil irrigated using polluted water from Asa river, Nigeria. Environ. Monit. Assess. 2015, 187, 281. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ming, R.; Zhang, J.; Tao, A.; Fang, P.; Qi, J. De novo transcriptome sequence and identification of major bast-related genes involved in cellulose biosynthesis in jute (Corchorus capsularis L.). BMC Genom. 2015, 16, 1062. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- NCBI. Available online: ftp://ftp.ncbi.nlm.nih.gov/blast/db/nr (accessed on 20 December 2016).
- NCBI. Available online: ftp://ftp.ncbi.nlm.nih.gov/blast/db/nt (accessed on 20 December 2016).
- KEGG. Available online: http://www.genome.jp/kegg/ (accessed on 20 December 2016).
- GO. Available online: http://www.geneontology.org/ (accessed on 20 December 2016).
- KOG. Available online: http://www.ncbi.nlm.nih.gov/COG/ (accessed on 20 December 2016).
- Pfam. Available online: http://pfam.sanger.ac.uk/ (accessed on 20 December 2016).
- SwissProt. Available online: http://www.ebi.ac.uk/uniprot/ (accessed on 20 December 2016).
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. Rsem: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. Degseq: An R package for identifying differentially expressed genes from RNA-Seq data. Bioinformatics 2009, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-Seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Ferdous, A.S.; Islam, M.T.; Alam, S.S.; Khan, H. Identification of stable reference genes for quantitative PCR in jute under different experimental conditions: An essential assessment for gene expression analysis. Aust. J. Crop Sci. 2015, 9, 646–655. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−δδct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Muthusamy, M.; Uma, S.; Backiyarani, S.; Saraswathi, M.S.; Chandrasekar, A. Transcriptomic changes of drought-tolerant and sensitive banana cultivars exposed to drought stress. Front. Plant Sci. 2016, 7, 1609. [Google Scholar] [CrossRef] [PubMed]
- Fracasso, A.; Trindade, L.M.; Amaducci, S. Drought stress tolerance strategies revealed by RNA-Seq in two sorghum genotypes with contrasting WUE. BMC Plant Biol. 2016, 16, 115. [Google Scholar] [CrossRef] [PubMed]
- Dalal, M.; Inupakutika, M. Transcriptional regulation of aba core signaling component genes in sorghum (Sorghum bicolor L. Moench). Mol. Breed. 2014, 34, 1517–1525. [Google Scholar] [CrossRef]
- Chen, X.; Han, H.; Jiang, P.; Nie, L.; Bao, H.; Fan, P.; Lv, S.; Feng, J.; Li, Y. Transformation of β-lycopene cyclase genes from Salicornia europaea and Arabidopsis conferred salt tolerance in Arabidopsis and Tobacco. Plant Cell Physiol. 2011, 52, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Shankar, R.; Bhattacharjee, A.; Jain, M. Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Sci. Rep. 2016, 6, 23719. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, H.; Shao, H.; Tang, X. Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 2016, 7, 67. [Google Scholar] [CrossRef] [PubMed]
- Mehta, R.H.; Ponnuchamy, M.; Kumar, J.; Reddy, N.R.R. Exploring drought stress-regulated genes in senna (Cassia angustifolia Vahl.): A transcriptomic approach. Funct. Integr. Genom. 2017, 17, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.C.; Chen, R.J.; Yu, R.R.; Zeng, H.L.; Zhang, D.P. Differential global genomic changes in rice root in response to low-, middle-, and high-osmotic stresses. Acta Physiol. Plant 2009, 31, 773–785. [Google Scholar] [CrossRef]
- Li, W.; Wang, T.; Zhang, Y.; Li, Y. Overexpression of soybean MIR172C confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenic Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 175–194. [Google Scholar] [CrossRef] [PubMed]
- Kumar, Y.; Zhang, L.; Panigrahi, P.; Dholakia, B.B.; Dewangan, V.; Chavan, S.G.; Kunjir, S.M.; Wu, X.; Li, N.; Rajmohanan, P.R.; et al. Fusarium oxysporum mediates systems metabolic reprogramming of chickpea roots as revealed by a combination of proteomics and metabolomics. Plant Biotechnol. J. 2016, 14, 1589–1603. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Sui, N.; Han, G. Salt-induced photoinhibition of PSII is alleviated in halophyte Thellungiella halophila by increases of unsaturated fatty acids in membrane lipids. Acta Physiol. Plant. 2014, 36, 983–992. [Google Scholar] [CrossRef]
| Genotypes | Tissues | Raw Reads (bp) | Clean Reads (bp) | Clean Bases (Gb) | Mapped Reads (%) | Q20 | Avg. GC (%) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | Salinity | Control | Salinity | Control | Salinity | Control | Salinity | (%) | |||
| NY | Leaf | 154,357,990 | 157,905,474 | 149,840,624 | 151,807,922 | 22.48 | 22.78 | 83.89% | 83.84% | >97 | 44.61 |
| Root | 170,983,540 | 150,144,930 | 165,732,832 | 145,221,480 | 24.86 | 21.78 | 80.72% | 80.98% | |||
| TC | Leaf | 154,625,422 | 146,504,736 | 149,883,254 | 140,337,978 | 22.48 | 21.06 | 83.47% | 83.22% | ||
| Root | 147,290,466 | 163,936,030 | 142,892,496 | 158,680,692 | 21.44 | 23.8 | 81.06% | 81.39% | |||
| Total | 1,245,748,588 | 1,204,397,278 (96.68%) | 180.68 | - | - | ||||||
| Parameters | Value |
|---|---|
| Number of unigenes | 68,961 |
| Number of transcripts | 134,793 |
| Average unigene length (bp) | 1322 |
| Average transcript length (bp) | 1600 |
| Unigene N50 (bp) | 1721 |
| transcript N50 (bp) | 2263 |
| Minimum unigene length (bp) | 201 |
| Minimum transcript length (bp) | 201 |
| Maximum unigene length (bp) | 15,890 |
| Maximum transcript length (bp) | 15,890 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Z.; Lu, R.; Dai, Z.; Yan, A.; Tang, Q.; Cheng, C.; Xu, Y.; Yang, W.; Su, J. Salt-Stress Response Mechanisms Using de Novo Transcriptome Sequencing of Salt-Tolerant and Sensitive Corchorus spp. Genotypes. Genes 2017, 8, 226. https://doi.org/10.3390/genes8090226
Yang Z, Lu R, Dai Z, Yan A, Tang Q, Cheng C, Xu Y, Yang W, Su J. Salt-Stress Response Mechanisms Using de Novo Transcriptome Sequencing of Salt-Tolerant and Sensitive Corchorus spp. Genotypes. Genes. 2017; 8(9):226. https://doi.org/10.3390/genes8090226
Chicago/Turabian StyleYang, Zemao, Ruike Lu, Zhigang Dai, An Yan, Qing Tang, Chaohua Cheng, Ying Xu, Wenting Yang, and Jianguang Su. 2017. "Salt-Stress Response Mechanisms Using de Novo Transcriptome Sequencing of Salt-Tolerant and Sensitive Corchorus spp. Genotypes" Genes 8, no. 9: 226. https://doi.org/10.3390/genes8090226
APA StyleYang, Z., Lu, R., Dai, Z., Yan, A., Tang, Q., Cheng, C., Xu, Y., Yang, W., & Su, J. (2017). Salt-Stress Response Mechanisms Using de Novo Transcriptome Sequencing of Salt-Tolerant and Sensitive Corchorus spp. Genotypes. Genes, 8(9), 226. https://doi.org/10.3390/genes8090226





