Genome-Wide Analysis of the Biosynthesis and Deactivation of Gibberellin-Dioxygenases Gene Family in Camellia sinensis (L.) O. Kuntze
Abstract
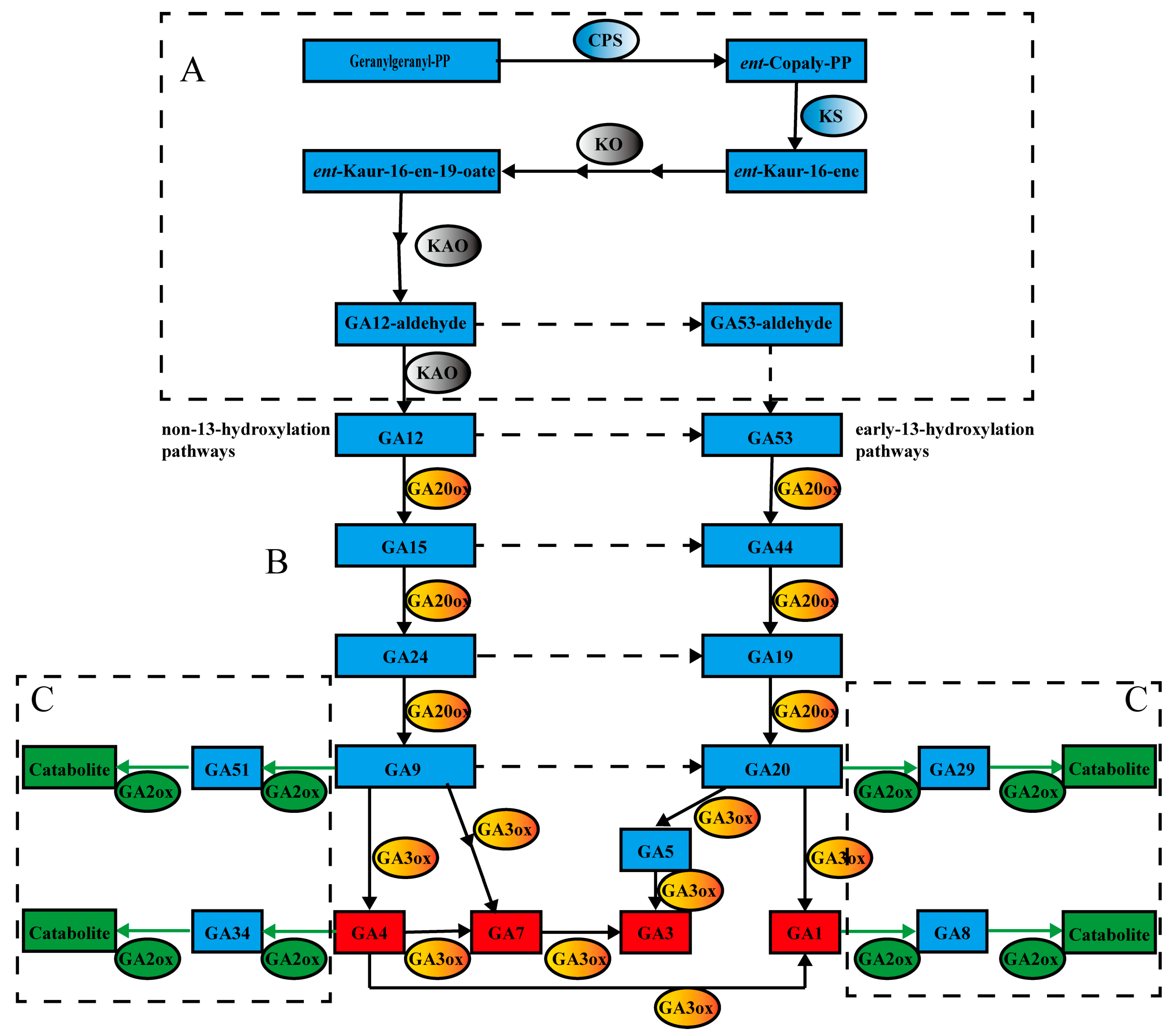
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Abiotic Treatments
2.2. RNA Isolation and cDNA Synthesis
2.3. Data Mining for GAox Protein Genes
2.4. Cloning the Full-Length of cDNA of GAox Protein Genes
2.5. Analysis of Sequences
2.6. Gene Expression Analysis by qRT-PCR
3. Results
3.1. Isolation, Identification and Annotation Information of the GAox Family Genes in Tea Plants
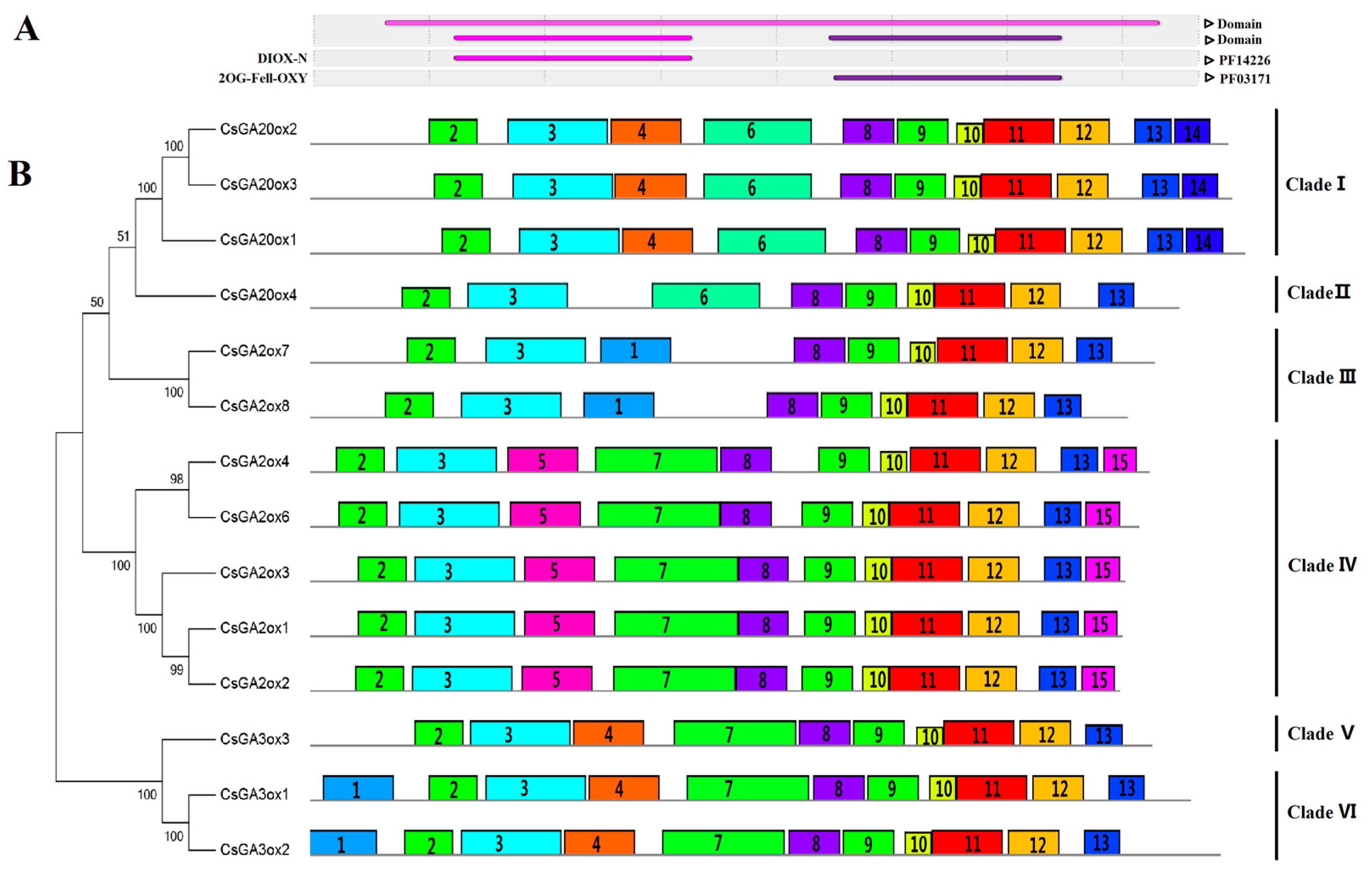
3.2. Similarity and Phylogenetic Gene Structure Analysis of the GAox Genes
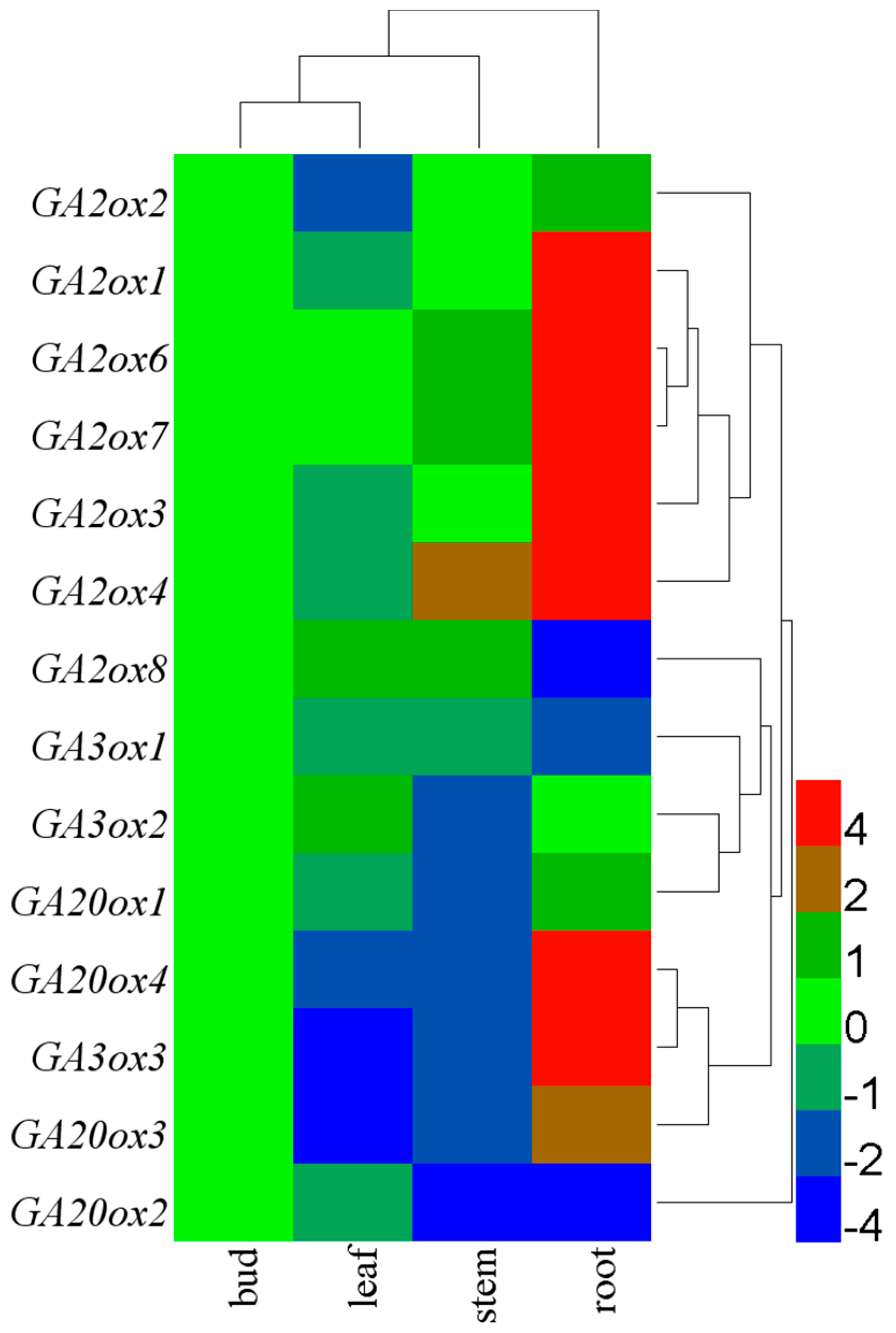
3.3. Tissue-Specific Expression of CsGAoxes
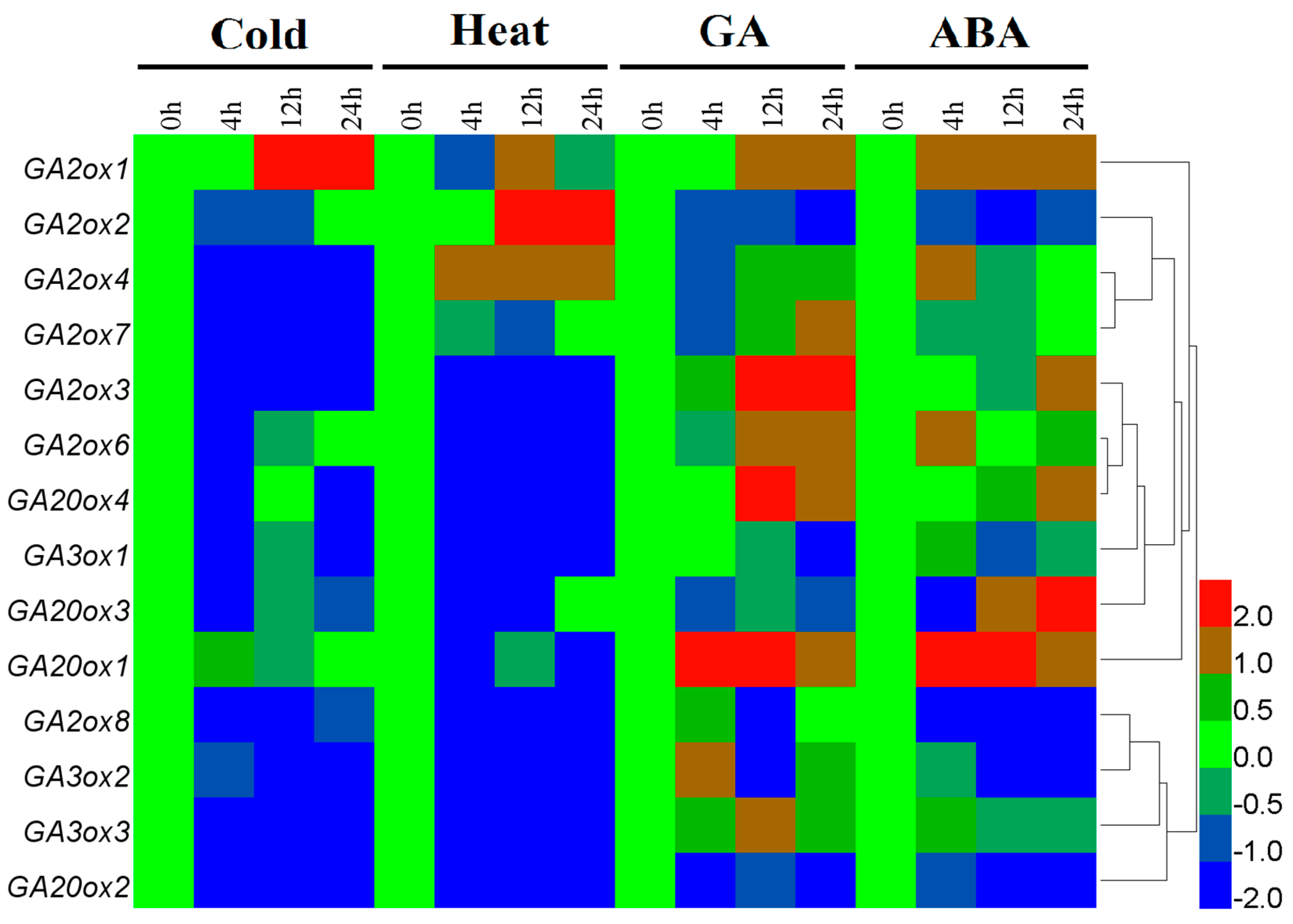
3.4. Differential Expression of CsGAox Genes under Abiotic Stresses
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gao, X.; Zhang, Y.; He, Z.; Fu, X. Hormone Metabolism & Signaling in Plants; Li, J.Y., Li, C.Y., Smith, S.M., Eds.; Academic Press: New York, NY, USA, 2017; pp. 107–160. [Google Scholar] [CrossRef]
- Hedden, P.; Kamiya, Y. Gibberellin biosynthesis: enzymes, genes and their regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 431–460. [Google Scholar] [CrossRef] [PubMed]
- Olszewski, N.; Gubler, F. Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 2002, 14, S61–S80. [Google Scholar] [PubMed]
- Vera-Sirera, F.; Gomez, M.D.; Perez-Amador, M.A. DELLA proteins, a group of GRAS transcription regulators that mediate gibberellin signaling. Plant Transcr. Factors. 2016, 313–328. [Google Scholar]
- Hedden, P.; Thomas, S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012, 444, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P.; Phillips, A.L. Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 2000, 5, 523–530. [Google Scholar] [CrossRef]
- Huerta, L.; Garcialor, A.; Garciamartinez, J.L. Characterization of gibberellin 20-oxidases in the citrus hybrid carrizo citrange. Tree Physiol. 2009, 29, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Sponsel, V.M.; Hedden, P. Plant Hormones. Biosynthesis, Signal Transduction, Action; Davies, P.J., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2004; pp. 63–94. [Google Scholar]
- De, C.E.; De, L.V. 2-oxoglutarate-dependent dioxygenase and related enzymes: biochemical characterization. Phytochemistry 1994, 36, 1093–1107. [Google Scholar]
- Phillips, A.L.; Ward, D.A.; Uknes, S.; Appleford, N.E.; Lange, T.; Huttly, A.K.; Gaskin, P.; Graebe, J.E.; Hedden, P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995, 108, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Schomburg, F.M.; Bizzell, C.M.; Lee, D.J.; Zeevaart, J.A.; Amasino, R.M. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 2003, 15, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Vidal, A.M.; Gisbert, C.; Talón, M.; Primomillo, E.; Lópezdíaz, I.; Garcíamartínez, J.L. The ectopic overexpression of a citrus gibberellin 20-oxidase enhances the non-13-hydroxylation pathway of gibberellin biosynthesis and induces an extremely elongated phenotype in tobacco. Physiol. Plant 2001, 112, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.L.; Li, L.; Wu, K.; Peeters, A.J.; Gage, D.A.; Zeevaart, J.A. The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc. Natl. Acad. Sci. USA 1995, 92, 6640–6644. [Google Scholar] [CrossRef] [PubMed]
- Lange, T.; Hedden, P.; Graebe, J.E. Expression cloning of a gibberellin 20-oxidase, a multifunctional enzyme involved in gibberellin biosynthesis. Proc. Natl. Acad. Sci. USA 1994, 91, 8552–8556. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Phillips, A.; Hedden, P. Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 1999, 96, 4698–4703. [Google Scholar] [CrossRef] [PubMed]
- Elliott, R.C.; Ross, J.J.; Smith, J.J.; Lester, D.R.; Reid, J.B. Feed-forward regulation of gibberellin deactivation in pea. J. Plant Growth Regul. 2001, 20, 87–94. [Google Scholar] [CrossRef]
- Barua, D.N. Seasonal Dormancy in Tea (Camellia sinensis L.). Nature 1969, 224, 514. [Google Scholar] [CrossRef]
- Nagar, P.K.; Kumar, A. Changes in endogenous gibberellin activity during winter dormancy in tea (Camellia sinensis (L.) O. Kuntze). Acta Physiol. Plant 2000, 22, 439–443. [Google Scholar] [CrossRef]
- Yue, C.; Zeng, J.M.; Zhang, Z.F.; Wang, X.C.; Cao, H.L. Research Progress in the Phytohormone of Tea Plant (Camellia sinensis). J. Tea Sci. 2012, 32, 382–392. [Google Scholar]
- Xia, E.H.; Zhang, H.B.; Sheng, J.; Li, K.; Zhang, Q.J.; Kim, C.; Zhang, Y.; Liu, Y.; Zhu, T.; Li, W.; et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant 2017, 10, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.N.; Tang, L.; Hou, Y.; Wang, P.; Yang, H.; Wei, C.L. Differential transcriptome analysis of leaves of tea plant (Camellia sinensis) provides comprehensive insights into the defense responses to Ectropis oblique attack using RNA-Seq. Funct. Integr. Genom. 2016, 16, 383–398. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.P. Textbook of Tea Cultivation; China Agriculture Press: Beijing, China, 2008; pp. 176–177. [Google Scholar]
- Li, H.; Huang, W.; Wang, G.L.; Wu, Z.J.; Zhuang, J. Expression profile analysis of ascorbic acid-related genes in response to temperature stress in the tea plant, Camellia sinensis (L.) O. Kuntze. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Li, X.W.; Feng, Z.G.; Yang, H.M.; Zhu, X.P.; Liu, J.; Yuan, H.Y. A novel cold-regulated gene from Camellia sinensis, CsCOR1, enhances salt- and dehydration-tolerance in tobacco. Biochem. Biophys. Res. Commun. 2010, 394, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Yue, C.; Cao, H.; Wang, L.; Zhou, Y.; Hao, X.; Zeng, J.; Wang, X.; Yang, Y. Molecular cloning and expression analysis of tea plant aquaporin (AQP) gene family. Plant Physiol. Biochem. 2014, 83, 65–76. [Google Scholar] [CrossRef] [PubMed]
- The University of California. Phytozome. Available online: https://phytozome.jgi.doe.gov/pz/portal.html (accessed on 9 May 2016).
- The Arabidopsis Biological Resource Center. TAIR. Available online: http://www.arabidopsis.org/ (accessed on 9 July 2017).
- Wang, D.; Pei, K.; Fu, Y.; Sun, Z.; Li, S.; Liu, H.; Tang, K.; Han, B.; Tao, Y. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 2007, 394, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, M.; Li, X.; Jiu, S.; Wang, C.; Fang, J. Genome-wide analysis of the sucrose synthase gene family in grape (Vitis vinifera): structure, evolution, and expression profiles. Genes 2017, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schafferi, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Altschul, S.F.; Wootton, J.C.; Gertz, E.M.; Agarwala, R.; Morgulis, A.; Schäffer, A.A.; Yu, Y.K. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005, 272, 5101–5109. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhard, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016, 44, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008, 36, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; William, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–532. [Google Scholar] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, k. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bataman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L.; et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009, 37, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Erik, V.B.; Asad, M.; Marc, D.L. Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR. BMC Mol. Biol. 2010, 11, 15. [Google Scholar] [CrossRef]
- Gohain, B.; Bandyopadhyay, T.; Borchetia, S.; Bharalee, R.; Gupta, S.; Bhorali, P.; Agarwala, N.; Das, S. Identification and validation of stable reference genes in Camellia Species. J. Bioteachnol. Pharm. Res. 2011, 2, 9–18. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Tang, D.; Shen, Y.; Qin, B.; Hong, L.; You, A.; Li, M.; Wang, X.; Yu, H.; Gu, M.; et al. Activation of gibberellin 2-oxidase 6 decreases active gibberellin levels and creates a dominant semi-dwarf phenotype in rice (Oryza sativa L.). J. Genet. Genom. 2010, 37, 23–36. [Google Scholar] [CrossRef]
- Lo, S.F.; Yang, S.Y.; Chen, K.T.; Hsing, Y.I.; Zeevaart, J.A.; Chen, L.J.; Yu, S.M. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 2008, 20, 2603–2618. [Google Scholar] [CrossRef] [PubMed]
- Shan, C.; Mei, Z.; Duan, J.; Chen, H.; Feng, H.; Cai, W. OsGA2ox5, a gibberellin metabolism enzyme, is involved in plant growth, the root gravity response and salt stress. PLoS One 2014, 9, e87110. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S.; Kamiya, Y. Gibberellin biosynthesis: its regulation by endogenous and environmental signals. Plant Cell Physiol. 2000, 41, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Lange, T.; Altpeter, F. Cloning and characterization of a cDNA encoding a multifunctional gibberellin 20-oxidase from perennial ryegrass (Lolium perenne L.). Plant Sci. 2002, 163, 147–155. [Google Scholar] [CrossRef]
- Lee, D.J.; Zeevaart, J.A. Molecular cloning of GA 2-oxidase3 from spinach and its ectopic expression in Nicotiana sylvestris. Plant Physiol. 2005, 138, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HemI: a toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.L.; Li, Q.; Deng, Y.W.; Lou, Y.G.; Wang, M.Y.; Zhou, G.X.; Zhang, Y.Y.; He, Z.H. Altered disease development in the eui mutants and Eui overexpressors indicates that gibberellins negatively regulate rice basal disease resistance. Mol. Plant 2008, 1, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Razem, F.A.; Baron, K.; Hill, R.D. Turning on gibberellin and abscisic acid signaling. Curr. Opin. Plant Biol. 2006, 9, 454–459. [Google Scholar] [CrossRef] [PubMed]
- Weiss, D.; Ori, N. Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 2007, 144, 1240–1246. [Google Scholar] [CrossRef] [PubMed]
- Bethke, P.C.; Jones, R.L. Gibberellin signaling. Curr. Opin. Plant Biol. 1998, 1, 440–446. [Google Scholar] [CrossRef]
- Phillips, A. Gibberellins in Arabidopsis. Plant Physiol. Biochem. 1998, 36, 115–124. [Google Scholar] [CrossRef]
- Venkatesh, J.; Park, S.W. Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato. Plant Physiol. Biochem. 2015, 94, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Liu, J.; Dong, C.; Cheng, Z.M. The CBL and CIPK gene family in grapevine (Vitis vinifera): genome-wide analysis and expression profiles in response to various abiotic stresses. Front. Plant Sci. 2017, 8, 978. [Google Scholar] [CrossRef] [PubMed]
- Chiang, H.; Hwang, l.; Goodma, H. Isolation of the Arabidopsis GA4 locus. Plant Cell 1995, 7, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Plackett, A.; Powers, S.; Fernandez-Garcia, N.; Urbanova, T.; Takebayashi, Y.; Seo, M.; Jikumaru, Y.; Benlloch, R.; Nilsson, O.; Ruiz-Rivero, O.; et al. Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell 2012, 24, 941–960. [Google Scholar] [CrossRef] [PubMed]
- Reinecke, D.M.; Wickramarathna, A.D.; Ozga, J.A.; Kurepin, L.V.; Jin, A.L.; Good, A.G.; Pharis, R.P. Gibberellin 3-oxidase gene expression patterns influence gibberellin biosynthesis, growth, and development in pea. Plant Physiol. 2013, 163, 929–945. [Google Scholar] [CrossRef] [PubMed]
- Sponsel, V.; Schmidt, F.; Porter, S.; Nakayama, M.; Kohlstruk, S.; Estelle, M. Characterization of new gibberellin-responsive semidwarf mutants of Arabidopsis. Plant Physiol. 1997, 115, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Coles, J.P.; Phillips, A.L.; Croker, S.J.; Garcia-Lepe, R.; Lewis, M.J.; Hedden, P. Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J. 1999, 17, 547–556. [Google Scholar] [PubMed]
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef] [PubMed]
- Itoh, H.; Ueguchi-Tanaka, M.; Sentoku, N.; Kitano, H.; Matsuoka, M.; Kobayashi, M. Cloning and functional analysis of two gibberellin 3β-hydroxylase genes that are differently expressed during the growth of rice. Proc. Natl. Acad. Sci. USA 2001, 98, 8909–8914. [Google Scholar] [CrossRef] [PubMed]
- Plackett, A.; Thomas, S.; Wilson, Z.; Hedden, P. Gibberellin control of stamen development: a fertile field. Trends Plant Sci. 2011, 16, 568–578. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S.; Smith, M.W.; Brown, R.G.S.; Kamiya, Y.; Sun, T. Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 1998, 10, 2115–2126. [Google Scholar] [PubMed]
- Huang, S.; Raman, A.S.; Ream, J.E.; Fujiwara, H.; Cerny, R.E.; Brown, S.M. Overexpression of 20-oxidase confers a gibberellin overproduction phenotype in Arabidopsis. Plant Physiol. 1998, 118, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Israelsson, M.; Mellerowicz, E.; Chono, M.; Gullberg, J.; Mortz, T. Cloning and overproduction of gibberellin 3-oxidase in hybrid aspen trees. Effects on gibberellin homeostasis and development. Plant Physiol. 2004, 135, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Kobayashi, M.; Itoh, H.; Tagiri, A.; Kayaka, H.; Lwahori, S.; Matsuoka, M. Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. 2001, 125, 1508–1516. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.J.; Zeevaart, J.A. Differential regulation of RNA levels of gibberellin dioxygenases by photoperiod in spinach. Plant Physiol. 2002, 130, 2085–2094. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Gong, F.; Cheminant, S.; Alioua, M.; Hedden, P.; Genschik, P. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 2008, 20, 2117–2129. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, Y.; Ogawa, M.; Kuwahara, A.; Hanada, A.; Kamiya, Y.; Yamaguchi, S. Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 2004, 16, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Toh, S.; Imamura, A.; Watanabe, A.; Nakabayashi, K.; Okamoto, M.; Jikumaru, Y.; Hanada, A.; Aso, Y.; Ishiyama, K.; Tamura, N.; et al. High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol. 2008, 146, 1368–1385. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Yamaguchi, S.; Hu, J.; Yusuke, J.; Jung, B.; Paik, L.; Lee, H.S.; Sun, T.P.; Kamiya, Y.; Choi, G. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 2007, 19, 1192–1208. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.; Hanada, A.; Kuwahara, A.; Endo, A.; Okamoto, M.; Yamauchi, Y.; North, H.; Marion-Poll, A.; Sun, T.P.; Koshiba, T.; et al. Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 2006, 48, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Zentella, R.; Zhang, Z.L.; Park, M.; Thomas, S.G.; Endo, A.; Murase, K.; Fleet, C.M.; Jikumary, Y.; Nambara, E.; Kamiya, Y.; Sun, T.P. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 2007, 19, 3037–3057. [Google Scholar] [CrossRef] [PubMed]
- Yaish, M.W.; El-Kereamy, A.; Zhu, T.; Beatty, P.H.; Good, A.G.; Bi, Y.M.; Rothstein, S.J. The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 2010, 6, e1001098. [Google Scholar] [CrossRef] [PubMed]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J. 2008, 56, 613–626. [Google Scholar] [CrossRef] [PubMed]


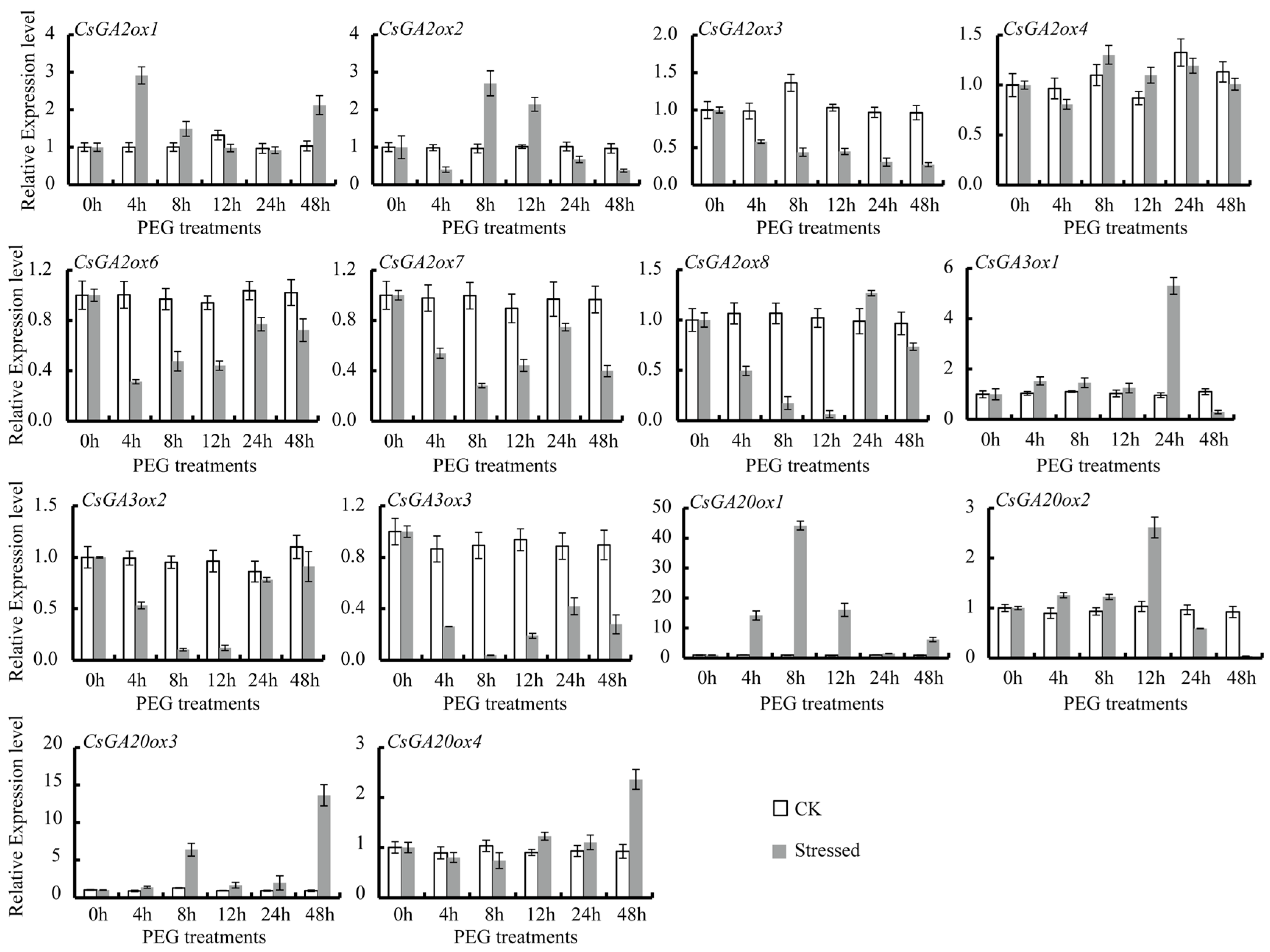
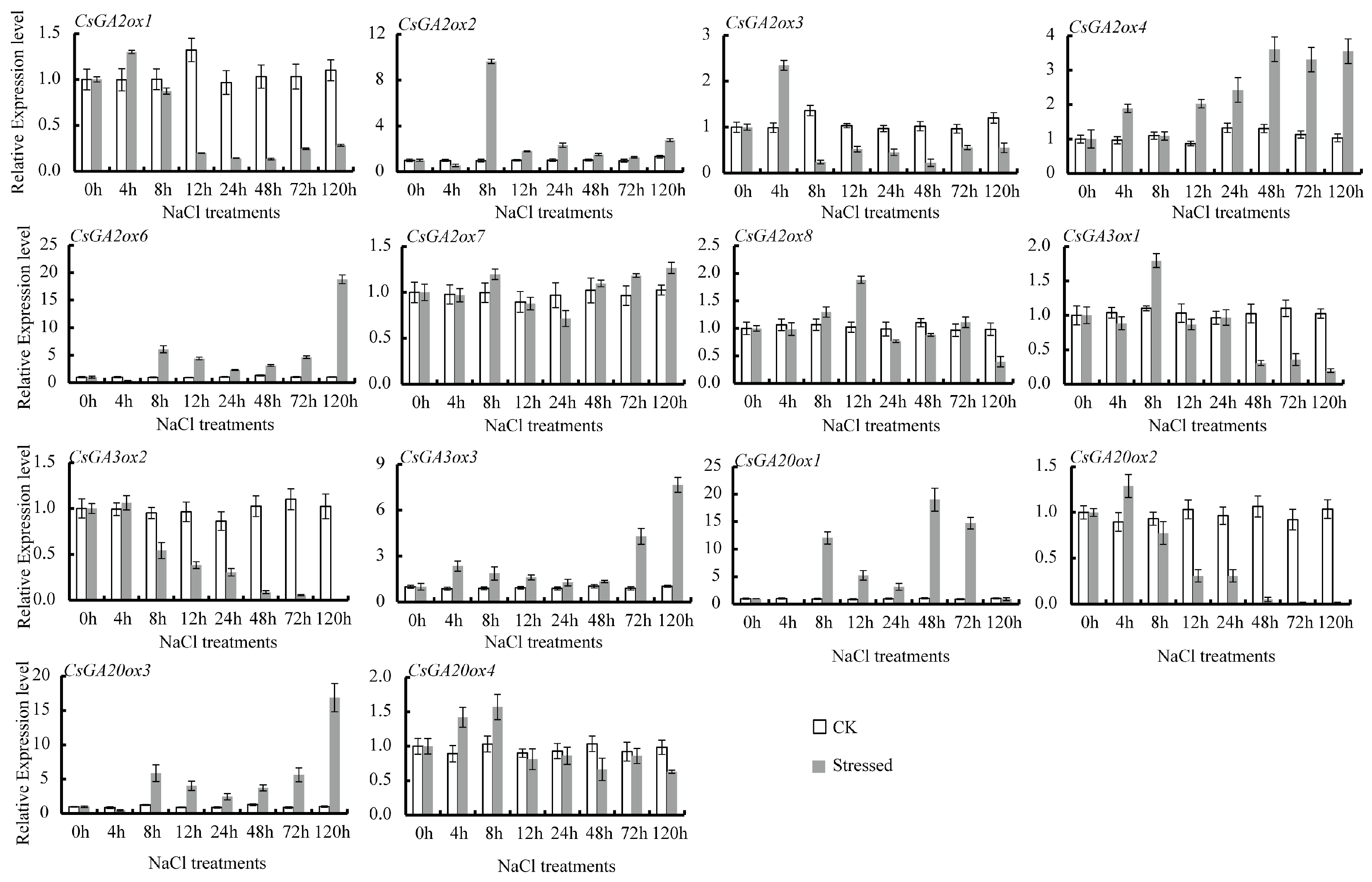
| Gene names | Accession Numbers | Gene ID a | Number of Deduced Amino Acid | Molecular Weight (kDa) | Subcellular Location (WoLF PSORT/TargetP) | Groups | Theoretical pI |
|---|---|---|---|---|---|---|---|
| CsGA20ox1 | KC193604 | CSA007392 | 383 | 42.94 | nucl | B | 6.06 |
| CsGA20ox2 | KY296366 | NF | 376 | 43.02 | nucl/cyto | B | 6.98 |
| CsGA20ox3 | MF370231 | CSA000105 | 378 | 43.03 | nucl/cyto | B | 6.94 |
| CsGA20ox4 | MF370232 | CSA002490 | 356 | 40.31 | cyto | B | 5.30 |
| CsGA2ox1 | KY296367 | CSA032124 | 333 | 37.18 | nucl/cyto | A1 | 5.93 |
| CsGA2ox2 | KY296368 | CSA026961 | 332 | 37.31 | cyto/nucl | A1 | 7.61 |
| CsGA2ox3 | KY296369 | CSA004444 | 334 | 37.56 | nucl/cyto/chlo | A1 | 5.47 |
| CsGA2ox4 | MF370234 | NF | 344 | 38.67 | nucl | A2 | 5.73 |
| CsGA2ox6 | MF370235 | CSA014596 | 340 | 38.02 | nucl/chol/mito | A2 | 7.94 |
| CsGA2ox7 | MF370236 | CSA013052 | 346 | 39.24 | cyto/nucl | A3 | 6.79 |
| CsGA2ox8 | MF370237 | CSA034015 | 335 | 38.53 | cyto/nucl | A3 | 5.33 |
| CsGA3ox1 | KF703743 | CSA002905 | 361 | 40.29 | chlo/mito/nucl | C | 6.67 |
| CsGA3ox2 | KF703744 | CSA020111 | 373 | 41.08 | nucl/cyto | C | 8.22 |
| CsGA3ox3 | MF370233 | CSA034282 | 345 | 38.72 | cyto/nucl | C | 5.66 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, C.; Tian, K.; Ban, Q.; Wang, L.; Sun, Q.; He, Y.; Yang, Y.; Pan, Y.; Li, Y.; Jiang, J.; et al. Genome-Wide Analysis of the Biosynthesis and Deactivation of Gibberellin-Dioxygenases Gene Family in Camellia sinensis (L.) O. Kuntze. Genes 2017, 8, 235. https://doi.org/10.3390/genes8090235
Pan C, Tian K, Ban Q, Wang L, Sun Q, He Y, Yang Y, Pan Y, Li Y, Jiang J, et al. Genome-Wide Analysis of the Biosynthesis and Deactivation of Gibberellin-Dioxygenases Gene Family in Camellia sinensis (L.) O. Kuntze. Genes. 2017; 8(9):235. https://doi.org/10.3390/genes8090235
Chicago/Turabian StylePan, Cheng, Kunhong Tian, Qiuyan Ban, Leigang Wang, Qilu Sun, Yan He, Yuanfei Yang, Yuting Pan, Yeyun Li, Jiayue Jiang, and et al. 2017. "Genome-Wide Analysis of the Biosynthesis and Deactivation of Gibberellin-Dioxygenases Gene Family in Camellia sinensis (L.) O. Kuntze" Genes 8, no. 9: 235. https://doi.org/10.3390/genes8090235
APA StylePan, C., Tian, K., Ban, Q., Wang, L., Sun, Q., He, Y., Yang, Y., Pan, Y., Li, Y., Jiang, J., & Jiang, C. (2017). Genome-Wide Analysis of the Biosynthesis and Deactivation of Gibberellin-Dioxygenases Gene Family in Camellia sinensis (L.) O. Kuntze. Genes, 8(9), 235. https://doi.org/10.3390/genes8090235





