Genome-Wide Analysis of the Sucrose Synthase Gene Family in Grape (Vitis vinifera): Structure, Evolution, and Expression Profiles
Abstract
:1. Introduction
2. Methods
2.1. Mining of Grape Sucrose Synthase Genes
2.2. Chromosomal Localization and Syntenic Analysis
2.3. Gene Structure and Phylogenetic Analysis
2.4. Ratio of Nonsynonymous to Synonymous Substitutions (dN/dS) and Relative Evolutionary Rate Test
2.5. Microarray Data Analysis
2.6. Plant Material and Treatments
2.7. RNA Extraction, cDNA Synthesis and Quantitative Real-Time PCR (qRT-PCR)
3. Results
3.1. Characteristics of the Grapevine Sucrose Synthase Gene Family
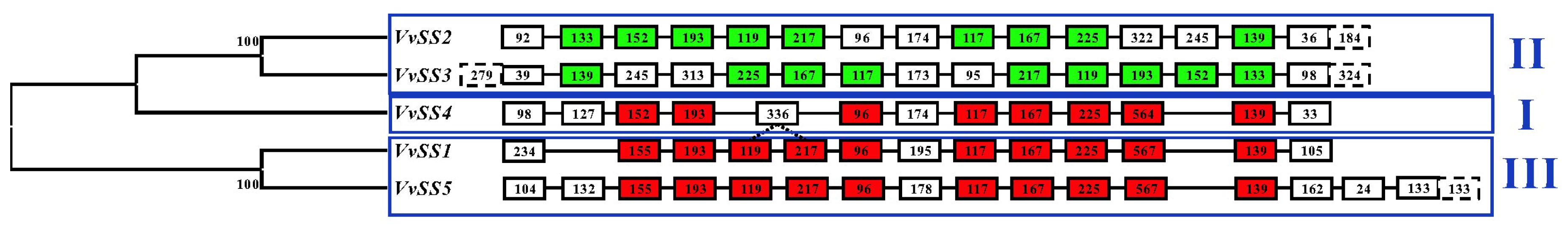
3.2. Exon/intron Organization of the Grapevine Sucrose Synthase Gene Family
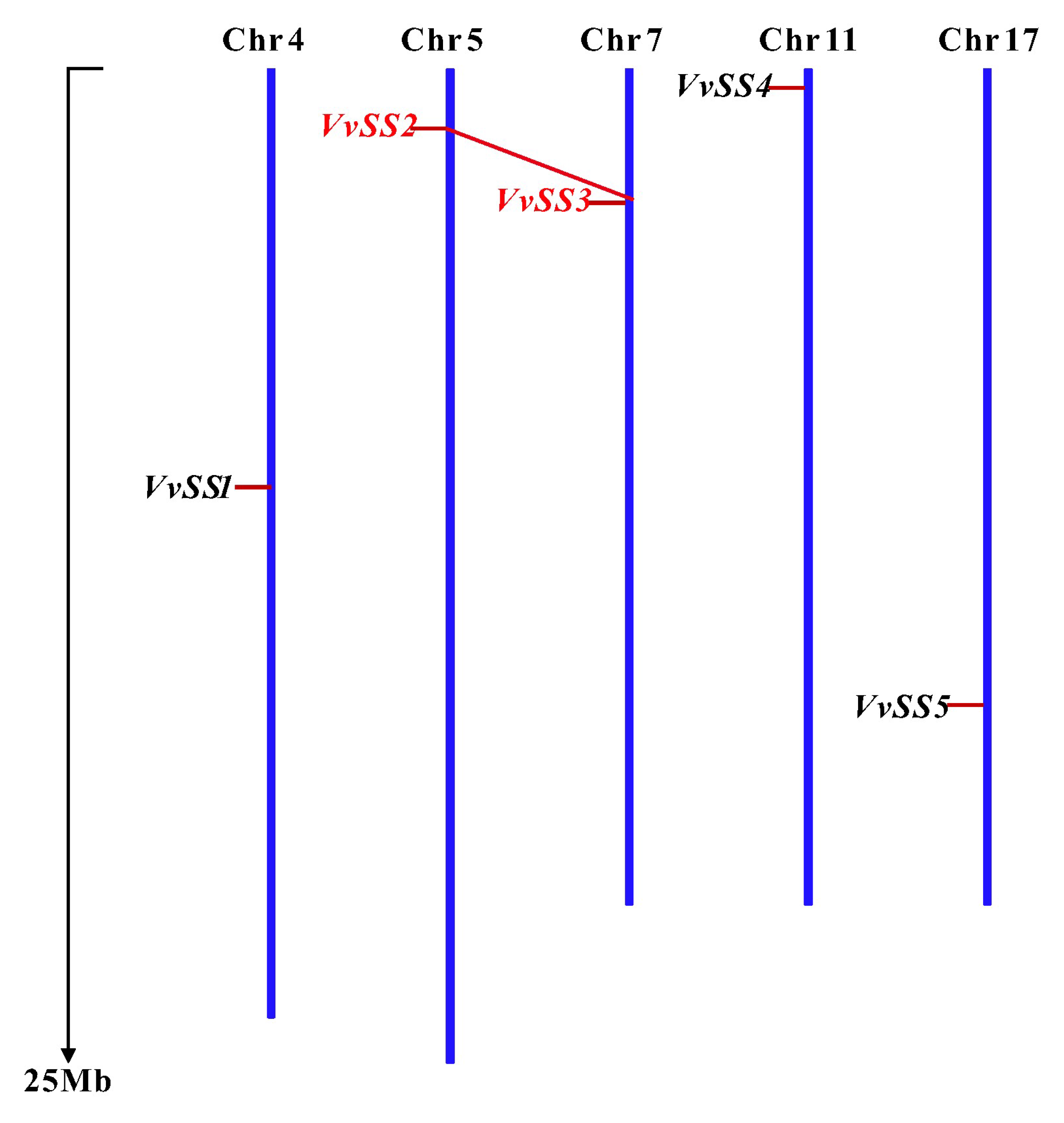
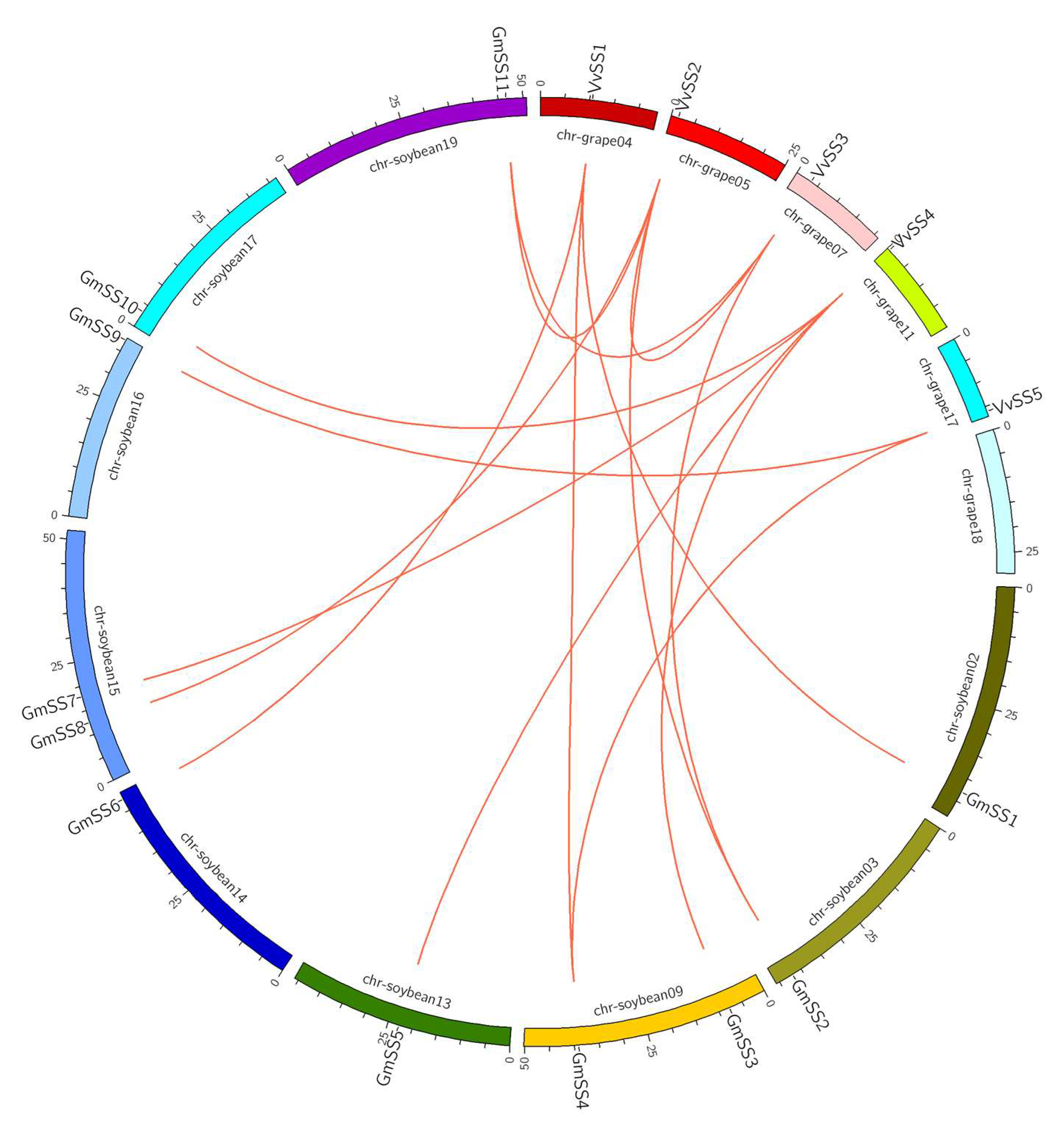
3.3. Gene Duplication and Syntenic Analysis of the Grapevine Sucrose Synthase Genes
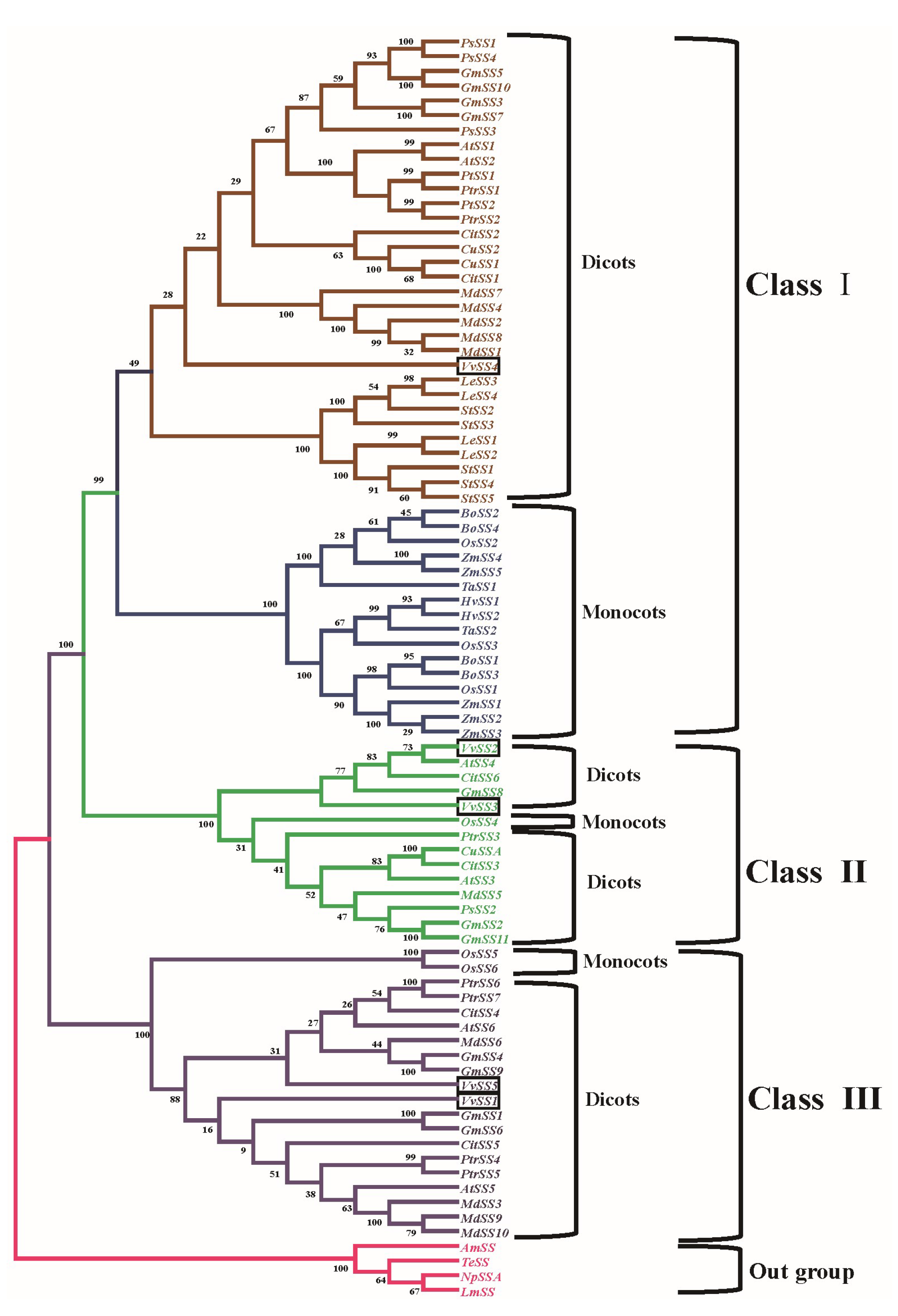
3.4. Phylogenetic Analysis of the Grapevine Sucrose Synthase Genes
3.5. Evolution of the Coding Sequences of the Grape SS Gene Pairs
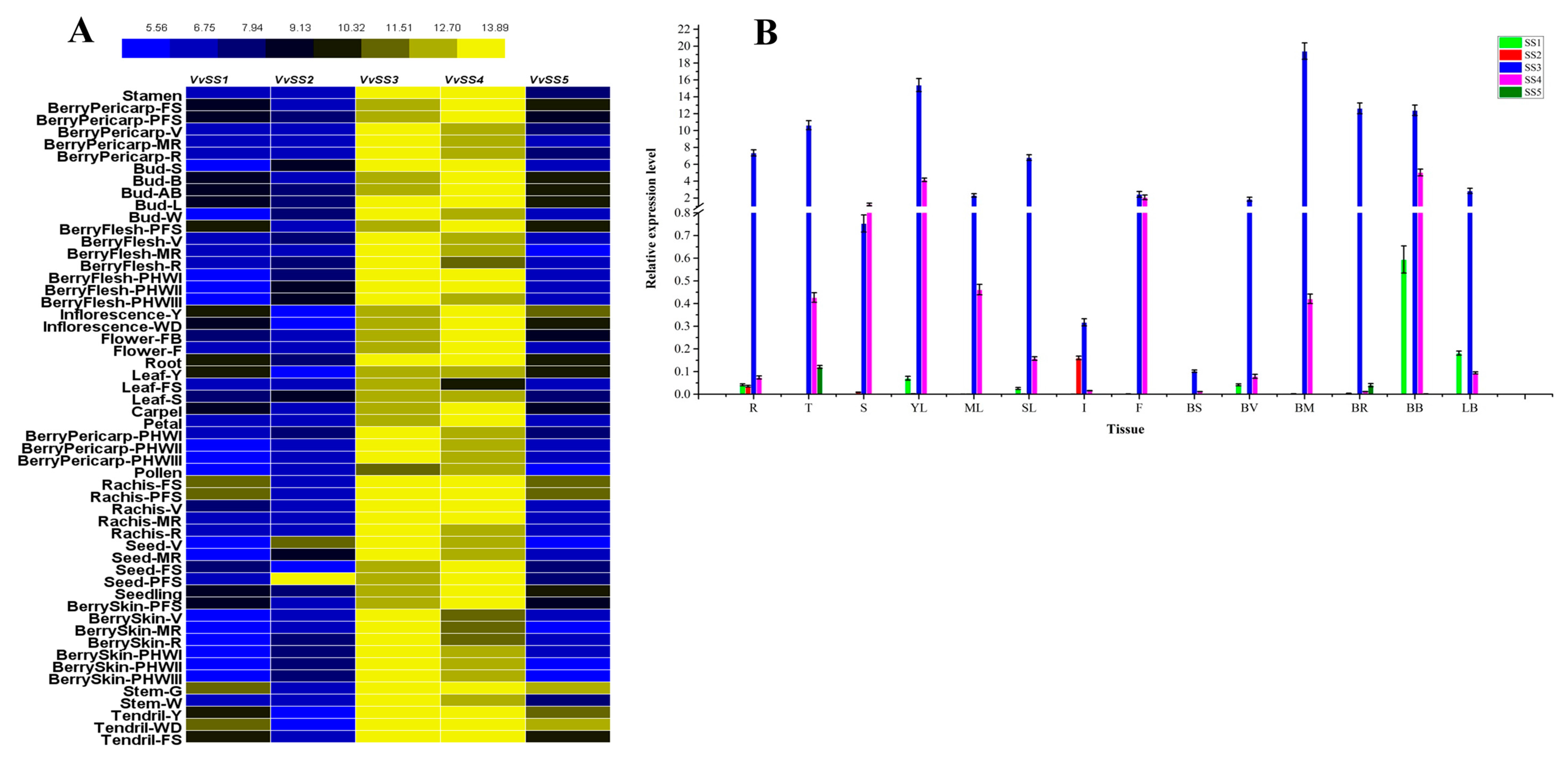
3.6. Expression Analysis of VvSS Genes in Different Organs, Tissues and Developmental Stages
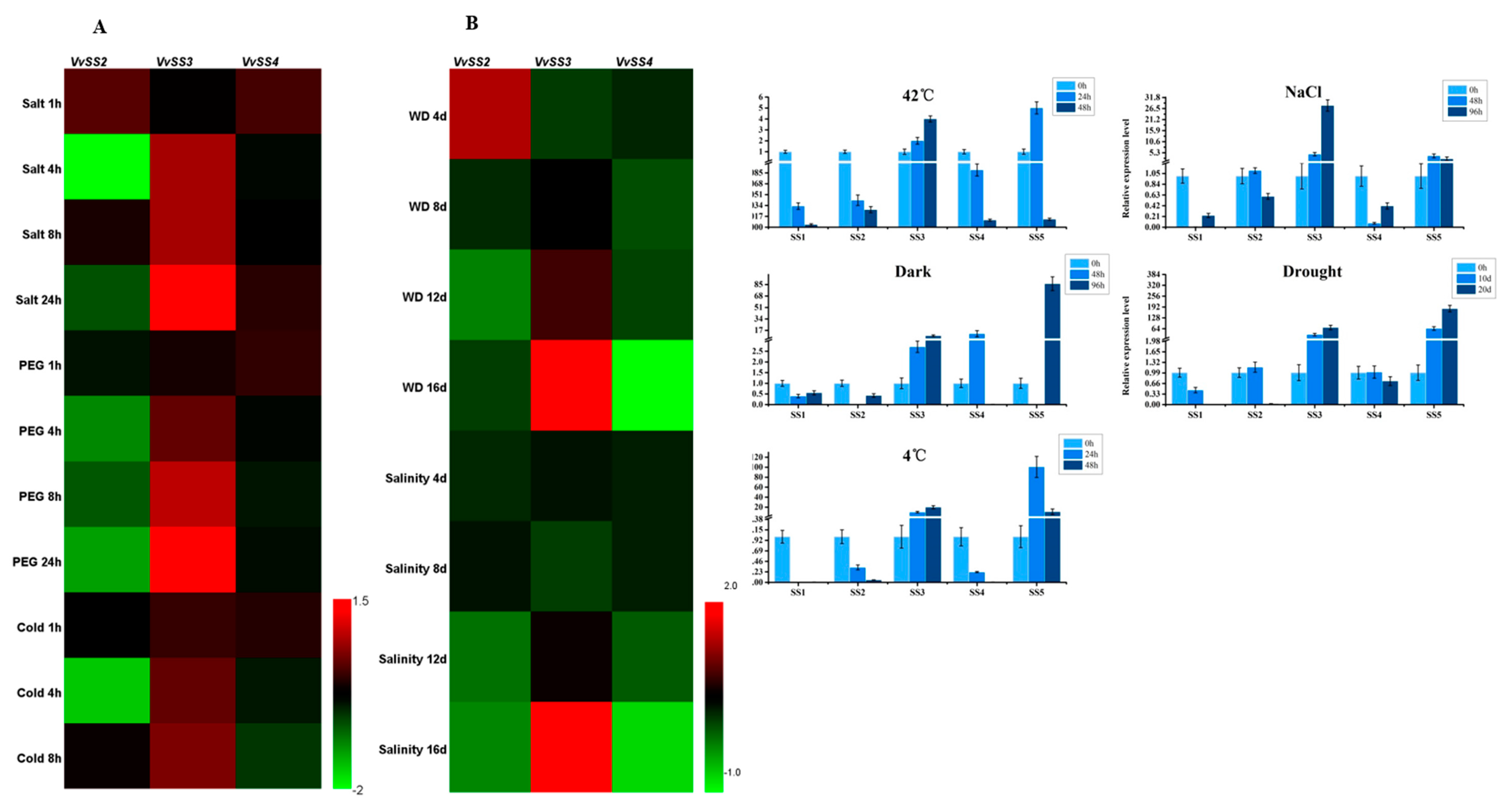
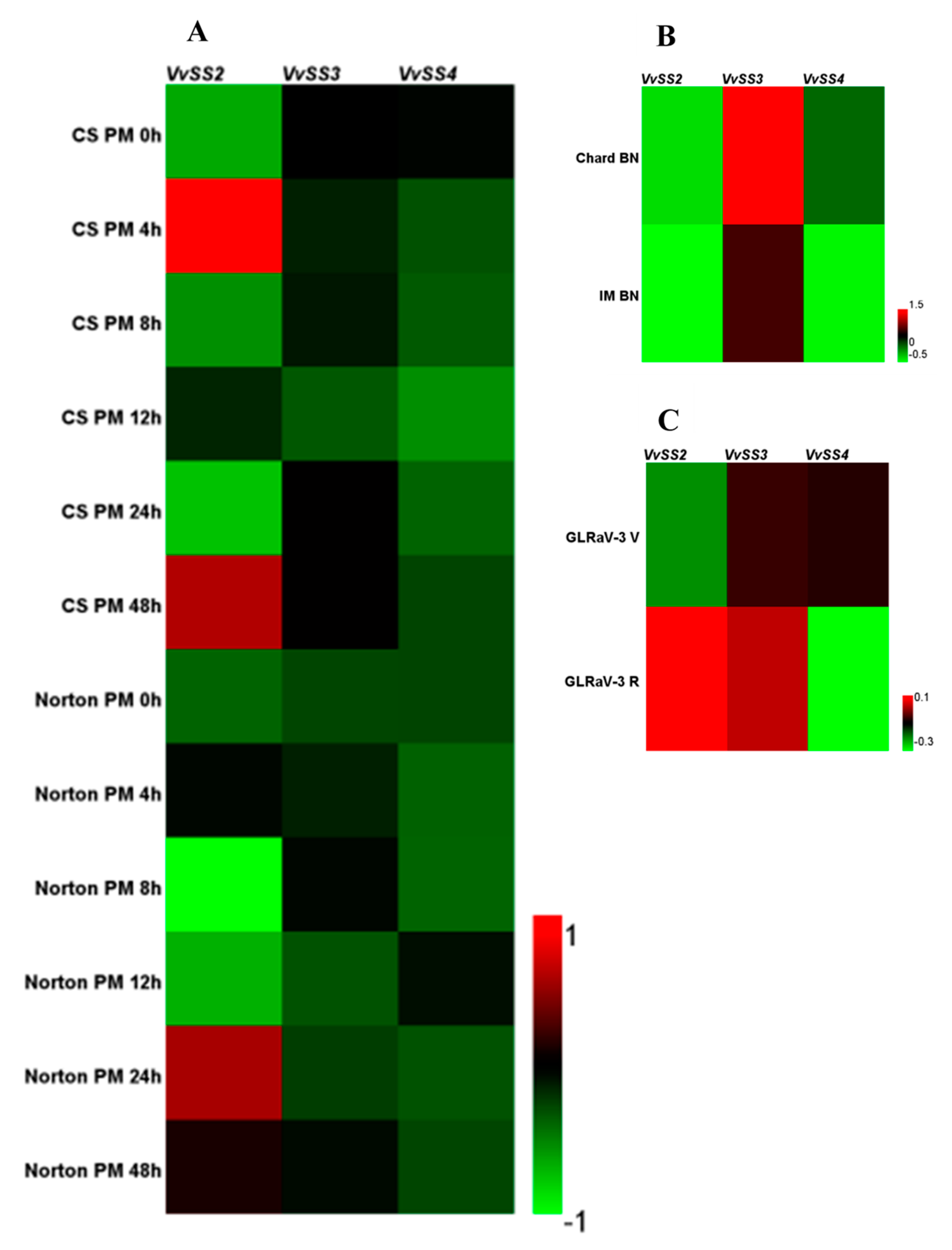
3.7. Expression Patterns of VvSS Genes in Response to Biotic and Abiotic Stresses
4. Discussion
4.1. Evolutionary Conservation and Divergence Among Grape SS Genes
4.2. VvSS Genes Involved in Grapevine Growth and Development
4.3. Stress Induced VvSS Expression in Grapevine
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lunn, J.E.; Furbank, R.T. Sucrose biosynthesis in C4 plants. New Phytol. 1999, 143, 221–237. [Google Scholar] [CrossRef]
- Chen, A.; He, S.; Li, F.; Li, Z.; Ding, M.; Liu, Q.; Rong, J. Analyses of the sucrose synthase gene family in cotton: Structure, phylogeny and expression patterns. BMC Plant Biol. 2012, 12, 85. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, J.; Wang, Z.; Zhu, Q. Activities of starch hydrolytic enzymes and sucrose-phosphate synthase in the stems of rice subjected to water stress during grain filling. J. Exp. Bot. 2001, 52, 2169–2179. [Google Scholar] [CrossRef] [PubMed]
- Strand, A.; Foyer, C.H.; Gustafsson, P.; Gardestrom, P.; Hurry, V. Altering flux through the sucrose biosynthesis pathway in transgenic Arabidopsis thaliana modifies photosynthetic acclimation at low temperatures and the development of freezing tolerance. Plant Cell Environ. 2003, 26, 523–536. [Google Scholar] [CrossRef]
- Ciereszko, I.; Johansson, H.; Kleczkowski, L.A. Sucrose and light regulation of a cold-inducible UDP-glucose pyrophosphorylase gene via a hexokinase-independent and abscisic acid-insensitive pathway in Arabidopsis. Biochem. J. 2001, 354, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Stitt, M.; Muller, C.; Matt, P.; Gibon, Y.; Carillo, P.; Morcuende, R.; Scheible, W.R.; Krapp, A. Steps towards an integrated view of nitrogen metabolism. J. Exp. Bot. 2002, 53, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, M.W.; Harrington, G.N.; Bush, D.R. Sucrose-mediated transcriptional regulation of sucrose symporter activity in the phloem. Proc. Natl. Acad. Sci. USA 2002, 99, 10876–10880. [Google Scholar] [PubMed]
- Zourelidou, M.; de Torres-Zabala, M.; Smith, C.; Bevan, M.W. Storekeeper defines a new class of plant-specific DNA-binding proteins and is a putative regulator of patatin expression. Plant J. 2002, 30, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Gaudin, V.; Lunness, P.A.; Fobert, P.R.; Towers, M.; Riou-Khamlichi, C.; Murray, J.A.; Coen, E.; Doonan, J.H. The expression of D-cyclin genes defines distinct developmental zones in snapdragon apical meristems and is locally regulated by the Cycloidea gene. Plant Physiol. 2000, 122, 1137–1148. [Google Scholar] [PubMed]
- Ohto, M.; Onai, K.; Furukawa, Y.; Aoki, E.; Araki, T.; Nakamura, K. Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol. 2001, 127, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Uggla, C.; Magel, E.; Moritz, T.; Sundberg, B. Function and dynamics of auxin and carbohydrates during earlywood/latewood transition in scots pine. Plant Physiol. 2001, 125, 2029–2039. [Google Scholar] [CrossRef] [PubMed]
- Iraqi, D.; Tremblay, F.M. Analysis of carbohydrate metabolism enzymes and cellular contents of sugars and proteins during spruce somatic embryogenesis suggests a regulatory role of exogenous sucrose in embryo development. J. Exp. Bot. 2001, 52, 2301–2311. [Google Scholar] [CrossRef] [PubMed]
- Rook, F.; Corke, F.; Card, R.; Munz, G.; Smith, C.; Bevan, M.W. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 2001, 26, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Geigenberger, P.; Stitt, M. Sucrose synthase catalyzes a readily reversible reaction in vivo in developing potato tubers and other plant tissues. Planta 1993, 189, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Koch, K. Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 2004, 7, 235–246. [Google Scholar] [PubMed]
- Jiang, N.; Jin, L.F.; da Silva, J.A.T.; Islam, M.D.Z.; Gao, H.W.; Liu, Y.Z.; Peng, S.A. Activities of enzymes directly related with sucrose and citric acid metabolism in citrus fruit in response to soil plastic film mulch. Sci. Hortic. 2014, 168, 73–80. [Google Scholar] [CrossRef]
- Xu, S.M.; Brill, E.; Llewellyn, D.J.; Furbank, R.T.; Ruan, Y.L. Overexpression of a potato sucrose synthase gene in cotton accelerates leaf expansion, reduces seed abortion, and enhances fiber production. Mol. Plant 2012, 5, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Hockema, B.R.; Etxeberria, E. Metabolic contributors to drought-enhanced accumulation of sugars and acids in oranges. J. Am. Soc. Hortic. Sci. 2001, 126, 599–605. [Google Scholar]
- Coleman, H.D.; Yan, J.; Mansfield, S.D. Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc. Natl. Acad. Sci. USA 2009, 106, 13118–13123. [Google Scholar] [CrossRef] [PubMed]
- Baroja-Fernández, E.; Muñoz, F.J.; Li, J.; Bahaji, A.; Almagro, G.; Montero, M.; Etxeberria, E.; Hidalgo, M.; Sesma, M.T.; Pozueta-Romero, J. Sucrose synthase activity in the sus1/sus2/sus3/sus4 Arabidopsis mutant is sufficient to support normal cellulose and starch production. Proc. Natl. Acad. Sci. USA 2012, 109, 321–326. [Google Scholar] [PubMed]
- Baier, M.C.; Keck, M.; Gödde, V.; Niehaus, K.; Küster, H.; Hohnjec, N. Knockdown of the symbiotic sucrose synthase MtSucS1 affects arbuscule maturation and maintenance in mycorrhizal roots of Medicago truncatula. Plant Physiol. 2010, 152, 1000–1014. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.H.; Tang, C.R.; Fang, Y.J.; Yang, M.; Zhou, B.H.; Qi, J.; Zhang, Y. Structure and expression profile of the sucrose synthase gene family in the rubber tree: Indicative of roles in stress response and sucrose utilization in the laticifers. FEBS J. 2014, 281, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Barratt, D.H.; Barber, L.; Kruger, N.J.; Smith, A.M.; Wang, T.L.; Martin, C. Multiple, distinct isoforms of sucrose synthase in pea. Plant Physiol. 2001, 127, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Duncan, K.A.; Hardin, S.C.; Huber, S.C. The three maize sucrose synthase isoforms differ in distribution, localization, and phosphorylation. Plant Cell Physiol. 2006, 47, 959–971. [Google Scholar] [CrossRef] [PubMed]
- Baud, S.; Vaultier, M.N.; Rochat, C. Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J. Exp. Bot. 2004, 55, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Horst, I.; Welham, T.; Kelly, S.; Kaneko, T.; Sato, S.; Tabata, S.; Parniske, M.; Wang, T.L. TILLING mutants of Lotus japonicus reveal that nitrogen assimilation and fixation can occur in the absence of nodule-enhanced sucrose synthase. Plant Physiol. 2007, 144, 806–820. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Scofield, G.N.; Terao, T. An expression analysis profile for the entire sucrose synthase gene family in rice. Plant Sci. 2008, 174, 534–543. [Google Scholar] [CrossRef]
- Zou, C.; Lu, C.; Shang, H.; Jing, X.; Cheng, H.; Zhang, Y.; Song, G. Genome-wide analysis of the Sus gene family in cotton. J. Integr. Plant Biol. 2013, 55, 643–653. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Chen, Z.; Wang, J.; Ye, M.; Ji, L.; Wang, J.; Liao, W.; Ma, H. Identification and characterization of the Populus sucrose synthase gene family. Gene 2014, 539, 58–67. [Google Scholar] [PubMed]
- Jiang, S.Y.; Chi, Y.H.; Wang, J.Z.; Zhou, J.X.; Cheng, Y.S.; Zhang, B.L.; Ma, A.; Vanitha, J.; Ramachandran, S. Sucrose metabolism gene families and their biological functions. Sci. Rep. 2015, 5, 17583. [Google Scholar] [CrossRef] [PubMed]
- Bieniawska, Z.; Paul Barratt, D.H.; Garlick, A.P.; Thole, V.; Kruger, N.J.; Martin, C.; Zrenner, R.; Smith, A.M. Analysis of the sucrose synthase gene family in Arabidopsis. Plant J. 2007, 49, 810–828. [Google Scholar] [PubMed]
- Wang, A.Y.; Kao, M.H.; Yang, W.H.; Sayion, Y.; Liu, L.F.; Lee, P.D.; Su, J.C. Differentially and developmentally regulated expression of three rice sucrose synthase genes. Plant Cell Physiol. 1999, 40, 800–807. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, A.; Moriguchi, T.; Koyama, K.; Omura, M.; Akihama, T. Analysis of sucrose synthase genes in citrus suggests different roles and phylogenetic relationships. J. Exp. Bot. 2002, 53, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Grimplet, J.; Adam-Blondon, A.F.; Bert, P.F.; Bitz, O.; Cantu, D.; Davies, C.; Delrot, S.; Pezzotti, M.; Rombauts, S.; Cramer, G.R. The grapevine gene nomenclature system. BMC Genom. 2014, 15, 1077. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, L.F.; Song, C.N.; Leng, X.P.; Kayesh, E.; Sun, X.; Fang, J.G. Mining and comparison of the genes encoding the key enzymes involved in sugar biosynthesis in apple, grape, and sweet orange. Sci. Hortic. 2014, 165, 311–318. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evo. 2007, 24, 1586–1591. [Google Scholar]
- Tajima, F. Unbiased estimation of evolutionary distance between nucleotide sequences. Mol. Biol. Evol. 1993, 10, 677–688. [Google Scholar] [PubMed]
- Vision, T.J.; Brown, D.G.; Tanksley, S.D. The origins of genomic duplications in Arabidopsis. Science 2000, 290, 2114–2117. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L. The Evolution of Functionally Novel Proteins after Gene Duplication. Proc. R. Soc. B-Biol. Sci. 1994, 256, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [CrossRef] [PubMed]
- Lunn, J.E. Evolution of sucrose synthesis. Plant Physiol. 2002, 128, 1490–1500. [Google Scholar] [PubMed]
- Yang, Z.; Nielsen, R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000, 17, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Fasoli, M.; Dal Santo, S.; Zenoni, S.; Tornielli, G.B.; Farina, L.; Zamboni, A.; Porceddu, A.; Venturini, L.; Bicego, M.; Murino, V.; et al. The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 2012, 24, 3489–3505. [Google Scholar] [CrossRef] [PubMed]
- Fung, R.W.; Gonzalo, M.; Fekete, C.; Kovacs, L.G.; He, Y.; Marsh, E.; McIntyre, L.M.; Schachtman, D.P.; Qiu, W. Powdery mildew induces defense-oriented reprogramming of the transcriptome in a susceptible but not in a resistant grapevine. Plant Physiol. 2008, 146, 236–249. [Google Scholar] [CrossRef] [PubMed]
- Albertazzi, G.; Milc, J.; Caffagni, A.; Francia, E.; Roncaglia, E.; Ferrari, F.; Tagliafico, E.; Stefania, E.; Pecchionia, N. Gene expression in grapevine cultivars in response to Bois Noir phytoplasma infection. Plant Sci. 2009, 176, 792–804. [Google Scholar] [CrossRef]
- Vega, A.; Gutierrez, R.A.; Pena-Neira, A.; Cramer, G.R.; Arce-Johnson, P. Compatible GLRaV-3 viral infections affect berry ripening decreasing sugar accumulation and anthocyanin biosynthesis in Vitis vinifera. Plant Mol. Biol. 2011, 77, 261–274. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.Z.; Hu, X.M.; Jin, L.F.; Liu, Y.Z.; Peng, S.A. Genome-wide identification and expression profile analysis of citrus sucrose synthase genes: Investigation of possible roles in the regulation of sugar accumulation. PLoS ONE 2014, 9, e113623. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wei, P.; Wu, M.; Xu, Y.; Li, F.; Luo, Z.; Zhang, J.; Chen, A.; Xie, X.; Cao, P.; et al. Analysis of the sucrose synthase gene family in tobacco: Structure, phylogeny, and expression patterns. Planta 2015, 242, 153–166. [Google Scholar] [PubMed]
- Frugoli, J.A.; McPeek, M.A.; Thomas, T.L.; McClung, C.R. Intron loss and gain during evolution of the catalase gene family in angiosperms. Genetics 1998, 149, 355–365. [Google Scholar] [PubMed]
- Lecharny, A.; Boudet, N.; Gy, I.; Aubourg, S.; Kreis, M. Introns in, introns out in plant gene families: A genomic approach of the dynamics of gene structure. J. Struct. Funct. Genom. 2003, 3, 111–116. [Google Scholar] [CrossRef]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Ghiurcuta, C.G.; Moret, B.M.E. Evaluating synteny for improved comparative studies. Bioinformatics 2014, 30, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.Q.; Xu, B.H.; Yang, X.H.; Zhang, Z.Y.; Li, B.L. The sucrose synthase gene family in Populus: Structure, expression, and evolution. Tree Genet Genomes 2011, 7, 443–456. [Google Scholar] [CrossRef]
- Flagel, L.E.; Wendel, J.F. Gene duplication and evolutionary novelty in plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Feng, F.; Cheng, L. Expression patterns of genes involved in sugar metabolism and accumulation during apple fruit development. PLoS ONE 2012, 7, e33055. [Google Scholar] [CrossRef] [PubMed]
- Barrero, S.C.; Hernando, A.S.; Gonzalez, M.P.; Carbonero, P. Structure, expression profile and subcellular localisation of four different sucrose synthase genes from barley. Planta 2011, 234, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Kleines, M.; Elster, R.C.; Rodrigo, M.J.; Blervacq, A.S.; Salamini, F.; Bartels, D. Isolation and expression analysis of two stress-responsive sucrose-synthase genes from the resurrection plant Craterostigma plantagineum (Hochst.). Planta 1999, 209, 13–24. [Google Scholar] [CrossRef] [PubMed]
| Name | Locus Id a | Genomic DNA Size (bp) | CDS Size (bp) | Number of Amino Acids | Predicted Mw (kDa) | Theoretical pI | Ai | GRAVY | Chromosome Location | Group | Functional Domains (Start–End, bp) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sucrose Synthase | Glycosyl Transferase | |||||||||||
| VvSS1 | VIT_204s0079g00230 | 4513 | 2457 | 840 | 95.4 | 8.28 | 83.49 | −0.339 | chr4:10519070-10523582 | II | 9-557 | 569-745 |
| VvSS2 | VIT_205s0077g01930 | 7914 | 2508 | 835 | 95.6 | 5.85 | 91.99 | −0.244 | chr5:1507610-1515524 | I | 8-553 | 566-765 |
| VvSS3 | VIT_207s0005g00750 | 9214 | 2436 | 811 | 92.4 | 5.73 | 90.49 | −0.254 | chr7:3380739-3389952 | I | 8-555 | 568-741 |
| VvSS4 | VIT_211s0016g00470 | 4229 | 2448 | 815 | 93.6 | 6.13 | 92.20 | −0.247 | chr11:489955-494383 | I | 9-553 | 565--740 |
| VvSS5 | VIT_217s0053g00700 | 4820 | 2610 | 906 | 102.7 | 7.55 | 81.19 | −0.359 | chr17:15994677-15999496 | II | 12-558 | 568-736 |
| VvSS1 | VvSS2 | VvSS3 | VvSS4 | VvSS5 | |
|---|---|---|---|---|---|
| VvSS1 | - | 51.79 | 53.81 | 53.45 | 70.56 |
| VvSS2 | 59.18 | - | 79.12 | 66.86 | 47.70 |
| VvSS3 | 59.93 | 77.55 | - | 71.15 | 50.00 |
| VvSS4 | 59.60 | 66.36 | 69.47 | - | 59.57 |
| VvSS5 | 70.50 | 54.85 | 56.60 | 56.85 | - |
| Gene 1 | Gene 2 | dN | dS | dN/dS |
|---|---|---|---|---|
| VvSS2 | VvSS3 | 0.3012 | 0.0957 | 3.1473 |
| VvSS1 | VvSS5 | 0.2759 | 0.1246 | 2.2143 |
| Testing Group | Mt b | M1 c | M2 d | X2 | P e |
|---|---|---|---|---|---|
| VvSS2/VvSS3 with GmSS2 | 586 | 11 | 30 | 8.91 | 0.00450 |
| VvSS1/VvSS5 with GmSS4 | 472 | 9 | 4 | 1.92 | 0.21690 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.; Wang, M.; Li, X.; Jiu, S.; Wang, C.; Fang, J. Genome-Wide Analysis of the Sucrose Synthase Gene Family in Grape (Vitis vinifera): Structure, Evolution, and Expression Profiles. Genes 2017, 8, 111. https://doi.org/10.3390/genes8040111
Zhu X, Wang M, Li X, Jiu S, Wang C, Fang J. Genome-Wide Analysis of the Sucrose Synthase Gene Family in Grape (Vitis vinifera): Structure, Evolution, and Expression Profiles. Genes. 2017; 8(4):111. https://doi.org/10.3390/genes8040111
Chicago/Turabian StyleZhu, Xudong, Mengqi Wang, Xiaopeng Li, Songtao Jiu, Chen Wang, and Jinggui Fang. 2017. "Genome-Wide Analysis of the Sucrose Synthase Gene Family in Grape (Vitis vinifera): Structure, Evolution, and Expression Profiles" Genes 8, no. 4: 111. https://doi.org/10.3390/genes8040111
APA StyleZhu, X., Wang, M., Li, X., Jiu, S., Wang, C., & Fang, J. (2017). Genome-Wide Analysis of the Sucrose Synthase Gene Family in Grape (Vitis vinifera): Structure, Evolution, and Expression Profiles. Genes, 8(4), 111. https://doi.org/10.3390/genes8040111






