Machine Learning Reveals Common Regulatory Mechanisms Mediated by Autophagy-Related Genes in the Development of Inflammatory Bowel Disease and Major Depressive Disorder
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Source
2.2. Identification of DE-ARGs and Immune Cell Infiltrations
2.3. Immune Cell Infiltrations
2.4. Machine Learning-Based Screening of Potential Biomarkers
2.5. Differential Analysis and Gene Set Variation Analysis (GSVA) Between Subgroups
2.6. Drug Prediction
2.7. Molecular Docking Analysis
2.8. scRNA-seq Analysis
2.9. Mendelian Randomization Analysis
2.10. Statistical Analysis
3. Results
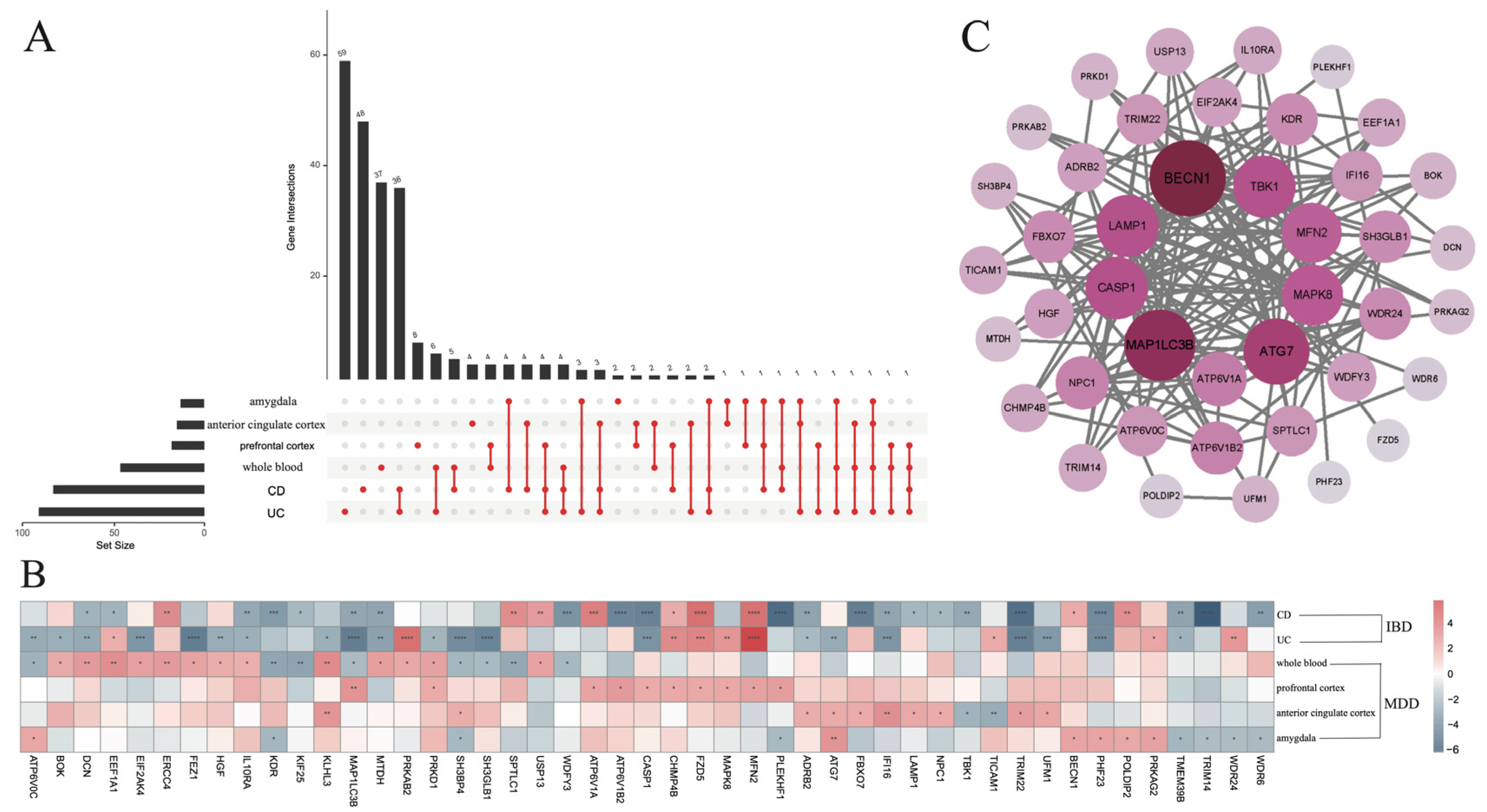
3.1. Detection of Co-DEGs Between IBD and MDD
3.2. Correlation Analysis of the 47 Co-DEGs
3.3. The Differential Infiltration of Immune Cells
3.4. Correlation Analysis Between 47 Co-DEGs and Immune Cells
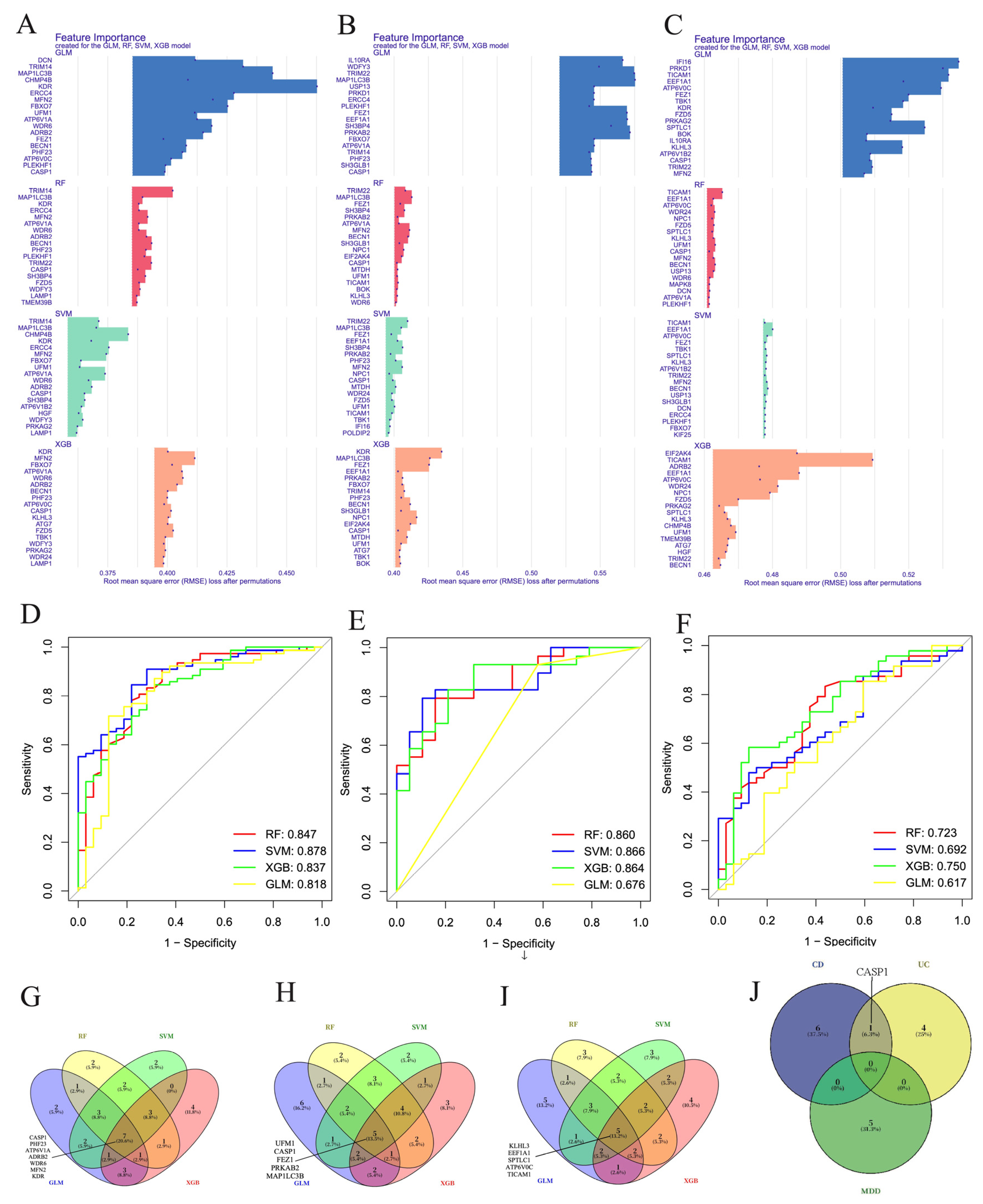
3.5. Machine Learning Screening for the Important Genes
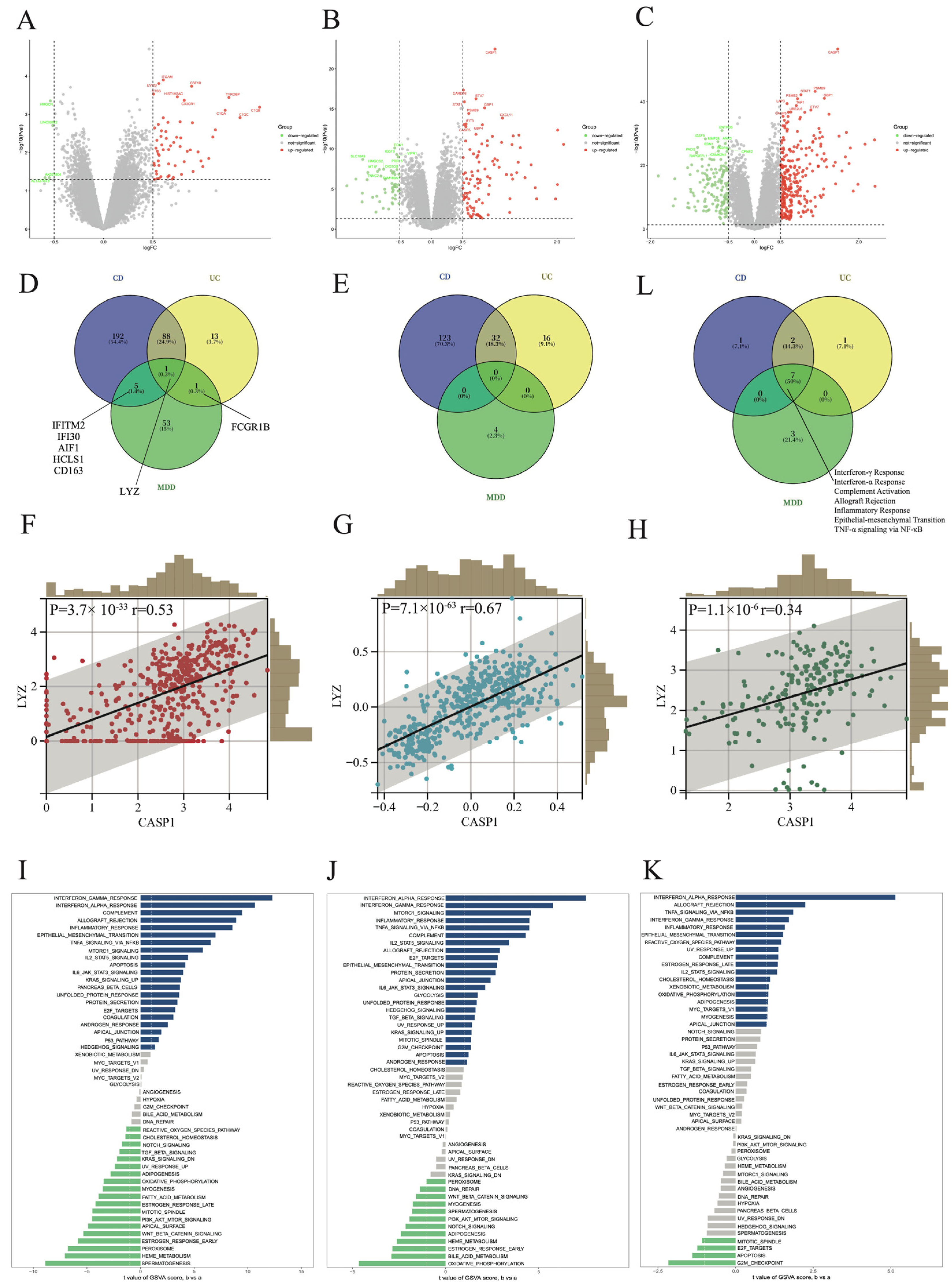
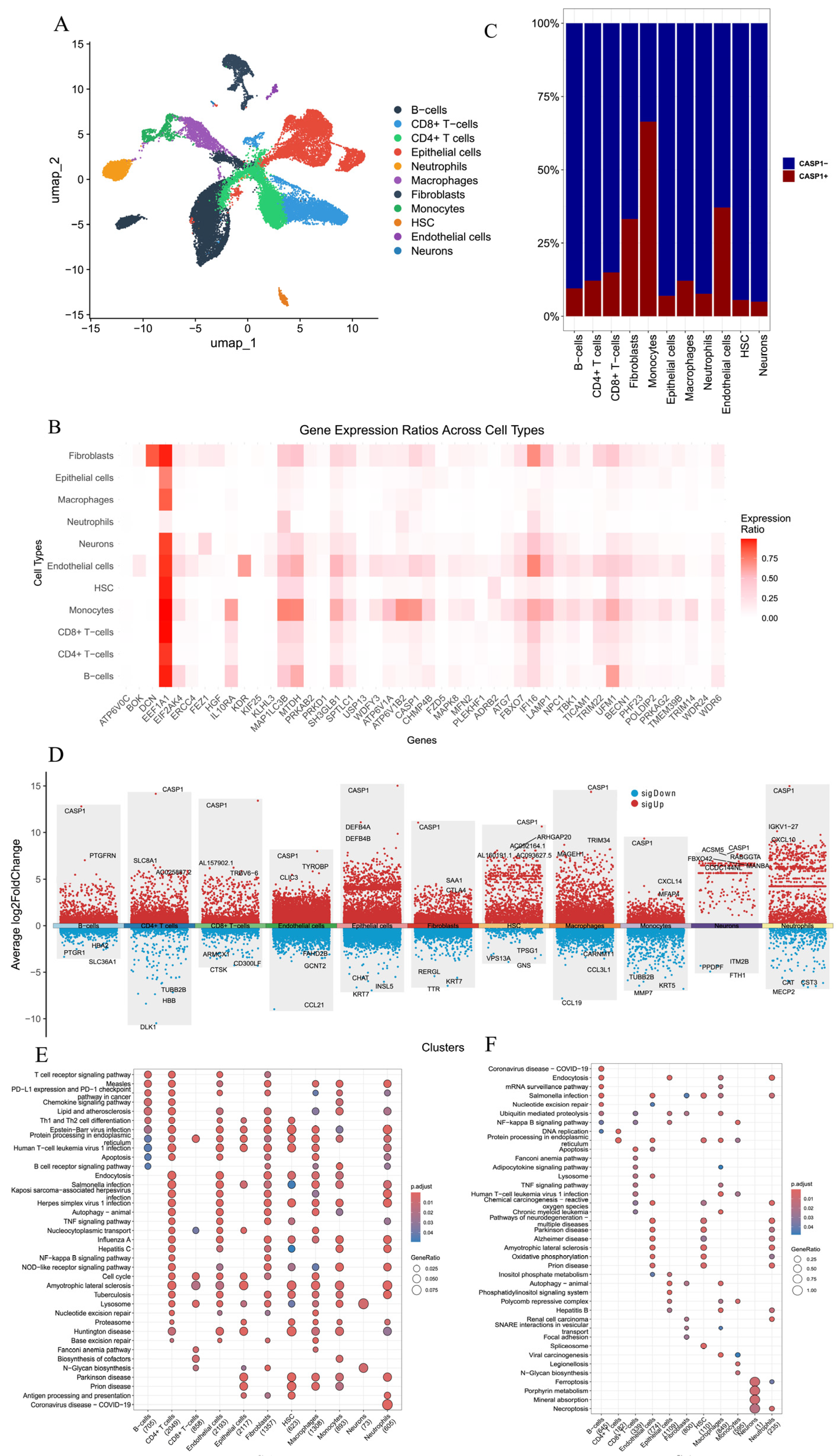
3.6. Comparison and Analysis of CASP1+ and CASP1− Groups
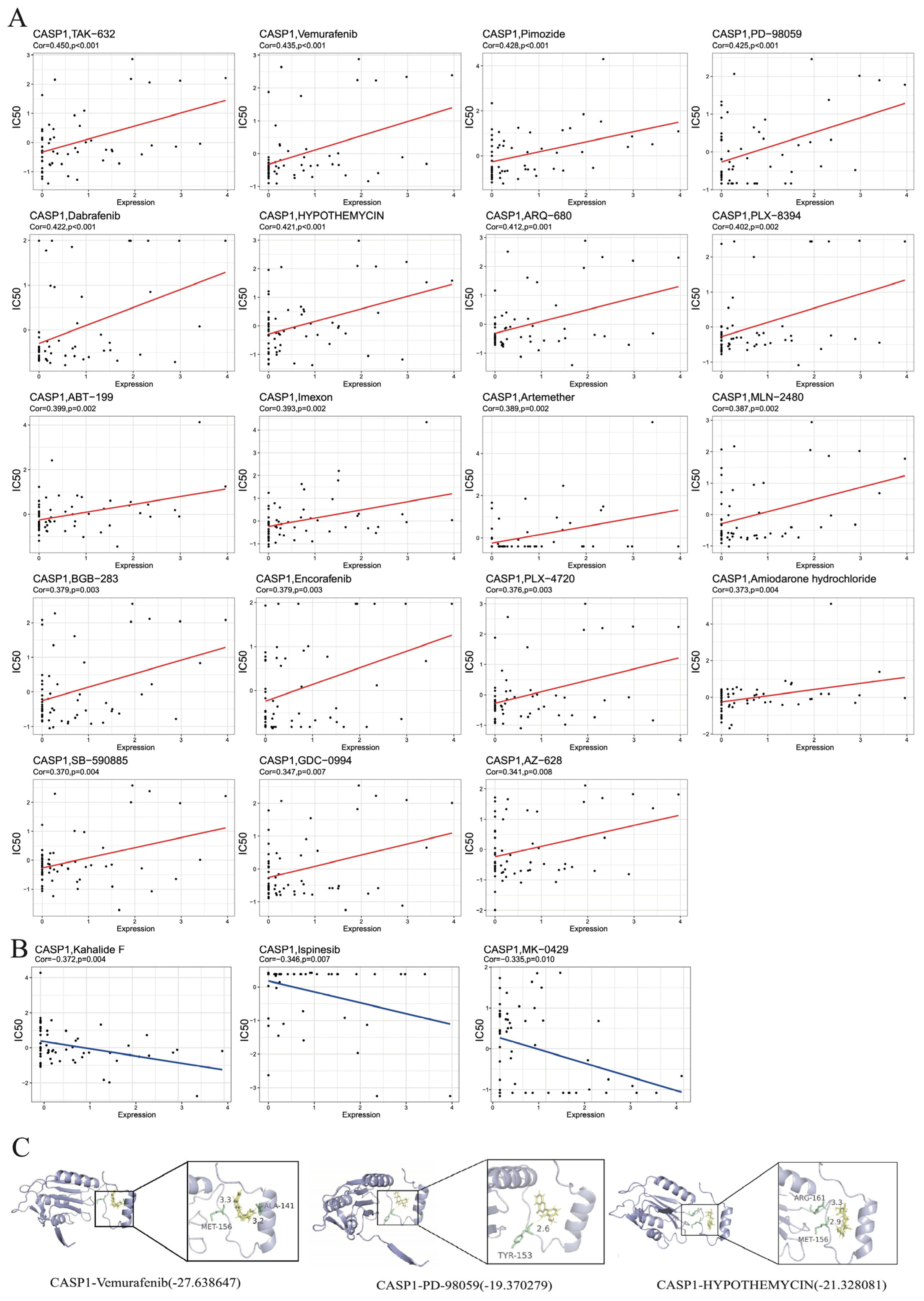
3.7. Drug Prediction and Molecular Docking Analysis
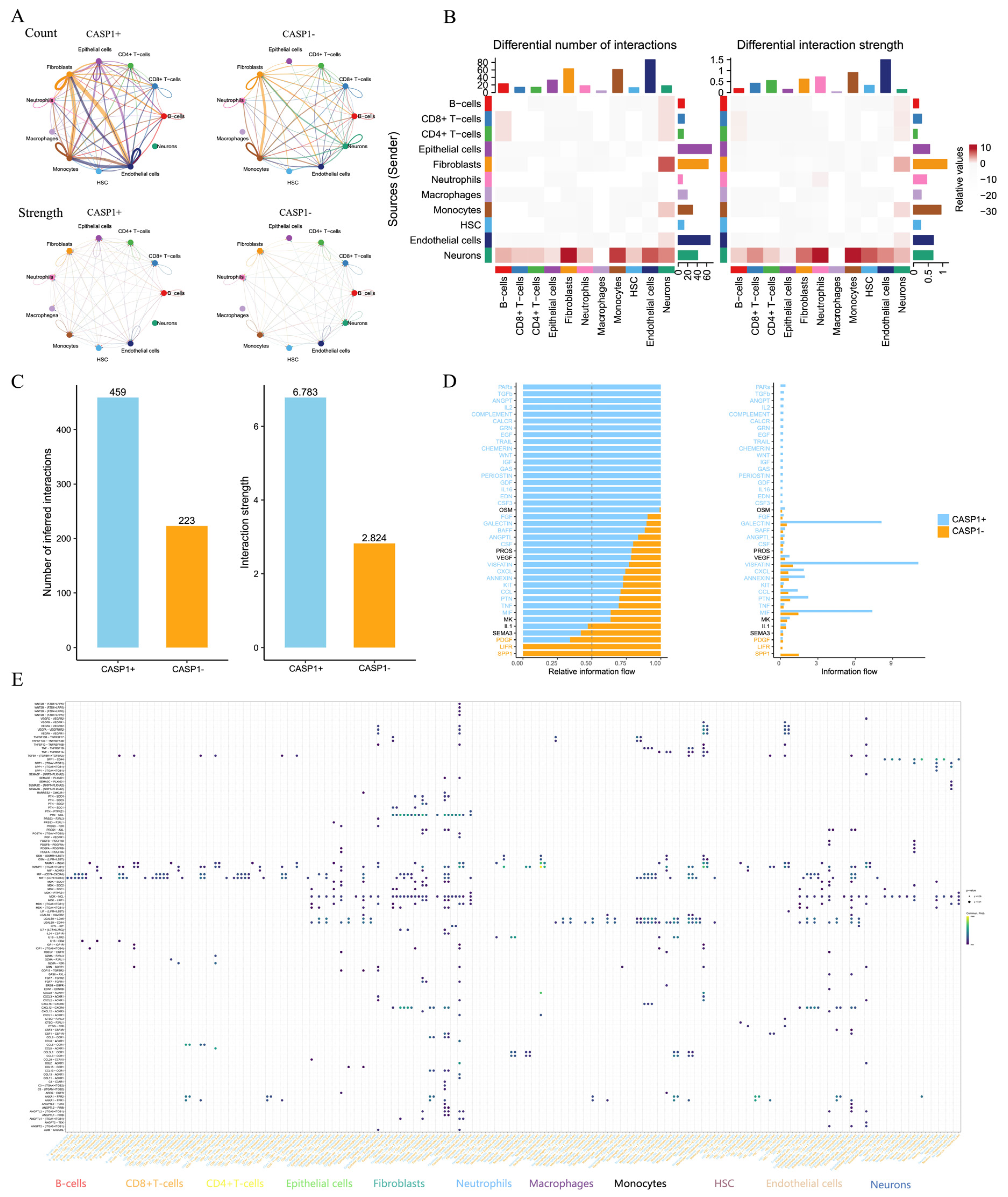
3.8. ScRNA-seq Analysis Revealed That CASP1 Regulates the Immune Microenvironment and Cell–Cell Communication in CD
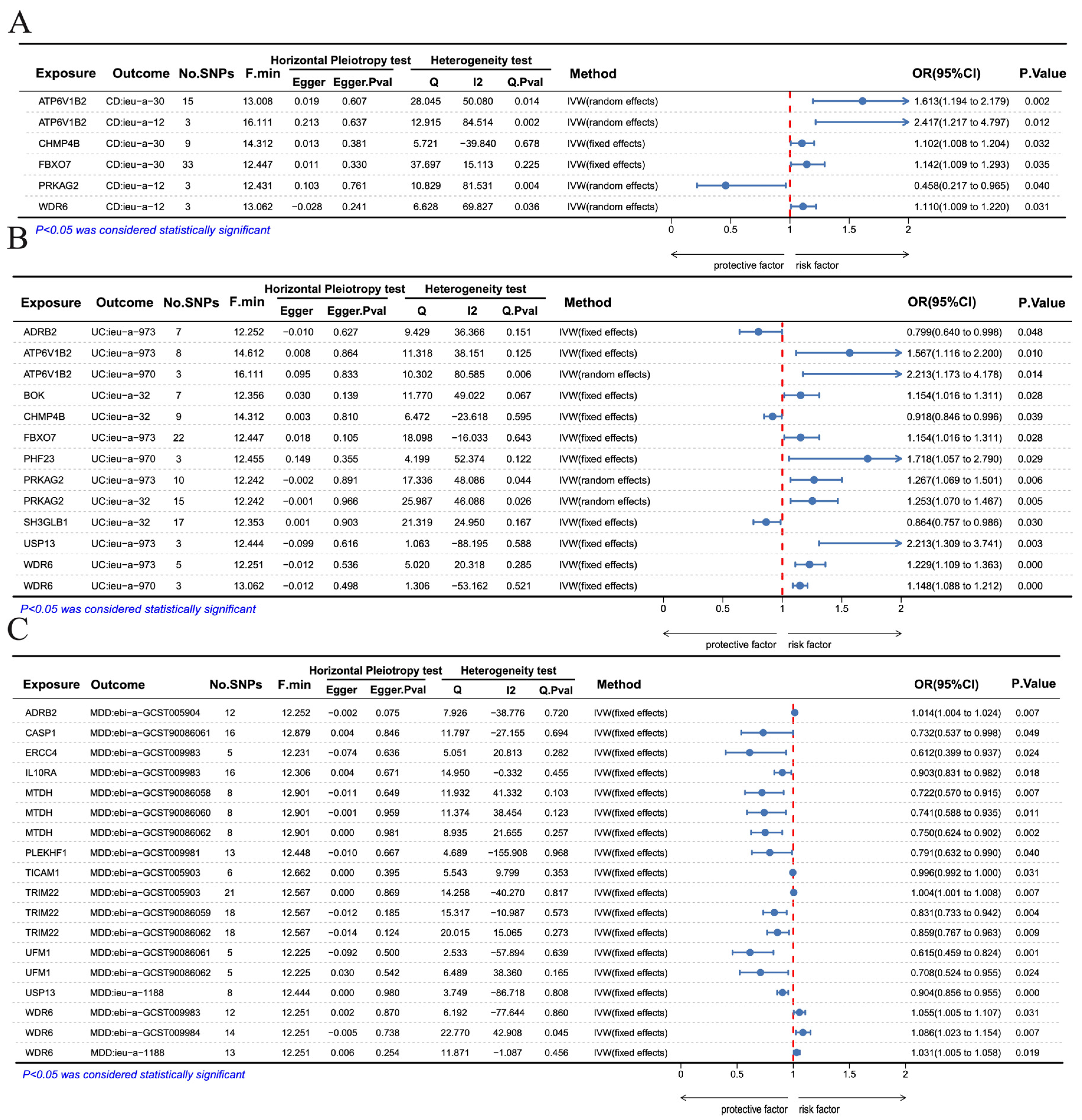
3.9. Mendelian Randomization Analysis
4. Discussion
5. Conclusions
6. Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MDD | Major Depressive Disorder |
| IBD | Inflammatory Bowel Disease |
| DE-ARGs | Differentially Expressed Autophagy-related Genes |
| Co-DEGs | Co-Differentially Expressed Genes |
| CD | Crohn’s disease |
| UC | Ulcerative colitis |
| GEO | Gene Expression Omnibus |
| PPI | Protein-protein Interaction |
| SVM | Support Vector Machine |
| RF | Random Forest |
| GLM | Generalized Linear Model |
| XGB | eXtreme Gradient Boosting |
| GSVA | Gene Set Variation Analysis |
| PCA | Principal Component Analysis |
| GWAS | Genome-wide Association Studies |
| IVs | Instrumental variables |
| IVW | Inverse-variance weighted |
| SSAG | Cross-disease shared susceptibility-associated gene |
References
- Dolinger, M.; Torres, J.; Vermeire, S. Crohn’s disease. Lancet 2024, 403, 1177–1191. [Google Scholar] [CrossRef] [PubMed]
- Buie, M.J.; Quan, J.; Windsor, J.W.; Coward, S.; Hansen, T.M.; King, J.A.; Kotze, P.G.; Gearry, R.B.; Ng, S.C.; Mak, J.W.; et al. Global Hospitalization Trends for Crohn’s Disease and Ulcerative Colitis in the 21st Century: A Systematic Review with Temporal Analyses. Clin. Gastroenterol. Hepatol. 2023, 21, 2211–2221. [Google Scholar] [CrossRef]
- Le Berre, C.; Honap, S.; Peyrin-Biroulet, L. Ulcerative colitis. Lancet 2023, 402, 571–584. [Google Scholar] [CrossRef]
- Rogler, G.; Singh, A.; Kavanaugh, A.; Rubin, D.T. Extraintestinal Manifestations of Inflammatory Bowel Disease: Current Concepts, Treatment, and Implications for Disease Management. Gastroenterology 2021, 161, 1118–1132. [Google Scholar] [CrossRef] [PubMed]
- Bisgaard, T.H.; Allin, K.H.; Keefer, L.; Ananthakrishnan, A.N.; Jess, T. Depression and anxiety in inflammatory bowel disease: Epidemiology, mechanisms and treatment. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 717–726. [Google Scholar] [CrossRef]
- Souza, P.B.; de Araujo Borba, L.; Castro de Jesus, L.; Valverde, A.P.; Gil-Mohapel, J.; Rodrigues, A.L.S. Major Depressive Disorder and Gut Microbiota: Role of Physical Exercise. Int. J. Mol. Sci. 2023, 24, 16870. [Google Scholar] [CrossRef]
- Hu, C.; Ge, M.; Liu, Y.; Tan, W.; Zhang, Y.; Zou, M.; Xiang, L.; Song, X.; Guo, H. From inflammation to depression: Key biomarkers for IBD-related major depressive disorder. J. Transl. Med. 2024, 22, 997. [Google Scholar] [CrossRef]
- Barberio, B.; Zamani, M.; Black, C.J.; Savarino, E.V.; Ford, A.C. Prevalence of symptoms of anxiety and depression in patients with inflammatory bowel disease: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2021, 6, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Caparrós, E.; Wiest, R.; Scharl, M.; Rogler, G.; Casbas, A.G.; Yilmaz, B.; Wawrzyniak, M.; Francés, R. Dysbiotic microbiota interactions in Crohn’s disease. Gut Microbes 2021, 13, 1949096. [Google Scholar] [CrossRef] [PubMed]
- Bethlehem, L.; Estevinho, M.M.; Grinspan, A.; Magro, F.; Faith, J.J.; Colombel, J.-F. Microbiota therapeutics for inflammatory bowel disease: The way forward. Lancet Gastroenterol. Hepatol. 2024, 9, 476–486. [Google Scholar] [CrossRef]
- Liu, S.; Yao, S.; Yang, H.; Liu, S.; Wang, Y. Autophagy: Regulator of cell death. Cell Death Dis. 2023, 14, 648. [Google Scholar] [CrossRef]
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef]
- Foerster, E.G.; Mukherjee, T.; Cabral-Fernandes, L.; Rocha, J.D.; Girardin, S.E.; Philpott, D.J. How autophagy controls the intestinal epithelial barrier. Autophagy 2022, 18, 86–103. [Google Scholar] [CrossRef] [PubMed]
- Caprilli, R.; Lapaquette, P.; Darfeuille-Michaud, A. Eating the enemy in Crohn’s disease: An old theory revisited. J. Crohns Colitis 2010, 4, 377–383. [Google Scholar] [CrossRef]
- Kong, J.; Xiang, Q.; Ge, W.; Wang, Y.; Xu, F.; Shi, G. Network pharmacology mechanisms and experimental verification of licorice in the treatment of ulcerative colitis. J. Ethnopharmacol. 2024, 324, 117691. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Liu, T.; Jiang, P.; Dang, R. The interaction between autophagy and neuroinflammation in major depressive disorder: From pathophysiology to therapeutic implications. Pharmacol. Res. 2021, 168, 105586. [Google Scholar] [CrossRef]
- Lei, L.; Chen, C.-Y.; Wang, Y.-F.; Zhang, Y. Identification of mitophagy-related genes and analysis of immune infiltration in the astrocytes based on machine learning in the pathogenesis of major depressive disorder. J. Affect. Disord. 2025, 368, 160–171. [Google Scholar] [CrossRef]
- Zou, Y.; Xie, J.; Zheng, S.; Liu, W.; Tang, Y.; Tian, W.; Deng, X.; Wu, L.; Zhang, Y.; Wong, C.-W.; et al. Leveraging diverse cell-death patterns to predict the prognosis and drug sensitivity of triple-negative breast cancer patients after surgery. Int. J. Surg. 2022, 107, 106936. [Google Scholar] [CrossRef]
- Noble, C.L.; Abbas, A.R.; Lees, C.W.; Cornelius, J.; Toy, K.; Modrusan, Z.; Clark, H.F.; Arnott, I.D.; Penman, I.D.; Satsangi, J.; et al. Characterization of intestinal gene expression profiles in Crohn’s disease by genome-wide microarray analysis. Inflamm. Bowel Dis. 2010, 16, 1717–1728. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Song, B.; Zhu, W.; Xu, X.; Gong, Q.Q.; Morando, C.; Dassopoulos, T.; Newberry, R.D.; Hunt, S.R.; Li, E. An ileal Crohn’s disease gene signature based on whole human genome expression profiles of disease unaffected ileal mucosal biopsies. PLoS ONE 2012, 7, e37139. [Google Scholar] [CrossRef]
- Keir, M.E.; Fuh, F.; Ichikawa, R.; Acres, M.; Hackney, J.A.; Hulme, G.; Carey, C.D.; Palmer, J.; Jones, C.J.; Long, A.K.; et al. Regulation and Role of αE Integrin and Gut Homing Integrins in Migration and Retention of Intestinal Lymphocytes during Inflammatory Bowel Disease. J. Immunol. 2021, 207, 2245–2254. [Google Scholar] [CrossRef] [PubMed]
- Bjerrum, J.T.; Hansen, M.; Olsen, J.; Nielsen, O.H. Genome-wide gene expression analysis of mucosal colonic biopsies and isolated colonocytes suggests a continuous inflammatory state in the lamina propria of patients with quiescent ulcerative colitis. Inflamm. Bowel Dis. 2010, 16, 999–1007. [Google Scholar] [CrossRef]
- Leday, G.G.R.; Vértes, P.E.; Richardson, S.; Greene, J.R.; Regan, T.; Khan, S.; Henderson, R.; Freeman, T.C.; Pariante, C.M.; Harrison, N.A.; et al. Replicable and Coupled Changes in Innate and Adaptive Immune Gene Expression in Two Case-Control Studies of Blood Microarrays in Major Depressive Disorder. Biol. Psychiatry 2018, 83, 70–80. [Google Scholar] [CrossRef]
- Spijker, S.; Van Zanten, J.S.; De Jong, S.; Penninx, B.W.; van Dyck, R.; Zitman, F.G.; Smit, J.H.; Ylstra, B.; Smit, A.B.; Hoogendijk, W.J. Stimulated gene expression profiles as a blood marker of major depressive disorder. Biol. Psychiatry 2010, 68, 179–186. [Google Scholar] [CrossRef]
- Chang, L.-C.; Jamain, S.; Lin, C.-W.; Rujescu, D.; Tseng, G.C.; Sibille, E. A conserved BDNF, glutamate- and GABA-enriched gene module related to human depression identified by coexpression meta-analysis and DNA variant genome-wide association studies. PLoS ONE 2014, 9, e90980. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef]
- He, S.; Ye, J.; Wang, Y.; Xie, L.Y.; Liu, S.Y.; Chen, Q.K. Identification and functional analysis of energy metabolism and pyroptosis-related genes in diabetic nephropathy. Heliyon 2025, 11, e42201. [Google Scholar] [CrossRef]
- Yang, X.; Yang, W.; Li, J.; Chen, C.; Chen, S.; Wang, H.; Wu, J.; Xue, H.; Liu, Y.; Lu, J.; et al. Major Facilitator Superfamily transporters balance sugar metabolism in peach. Plant Physiol. 2025, 198, kiaf192. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, J.; Wang, N.; Liu, C.; Wang, S.; Dong, X.; Yang, L.; Bao, X.; Nie, X.; Li, J. Bioinformatics-based identification of CTSS, DOK2, and ENTPD1 as potential blood biomarkers of schizophrenia. BMC Psychiatry 2025, 25, 157. [Google Scholar] [CrossRef]
- Wei, X.; Deng, W.; Dong, Z.; Xie, Z.; Zhang, J.; Wang, R.; Zhang, R.; Na, N.; Zhou, Y. Identification of Subtypes and a Delayed Graft Function Predictive Signature Based on Ferroptosis in Renal Ischemia-Reperfusion Injury. Front. Cell Dev. Biol. 2022, 10, 800650. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Zhou, J.; Wang, Z.; Liu, D.; Zhang, H.; Xie, S.; Wu, K. Epidemiology, pathogenesis, diagnosis, and treatment of inflammatory bowel disease: Insights from the past two years. Chin. Med. J. 2025, 138, 763–776. [Google Scholar] [CrossRef] [PubMed]
- Yao, P.; Han, H. Advances in Autophagy-Lysosomal Pathway and Neurodegeneration via Brain-Gut Axis. Biomedicines 2025, 13, 1390. [Google Scholar] [CrossRef] [PubMed]
- McGillis, L.; Bronte-Tinkew, D.M.; Dang, F.; Capurro, M.; Prashar, A.; Ricciuto, A.; Greenfield, L.; Lozano-Ruf, A.; Siddiqui, I.; Hsieh, A.; et al. Vitamin D deficiency enhances expression of autophagy-regulating miR-142-3p in mouse and “involved” IBD patient intestinal tissues. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 321, G171–G184. [Google Scholar] [CrossRef]
- Yang, L.; Guo, C.; Zheng, Z.; Dong, Y.; Xie, Q.; Lv, Z.; Li, M.; Lu, Y.; Guo, X.; Deng, R.; et al. Stress dynamically modulates neuronal autophagy to gate depression onset. Nature 2025, 641, 427–437. [Google Scholar] [CrossRef]
- Tang, J.; Chen, Q.; Xiang, L.; Tu, T.; Zhang, Y.; Ou, C. TRIM28 Fosters Microglia Ferroptosis via Autophagy Modulation to Enhance Neuropathic Pain and Neuroinflammation. Mol. Neurobiol. 2024, 61, 9459–9477. [Google Scholar] [CrossRef]
- Qi, Y.; Zhu, H.; Chen, Y.; Zhang, Y.; Jin, S.; Xu, X.; Ma, X.; Chen, L.; Zhao, M.; Zhu, H.; et al. 4-Hydroxydictyolactone alleviates cerebral ischemia injury by regulating neuroinflammation and autophagy via AMPK signaling pathway. Phytomedicine 2024, 135, 156157. [Google Scholar] [CrossRef]
- Guo, J.; Li, Y.; Ma, K.; Su, G. Enhancing autophagy mitigates LPS-induced neuroinflammation by inhibiting microglial M1 polarization and neuronophagocytosis. Front. Cell Neurosci. 2025, 19, 1546848. [Google Scholar] [CrossRef]
- Lee, H.M.; Kim, J.; Kim, J.Y.; An, M.J.; Shin, G.S.; Jo, A.R.; Park, Y.; Kim, C.H.; Hwangbo, Y.; Lee, J.H.; et al. Retained introns in phototransduction genes of 5xFAD mouse retina suggest vision impairment as an early diagnostic marker for Alzheimer’s disease. Sci. Rep. 2025, 15, 25523. [Google Scholar] [CrossRef]
- Gui, J.; Chu, H.; Zhang, J.; Li, X.; Liu, W.; Li, R.; Zhang, F.; Dong, M.; Gao, K.; Luo, H.; et al. Proteome-Wide and Immune Cell Phenotype Mendelian Randomization Highlights Immune Involvement in Genetic Generalized Epilepsy. Brain Behav. 2025, 15, e70625. [Google Scholar] [CrossRef] [PubMed]
- Bespalov, D.; Pino, D.; Vidal-Guirao, S.; Franquesa, J.; Lopez-Ramajo, D.; Filgaira, I.; Wan, L.; O’Sullivan, P.A.; Ley, S.C.; Forcales, S.V.; et al. Bioinformatic analysis of molecular characteristics and oncogenic features of CARD14 in human cancer. Sci. Rep. 2024, 14, 22972. [Google Scholar] [CrossRef]
- Wen, Y.; He, P.; Huang, Z.; Ding, C.; Zhang, T.; Zhang, L.; Zheng, J.; Chen, M.; Chen, C.; Liu, Y.; et al. The Epigenetic Reader PHF23 Is Required for Embryonic Neurogenesis. J. Neurosci. 2025, 45, e2090242025. [Google Scholar] [CrossRef]
- Sarah, C.; Kieran, G.; Brenda, A.; Andre, T.L.; Elaine, K.; Paul, C.; Derek, W.M.; Daniel, G.B.; O’Farrelly, C. Avian resistance to Campylobacter jejuni colonization is associated with an intestinal immunogene expression signature identified by mRNA sequencing. PLoS ONE 2012, 7, e40409. [Google Scholar]
- Chen, Y.; Qiu, X.; Liu, R. Comprehensive characterization of immunogenic cell death in acute myeloid leukemia revealing the association with prognosis and tumor immune microenvironment. BMC Med Genomics 2024, 17, 107. [Google Scholar] [CrossRef] [PubMed]
- Suzaan, M.; Rachel, P.J.L.; Katalin, A.W.; Graeme, M.; O’Garra, A.; Robert, J.W. Inflammasome Activation Underlying Central Nervous System Deterioration in HIV-Associated Tuberculosis. J. Infect. Dis. 2017, 215, 677–686. [Google Scholar]
- Ling, W.; Chengfu, Z.; Jianfeng, W.; Shikun, C.; Zedan, T.; Quan, D. Corticosterone Inhibits LPS-Induced NLRP3 Inflammasome Priming in Macrophages by Suppressing Xanthine Oxidase. Mediators Inflamm. 2020, 2020, 6959741. [Google Scholar]
- Zhang, Z.; Zhu, Q. WD Repeat and HMG Box DNA Binding Protein 1, An Oncoprotein at the Hub of Tumorigenesis and a Novel Therapeutic Target. Int. J. Mol. Sci. 2023, 24, 12494. [Google Scholar] [CrossRef]
- Gao, B.; Zhang, D.; Li, X.; Yang, H.; Wood, A.J. De novo assembly and characterization of the transcriptome in the desiccation-tolerant moss Syntrichia caninervis. BMC Res. Notes 2014, 7, 490. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, Y.; Fan, H.; Yuan, R.; Cai, J.; Zhong, B.; Qin, Q.; Zhang, Z.; Zhang, Y.; Cheng, S. Investigating the shared genetic architecture between anxiety and stroke. Behav. Brain Res. 2025, 480, 115400. [Google Scholar] [CrossRef]
| CD | UC | |||||
|---|---|---|---|---|---|---|
| Cell Type | Normal Samples | Disease Samples | p | Normal Samples | Disease Samples | p |
| B.cells.naive | 0.0495 ± 0.0524 | 0.0252 ± 0.0431 | 0.0850 | 0.0303 ± 0.0429 | 0.0220 ± 0.0293 | 0.4852 |
| B.cells.memory | 0.0874 ± 0.0963 | 0.0393 ± 0.0545 | 0.0604 | 0.0877 ± 0.0815 | 0.0533 ± 0.0466 | 0.1366 |
| Plasma.cells | 0.0009 ± 0.0037 | 0.0053 ± 0.0119 | 0.0036 | 0.0010 ± 0.0030 | 0.0059 ± 0.0168 | 0.0426 |
| T.cells.CD8 | 0.0205 ± 0.0266 | 0.0203 ± 0.0300 | 0.9877 | 0.0613 ± 0.0506 | 0.0508 ± 0.0473 | 0.4777 |
| T.cells.CD4.naive | 0.0000 ± 0.0000 | 0.0005 ± 0.0028 | 0.1050 | 0.0000 ± 0.0000 | 0.0000 ± 0.0000 | NA |
| T.cells.CD4.memory.resting | 0.1289 ± 0.0545 | 0.0873 ± 0.0571 | 0.0085 | 0.1177 ± 0.0735 | 0.0926 ± 0.0560 | 0.2321 |
| T.cells.CD4.memory.activated | 0.0847 ± 0.0433 | 0.1374 ± 0.0787 | <0.001 | 0.1082 ± 0.0683 | 0.1217 ± 0.0697 | 0.5055 |
| T.cells.follicular.helper | 0.0015 ± 0.0063 | 0.0019 ± 0.0077 | 0.8196 | 0.0021 ± 0.0081 | 0.0036 ± 0.0103 | 0.5556 |
| T.cells.regulatory.Tregs. | 0.0255 ± 0.0309 | 0.0255 ± 0.0305 | 0.9935 | 0.0403 ± 0.0308 | 0.0299 ± 0.0275 | 0.2485 |
| T.cells.gamma.delta | 0.0365 ± 0.0327 | 0.0162 ± 0.0253 | 0.0248 | 0.0263 ± 0.0225 | 0.0220 ± 0.0293 | 0.5412 |
| NK.cells.resting | 0.0056 ± 0.0134 | 0.0215 ± 0.0277 | <0.0010 | 0.0022 ± 0.0087 | 0.0198 ± 0.0235 | <0.0010 |
| NK.cells.activated | 0.0352 ± 0.0323 | 0.0297 ± 0.0334 | 0.5263 | 0.0321 ± 0.0223 | 0.0225 ± 0.0394 | 0.2227 |
| Monocytes | 0.0000 ± 0.0000 | 0.0046 ± 0.0101 | <0.0010 | 0.0015 ± 0.0059 | 0.0023 ± 0.0072 | 0.6884 |
| Macrophages.M0 | 0.1455 ± 0.0519 | 0.1420 ± 0.0754 | 0.8155 | 0.1212 ± 0.0384 | 0.1425 ± 0.0722 | 0.1275 |
| Macrophages.M1 | 0.0996 ± 0.0401 | 0.1003 ± 0.0368 | 0.9527 | 0.1019 ± 0.0431 | 0.0883 ± 0.0345 | 0.2726 |
| Macrophages.M2 | 0.0901 ± 0.0412 | 0.0634 ± 0.0440 | 0.0231 | 0.0835 ± 0.0364 | 0.0798 ± 0.0485 | 0.7498 |
| Dendritic.cells.resting | 0.0210 ± 0.0265 | 0.0210 ± 0.0313 | 0.9997 | 0.0208 ± 0.0139 | 0.0146 ± 0.0226 | 0.1910 |
| Dendritic.cells.activated | 0.0049 ± 0.0107 | 0.0058 ± 0.0125 | 0.7626 | 0.0075 ± 0.0216 | 0.0100 ± 0.0155 | 0.6853 |
| Mast.cells.resting | 0.0813 ± 0.0527 | 0.1056 ± 0.1105 | 0.1590 | 0.0665 ± 0.0575 | 0.0426 ± 0.0580 | 0.1658 |
| Mast.cells.activated | 0.0385 ± 0.0342 | 0.0604 ± 0.0771 | 0.0600 | 0.0366 ± 0.0344 | 0.0965 ± 0.0799 | <0.0010 |
| Eosinophils | 0.0000 ± 0.0000 | 0.0028 ± 0.0136 | 0.0426 | 0.0019 ± 0.0063 | 0.0050 ± 0.0131 | 0.1970 |
| Neutrophils | 0.0430 ± 0.0242 | 0.0839 ± 0.0541 | <0.0010 | 0.0493 ± 0.0269 | 0.0744 ± 0.0474 | 0.0103 |
| Blood | Prefrontal Cortex | Anterior Cingulate Cortex | Anterior Amygdala | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell Type | Normal Samples | Disease Samples | p | Normal Samples | Disease Samples | p | Normal Samples | Disease Samples | p | Normal Samples | Disease Samples | p |
| B.cells.naive | 0.0153 ± 0.0159 | 0.0168 ± 0.0173 | 0.4336 | 0.0172 ± 0.0349 | 0.0218 ± 0.0323 | 0.7110 | 0.0184 ± 0.0324 | 0.0218 ± 0.0336 | 0.7930 | 0.0096 ± 0.0229 | 0.0117 ± 0.0238 | 0.7714 |
| B.cells.memory | 0.0181 ± 0.0168 | 0.0153 ± 0.0167 | 0.1426 | 0.0784 ± 0.0641 | 0.0679 ± 0.0838 | 0.7030 | 0.1112 ± 0.0958 | 0.1014 ± 0.1043 | 0.8060 | 0.0470 ± 0.0553 | 0.0539 ± 0.0750 | 0.7377 |
| Plasma.cells | 0.0008 ± 0.0024 | 0.0010 ± 0.0032 | 0.6447 | 0.0445 ± 0.0274 | 0.0452 ± 0.0188 | 0.9350 | 0.0454 ± 0.0324 | 0.0493 ± 0.0331 | 0.7660 | 0.1305 ± 0.0900 | 0.0819 ± 0.0789 | 0.0704 |
| T.cells.CD8 | 0.2383 ± 0.0760 | 0.2368 ± 0.0917 | 0.8716 | 0.2689 ± 0.0542 | 0.2401 ± 0.1016 | 0.3440 | 0.2857 ± 0.0598 | 0.2790 ± 0.0836 | 0.8150 | 0.0953 ± 0.0717 | 0.0737 ± 0.0906 | 0.3972 |
| T.cells.CD4.naive | 0.0386 ± 0.0397 | 0.0377 ± 0.0354 | 0.8333 | 0.0541 ± 0.0515 | 0.0712 ± 0.0503 | 0.3640 | 0.0499 ± 0.0426 | 0.0439 ± 0.0314 | 0.6880 | 0.0096 ± 0.0283 | 0.0033 ± 0.0144 | 0.3653 |
| T.cells.CD4.memory.resting | 0.0120 ± 0.0316 | 0.0087 ± 0.0269 | 0.3254 | 0.0005 ± 0.0018 | 0.0130 ± 0.0446 | 0.2940 | 0.0043 ± 0.0093 | 0.0152 ± 0.0353 | 0.3020 | 0.1900 ± 0.1107 | 0.2308 ± 0.1135 | 0.2448 |
| T.cells.CD4.memory.activated | 0.0445 ± 0.0379 | 0.0468 ± 0.0377 | 0.5918 | 0.0000 ± 0.0000 | 0.0000 ± 0.0000 | 0.0000 | 0.0000 ± 0.0000 | 0.0000 ± 0.0000 | 0.0000 | 0.0000 ± 0.0000 | 0.0014 ± 0.0045 | 0.1791 |
| T.cells.follicular.helper | 0.0000 ± 0.0000 | 0.0000 ± 0.0002 | 0.3003 | 0.0414 ± 0.0505 | 0.0359 ± 0.0335 | 0.7300 | 0.0337 ± 0.0374 | 0.0505 ± 0.0438 | 0.3050 | 0.0494 ± 0.0432 | 0.0540 ± 0.0557 | 0.7670 |
| T.cells.regulatory..Tregs. | 0.0088 ± 0.0135 | 0.0085 ± 0.0137 | 0.8165 | 0.1014 ± 0.0504 | 0.0881 ± 0.0326 | 0.3980 | 0.0929 ± 0.0441 | 0.0992 ± 0.0498 | 0.7360 | 0.0233 ± 0.0470 | 0.0145 ± 0.0261 | 0.4562 |
| T.cells.gamma.delta | 0.0003 ± 0.0025 | 0.0005± 0.0050 | 0.5833 | 0.0000 ± 0.0000 | 0.0000 ± 0.0000 | 0.0000 | 0.0000 ± 0.0000 | 0.0000 ± 0.0000 | 0.0000 | 0.0023 ± 0.0077 | 0.0019 ± 0.0087 | 0.8670 |
| NK.cells.resting | 0.0670 ± 0.0315 | 0.0706 ± 0.031 | 0.3097 | 0.0353 ± 0.0461 | 0.0481 ± 0.0572 | 0.5050 | 0.0374 ± 0.0392 | 0.0316 ± 0.0434 | 0.7250 | 0.0378 ± 0.0457 | 0.0420 ± 0.0435 | 0.7630 |
| NK.cells.activated | 0.0290 ± 0.0314 | 0.0233 ± 0.0271 | 0.0934 | 0.0690 ± 0.0580 | 0.0722 ± 0.0802 | 0.9040 | 0.0497 ± 0.0596 | 0.0584 ± 0.0805 | 0.7580 | 0.0346 ± 0.0487 | 0.0522 ± 0.0636 | 0.3226 |
| Monocytes | 0.1650 ± 0.0590 | 0.1575 ± 0.0588 | 0.2598 | 0.0193 ± 0.0234 | 0.0206 ± 0.0274 | 0.8950 | 0.0182 ± 0.0223 | 0.0179 ± 0.0186 | 0.9700 | 0.0285 ± 0.0354 | 0.0532 ± 0.0499 | 0.0724 |
| Macrophages.M0 | 0.0222 ± 0.0211 | 0.0315 ± 0.0254 | <0.0010 | 0.0265 ± 0.0354 | 0.0340 ± 0.0346 | 0.5590 | 0.0137 ± 0.0228 | 0.0350 ± 0.0394 | 0.1090 | 0.0131 ± 0.0277 | 0.0324 ± 0.0483 | 0.1208 |
| Macrophages.M1 | 0.0041 ± 0.0075 | 0.0041 ± 0.0092 | 0.9813 | 0.0417 ± 0.0325 | 0.0391 ± 0.0328 | 0.8280 | 0.0421 ± 0.0286 | 0.0238 ± 0.0279 | 0.1100 | 0.0074 ± 0.0153 | 0.0090 ± 0.0185 | 0.7628 |
| Macrophages.M2 | 0.0066 ± 0.0098 | 0.0070 ± 0.0098 | 0.7026 | 0.1208 ± 0.0441 | 0.1072 ± 0.0374 | 0.3710 | 0.1247 ± 0.0360 | 0.1052 ± 0.0368 | 0.1850 | 0.2059 ± 0.0974 | 0.1466 ± 0.1161 | 0.0807 |
| Dendritic.cells.resting | 0.0001± 0.0006 | 0.0000 ± 0.0002 | 0.0950 | 0.0288 ± 0.0241 | 0.0202 ± 0.0204 | 0.3060 | 0.0186 ± 0.0172 | 0.0108 ± 0.0192 | 0.2860 | 0.0000 ± 0.0000 | 0.0005 ± 0.0023 | 0.3293 |
| Dendritic.cells.activated | 0.0033 ± 0.0072 | 0.0027 ± 0.0058 | 0.4366 | 0.0002 ± 0.0008 | 0.0001 ± 0.0003 | 0.5230 | 0.0000 ± 0.0000 | 0.0000 ± 0.0000 | 0.0000 | 0.0120 ± 0.0215 | 0.0095 ± 0.0154 | 0.6694 |
| Mast.cells.resting | 0.0115 ± 0.0138 | 0.0093 ± 0.0128 | 0.1598 | 0.0277 ± 0.0298 | 0.0435 ± 0.0379 | 0.2150 | 0.0224 ± 0.0321 | 0.0369 ± 0.0426 | 0.3400 | 0.0667 ± 0.0822 | 0.0980 ± 0.0871 | 0.2377 |
| Mast.cells.activated | 0.0354 ± 0.0549 | 0.0247 ± 0.0436 | 0.0641 | 0.0011 ± 0.0029 | 0.0025 ± 0.0096 | 0.6050 | 0.0063 ± 0.0180 | 0.0006 ± 0.0022 | 0.2780 | 0.0091 ± 0.0269 | 0.0120 ± 0.0336 | 0.7558 |
| Eosinophils | 0.0077 ± 0.0130 | 0.0104 ± 0.0152 | 0.0945 | 0.0110 ± 0.0217 | 0.0105 ± 0.0202 | 0.9410 | 0.0090 ± 0.0145 | 0.0033 ± 0.0082 | 0.2270 | 0.0124 ± 0.0213 | 0.0075 ± 0.0144 | 0.3856 |
| Neutrophils | 0.2714 ± 0.0970 | 0.2868 ± 0.1079 | 0.1806 | 0.0124 ± 0.0109 | 0.0190 ± 0.0287 | 0.4160 | 0.0162 ± 0.0126 | 0.0164 ± 0.0192 | 0.9790 | 0.0154 ± 0.0174 | 0.0100 ± 0.0215 | 0.3799 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Wang, G.; Wu, L.; Shi, J.; Sang, M.; Mao, L. Machine Learning Reveals Common Regulatory Mechanisms Mediated by Autophagy-Related Genes in the Development of Inflammatory Bowel Disease and Major Depressive Disorder. Genes 2026, 17, 4. https://doi.org/10.3390/genes17010004
Wang G, Wu L, Shi J, Sang M, Mao L. Machine Learning Reveals Common Regulatory Mechanisms Mediated by Autophagy-Related Genes in the Development of Inflammatory Bowel Disease and Major Depressive Disorder. Genes. 2026; 17(1):4. https://doi.org/10.3390/genes17010004
Chicago/Turabian StyleWang, Gengxian, Luojin Wu, Jiyuan Shi, Mengmeng Sang, and Liming Mao. 2026. "Machine Learning Reveals Common Regulatory Mechanisms Mediated by Autophagy-Related Genes in the Development of Inflammatory Bowel Disease and Major Depressive Disorder" Genes 17, no. 1: 4. https://doi.org/10.3390/genes17010004
APA StyleWang, G., Wu, L., Shi, J., Sang, M., & Mao, L. (2026). Machine Learning Reveals Common Regulatory Mechanisms Mediated by Autophagy-Related Genes in the Development of Inflammatory Bowel Disease and Major Depressive Disorder. Genes, 17(1), 4. https://doi.org/10.3390/genes17010004






