Fine Mapping of a Major Locus for Leaf Sheath Hairiness in Wheat Identifies TaSAIN1-4D as a Candidate Gene
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Field Trial and Growth Conditions
2.3. Bulk Construction and Targeted Genotyping by Sequencing
2.4. Data Processing and Variant Calling
2.5. BSA-Seq Analysis
2.6. KASP Marker Development and Fine Mapping
2.7. Molecular Cloning and Sequence Analysis of Candidate Genes
3. Results
3.1. Phenotypic Evaluation
3.2. Characterization of Targeted Capture Sequencing Data
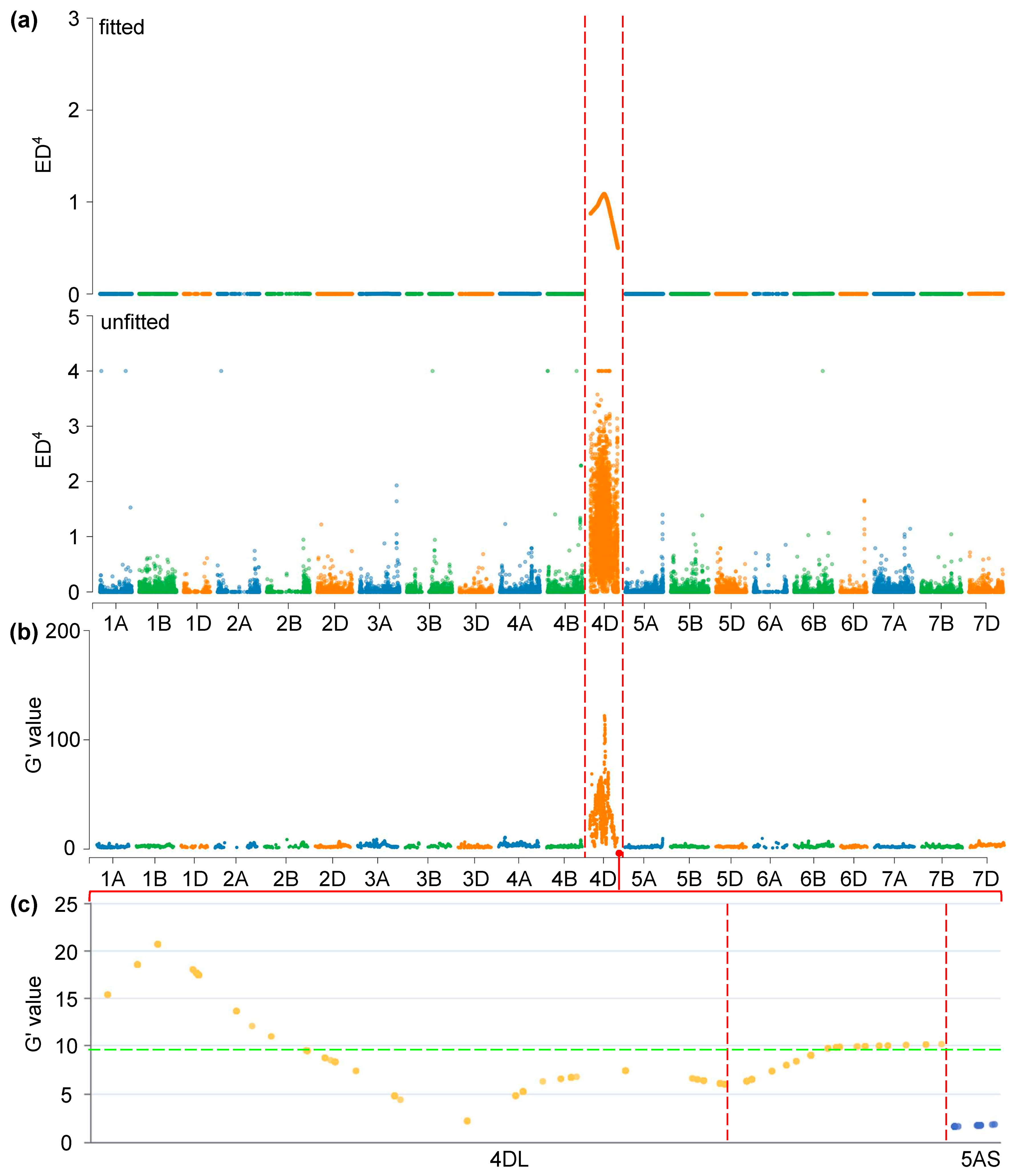
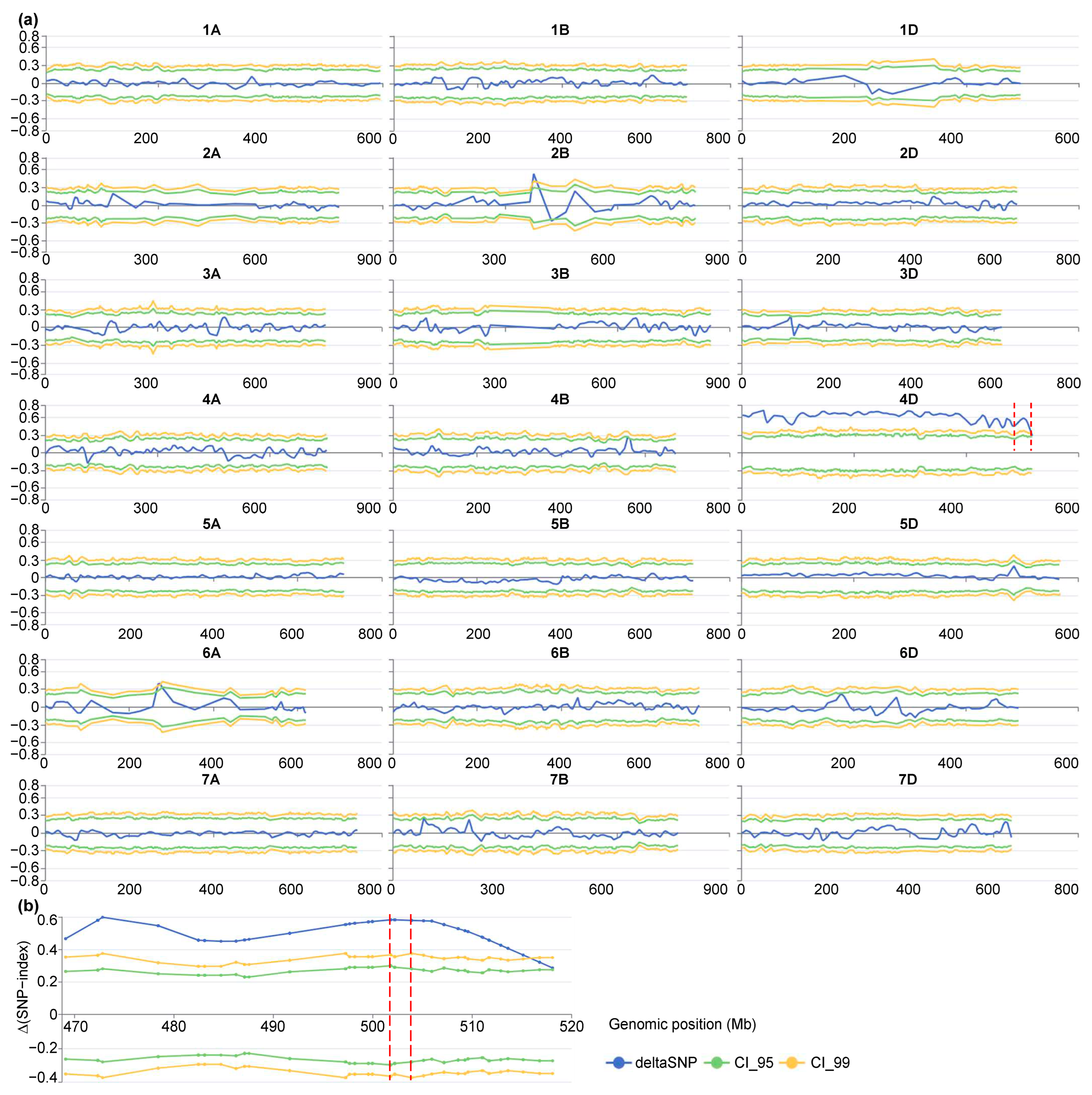
3.3. BSA-Seq Analysis and QTL Identification
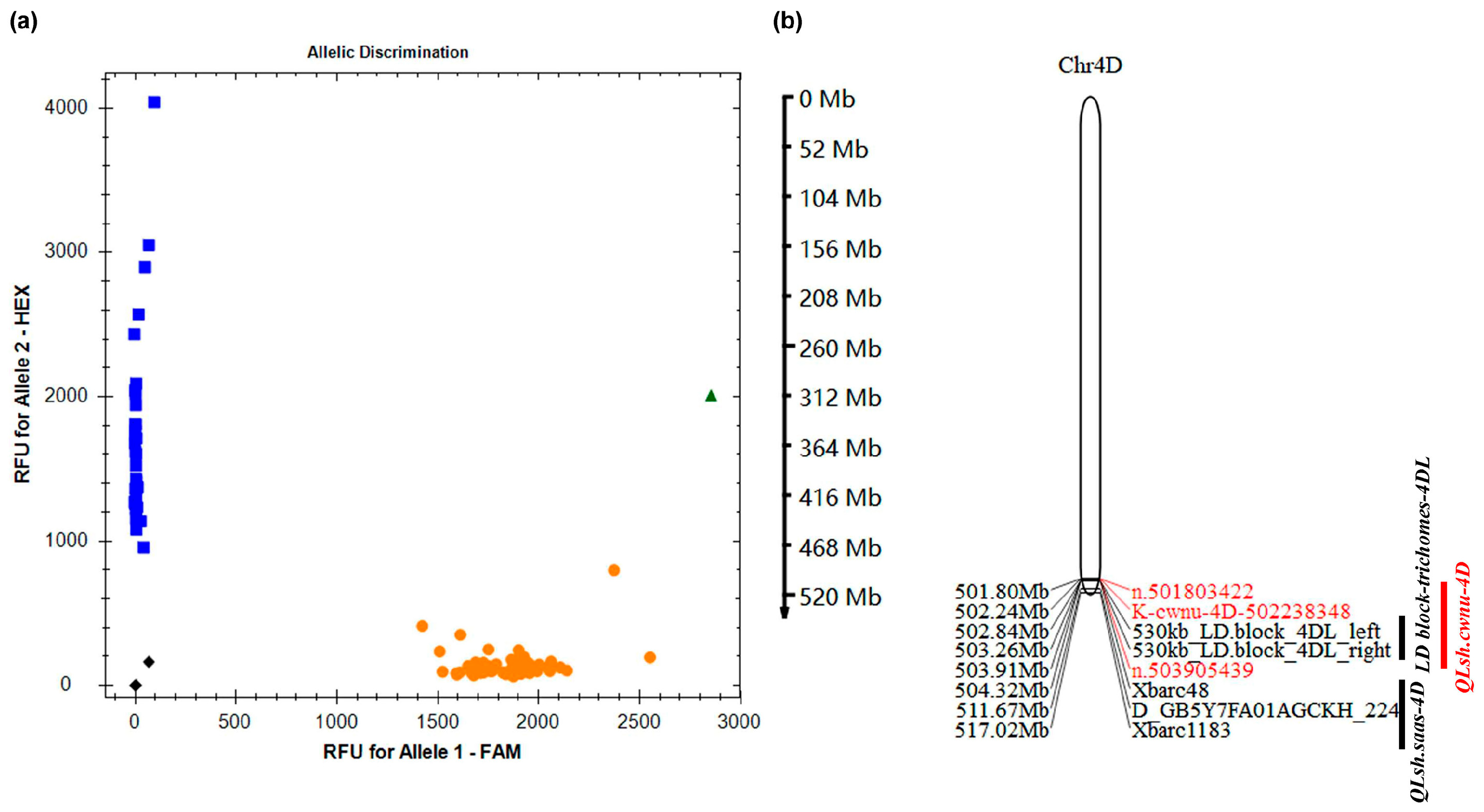
3.4. KASP Marker-Based Fine Mapping of LSH Traits
3.5. Gene Prediction Within QLsh.cwnu-4D
3.6. Structural Variation and Amplification Analysis Within QLsh.cwnu-4D
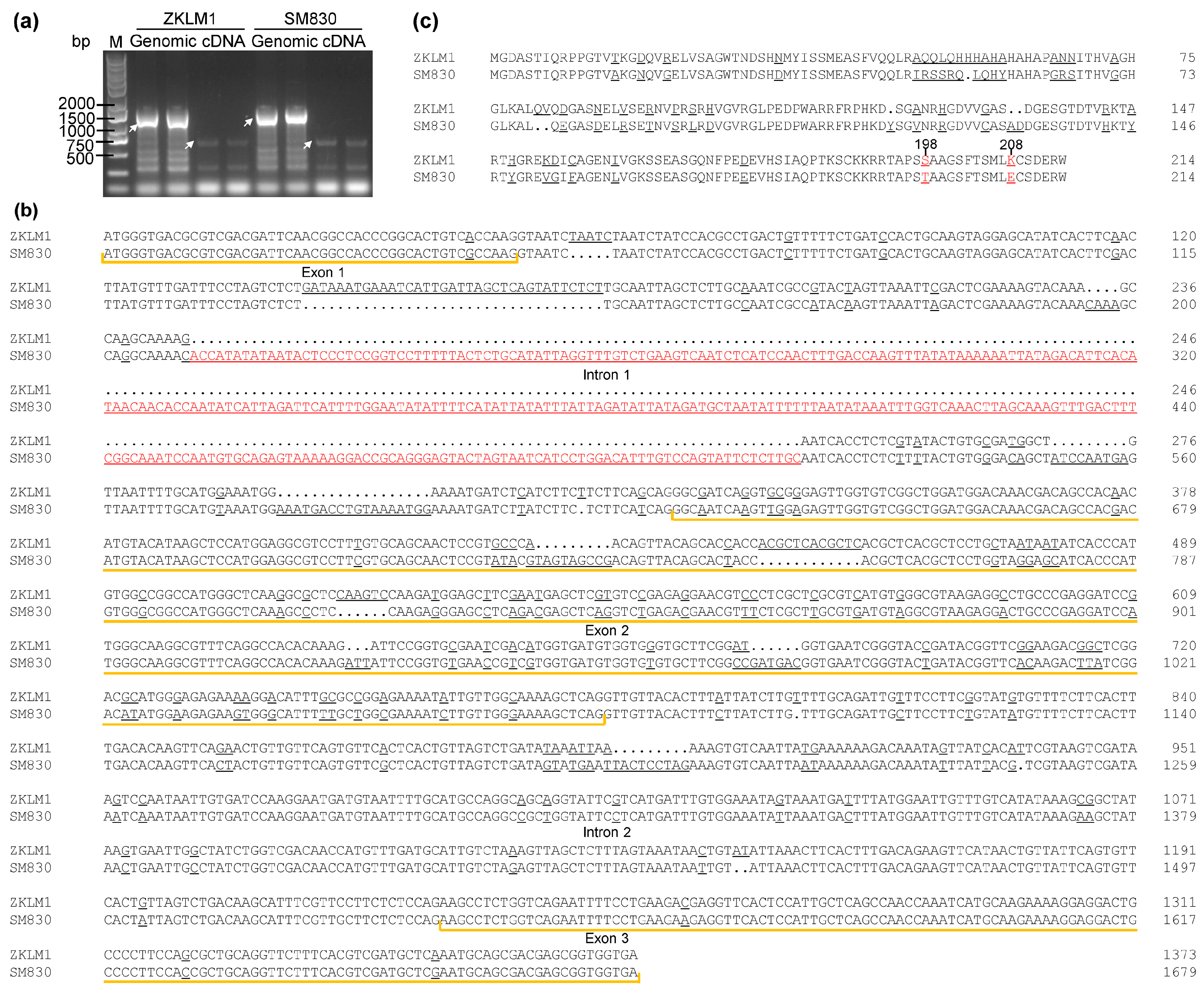
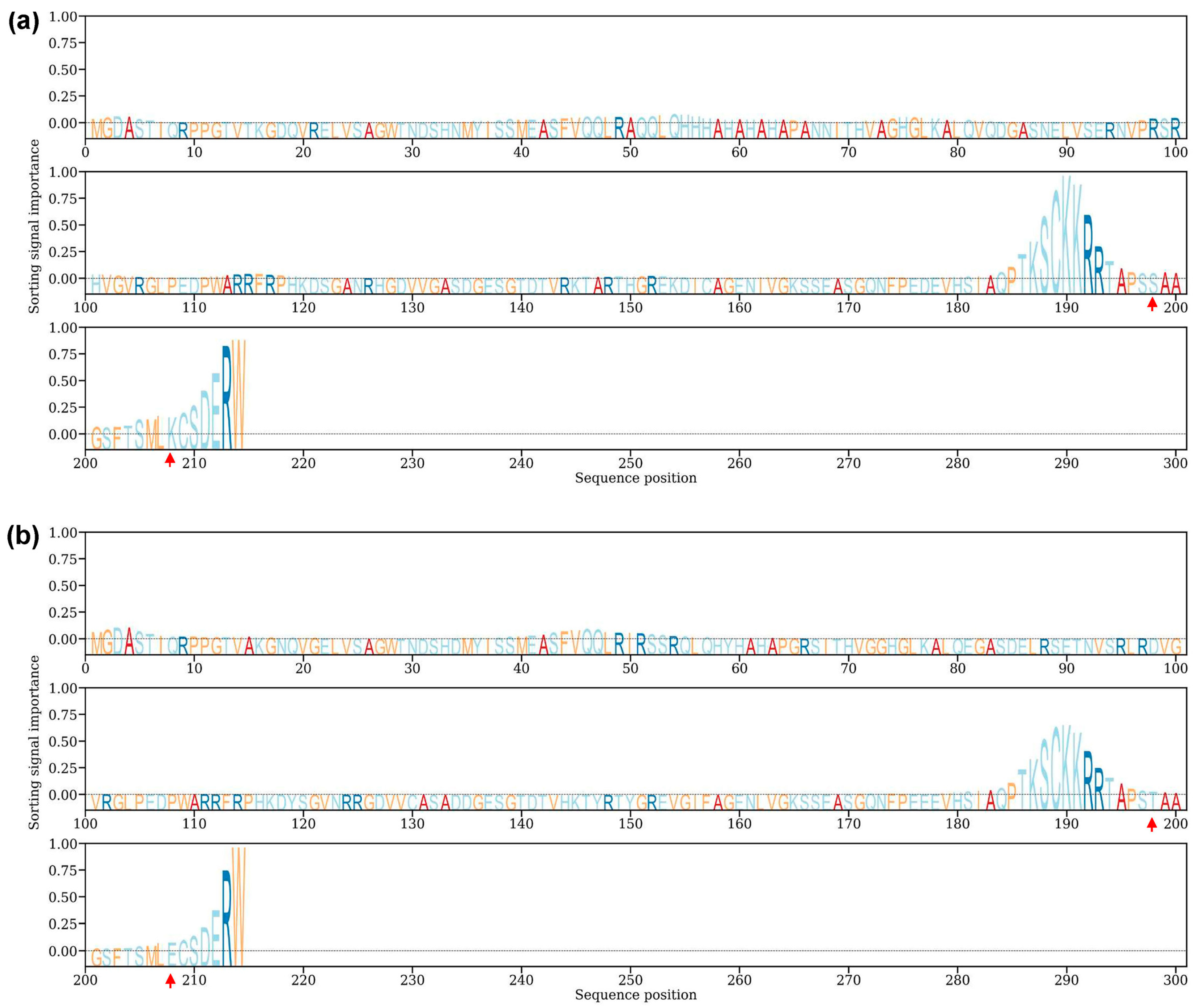
3.7. Identification and Sequence Analysis of TaSAIN1-4D
3.8. Predicted Subcellular Localization of TaSAIN1-4D
3.9. Comparative Analysis of TaSAIN1-4D and Known Trichome-Related Genes/QTLs
4. Discussion
4.1. Genetic Basis of Leaf Sheath Hairiness in Wheat
4.2. Comparative Genomics
4.3. Relationship to Stress Tolerance
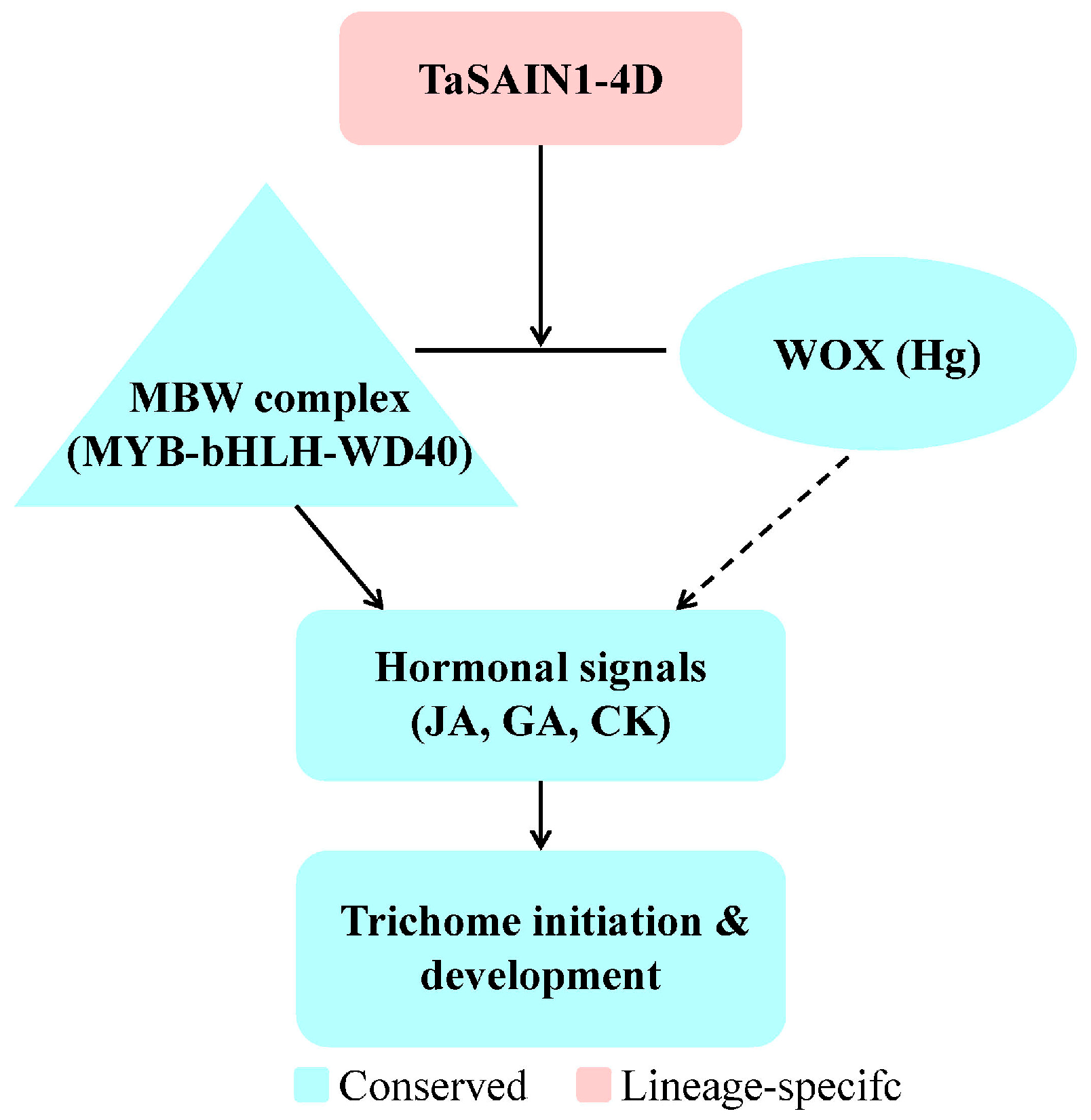
4.4. Proposed Mechanism
4.5. Implications for Wheat Breeding
4.6. Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BSA-seq | Bulk segregant analysis sequencing |
| QTL | Quantitative trait locus |
| GWAS | Genome wide association study |
| ED | Euclidean distance |
| KASP | Kompetitive Allele Specific PCR |
| MAS | Marker-assisted selection |
| SNP | Single nucleotide polymorphism |
| InDel | Insertion and Deletion |
| CDS | coding sequence |
| cDNA | complementary DNA |
| LD block | linkage disequilibrium block |
| RILs | Recombinant inbred lines |
References
- Vavilov, N.I. Scientific Foundations of Wheat Breeding; Selkhozgiz Publishers: Moscow, Russia, 1935; pp. 70–87. (In Russian) [Google Scholar]
- Kaur, J.; Kariyat, R. Role of trichomes in plant stress biology. In Evolutionary Ecology of Plant-Herbivore Interaction; Núñez-Farfán, J., Valverde, P., Eds.; Springer: Berlin/Heidelberg, Germany, 2020; pp. 15–35. [Google Scholar] [CrossRef]
- Duffey, S.S. Plant glandular trichomes: Their role in partial defense against insects. In Insects and the Plant Surface; Southwood, S.R., Juniper, B.E., Eds.; Edward Arnold Publishers: London, UK, 1986; pp. 151–172. [Google Scholar]
- Pshenichnikova, T.A.; Doroshkov, A.V.; Osipova, S.V.; Permyakov, A.V.; Permyakova, M.D.; Efimov, V.M.; Afonnikov, D.A. Quantitative characteristics of pubescence in wheat (Triticum aestivum L.) are associated with photosynthetic parameters under conditions of normal and limited water supply. Planta 2019, 249, 839–847. [Google Scholar] [CrossRef]
- Osipova, S.V.; Rudikvskii, A.V.; Permyakov, A.V.; Rudikovskaya, E.G.; Permyakova, M.D.; Verkhoturov, V.V.; Pshenichnikova, T.A. Physiological responses to water deficiency in bread wheat (Triticum aestivum L.) lines with genetically different leaf pubescence. Vavilov J. Genet. Breed. 2020, 24, 813–820. [Google Scholar] [CrossRef]
- Maistrenko, O.I. Identification and localization of genes controlling the pubescence of the leaf of young soft wheat plants. Genetika 1976, 12, 5–15. (In Russian) [Google Scholar]
- Dobrovolskaya, O.B.; Pshenichnikova, T.A.; Arbuzova, V.S.; Lohwasser, U.; Röder, M.S.; Börner, A. Molecular mapping of genes determining hairy leaf character in common wheat with respect to other species of the Triticeae. Euphytica 2007, 155, 285–293. [Google Scholar] [CrossRef]
- Wan, H.; Yang, Y.; Li, J.; Zhang, Z.; Yang, W. Mapping a major QTL for hairy leaf sheath introgressed from Aegilops tauschii and its association with enhanced grain yield in bread wheat. Euphytica 2015, 205, 275–285. [Google Scholar] [CrossRef]
- Shvachko, N.A.; Semilet, T.V.; Tikhonova, N.G. Trichomes in higher plants: Homological series in hereditary variability and molecular genetic mechanisms. Russ. J. Genet. 2020, 56, 1359–1370. [Google Scholar] [CrossRef]
- Saade, S.; Kutlu, B.; Draba, V.; Förster, K.; Schumann, E.; Tester, M.; Pillen, K.; Maurer, A. A donor-specific QTL, exhibiting allelic variation for leaf sheath hairiness in a nested association mapping population, is located on barley chromosome 4H. PLoS ONE 2017, 12, e0189446. [Google Scholar] [CrossRef]
- Korzun, V.; Malyshev, S.; Pickering, R.A.; Börner, A. RFLP mapping of a gene for hairy leaf sheath using a recombinant line from Hordeum vulgare L. × Hordeum bulbosum L. cross. Genome 1999, 42, 960–963. [Google Scholar] [CrossRef]
- Devos, K.M.; Atkinson, M.D.; Chinoy, C.N.; Francis, H.A.; Harcourt, R.L.; Koebner, R.M.D.; Liu, C.J.; Masojć, P.; Xie, D.X.; Gale, M.D. Chromosomal rearrangements in the rye genome relative to that of wheat. Theor. Appl. Genet. 1993, 85, 673–680. [Google Scholar] [CrossRef]
- Morihiro, H.; Takumi, S. Natural variation of trichome density on leaf in wild wheat Aegilops tauschii Coss. Wheat Inf. Serv. 2010, 109, 2010. [Google Scholar]
- Nishijima, R.; Okamoto, Y.; Hatano, H.; Takumi, S. Quantitative trait locus analysis for spikelet shape-related traits in wild wheat progenitor Aegilops tauschii: Implications for intraspecific diversification and subspecies differentiation. PLoS ONE 2017, 12, e0173210. [Google Scholar] [CrossRef]
- Gaurav, K.; Arora, S.; Silva, P.; Sánchez-Martín, J.; Horsnell, R.; Gao, L.; Brar, G.S.; Widrig, V.; Raupp, W.J.; Singh, N.; et al. Population genomic analysis of Aegilops tauschii identifies targets for bread wheat improvement. Nat. Biotechnol. 2022, 40, 422–431. [Google Scholar] [CrossRef]
- Simonov, A.V.; Gordeeva, E.I.; Genaev, M.A.; Li, W.; Bulatov, I.O.; Pshenichnikova, T.A. A new leaf pubescence gene, Hl1th, introgressed into bread wheat from Thinopyrum ponticum and its phenotypic manifestation under homoeologous chromosomal substitutions. Vavilov J. Genet. Breed. 2024, 28, 602–609. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Zhu, T.; Wang, L.; Rimbert, H.; Rodriguez, J.C.; Deal, K.R.; De Oliveira, R.; Choulet, F.; Keeble-Gagnère, G.; Tibbits, J.; Rogers, J.; et al. Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J. 2021, 107, 303–314. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Genomics 2013, 1303, 3997. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms. SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Hill, J.T.; Demarest, B.L.; Bisgrove, B.W.; Gorsi, B.; Su, Y.C.; Yost, H.J. MMAPPR: Mutation mapping analysis pipeline for pooled RNA-seq. Genome Res. 2013, 23, 687–697. [Google Scholar] [CrossRef]
- Magwene, P.M.; Willis, J.H.; Kelly, J.K. The statistics of bulk segregant analysis using next generation sequencing. PLoS Comput. Biol. 2011, 7, e1002255. [Google Scholar] [CrossRef]
- Takagi, H.; Abe, A.; Yoshida, K.; Kosugi, S.; Natsume, S.; Mitsuoka, C.; Uemura, A.; Utsushi, H.; Tamiru, M.; Takuno, S.; et al. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013, 74, 174–183. [Google Scholar] [CrossRef]
- Mansfeld, B.N.; Grumet, R. QTLseqr: An R package for bulk segregant analysis with next-generation sequencing. Plant Genome 2018, 11, 180006. [Google Scholar] [CrossRef]
- Liu, Q.; Xuan, Q.; Lan, Y.; Xie, X.; Chen, B.; You, J.; Su, L.; Lohani, M.N.; Wu, L.; Hu, X.; et al. Genetic identification and characterization of a novel locus for wheat kernel length. J. Integr. Agric. 2024; in press. [Google Scholar] [CrossRef]
- Hu, X.K.; Dai, S.F.; Ouellet, T.; Balcerzak, M.; Rocheleau, H.; Khanizadeh, S.; Pu, Z.J.; Yan, Z.H. Characterization of novel D-hordeins from Psathyrostachys juncea. Biol. Plant. 2018, 62, 369–378. [Google Scholar] [CrossRef]
- Liu, S.; Li, K.; Dai, X.; Qin, G.; Lu, D.; Gao, Z.; Li, X.; Song, B.; Bian, J.; Ren, D.; et al. A telomere-to-telomere genome assembly coupled with multi-omic data provides insights into the evolution of hexaploid bread wheat. Nat. Genet. 2025, 57, 1008–1020. [Google Scholar] [CrossRef]
- Oppenheimer, D.G.; Herman, P.L.; Sivakumaran, S.; Esch, J.; Marks, M.D. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 1991, 67, 483–493. [Google Scholar] [CrossRef]
- Zhang, F.; Gonzalez, A.; Zhao, M.; Payne, C.T.; Lloyd, A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 2003, 130, 4859–4869. [Google Scholar] [CrossRef]
- Walker, A.R.; Davison, P.A.; Bolognesi-Winfield, A.C.; James, C.M.; Srinivasan, N.; Blundell, T.L.; Esch, J.J.; Marks, M.D.; Gray, J.C. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 1999, 11, 1337–1350. [Google Scholar] [CrossRef]
- Schellmann, S.; Schnittger, A.; Kirik, V.; Wada, T.; Okada, K.; Beermann, A.; Thumfahrt, J.; Jürgens, G.; Hülskamp, M. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 2002, 21, 5036–5046. [Google Scholar] [CrossRef]
- Morran, S.; Eini, O.; Pyvovarenko, T.; Parent, B.; Singh, R.; Ismagul, A.; Eliby, S.; Shirley, N.; Langridge, P.; Lopato, S. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 2011, 9, 230–249. [Google Scholar] [CrossRef]
- Pshenichnikova, T.A.; Doroshkov, A.V.; Simonov, A.V.; Afonnikov, D.A.; Börner, A. Diversity of leaf pubescence in bread wheat and relative species. Genet. Resour. Crop Evol. 2017, 64, 1761–1773. [Google Scholar] [CrossRef]
- Zheng, Y.; Khine, E.; Thi, K.; Nyein, E.; Huang, L.; Lin, L.; Xie, X.; Lin, M.H.W.; Oo, K.T.; Moe, M.M.; et al. Multi-environment BSA-seq using large F3 populations is able to achieve reliable QTL mapping with high power and resolution: An experimental demonstration in rice. Crop J. 2024, 12, 549–557. [Google Scholar] [CrossRef]
- Du, H.; Zhu, J.; Su, H.; Huang, M.; Wang, H.; Ding, S. Bulked segregant RNA-seq reveals differential expression and SNPs of candidate genes associated with waterlogging tolerance in maize. Front. Plant Sci. 2017, 8, 1022. [Google Scholar] [CrossRef]
- Zhao, J.; Xingwei, Z.; Ling, Q.; Chenkang, Y.; Bangbang, W.; Ziming, H.; Yuqing, T.; Li, G.; Yang, Z.; Zheng, J.; et al. Genome wide association study reveals structural chromosome variations (SCVs) with phenotypic effects in wheat (Triticum aestivum L.). Plant J. 2022, 112, 1447–1461. [Google Scholar] [CrossRef]
- Li, W.; Huang, L.; Gill, B. Recurrent deletions of puroindoline genes at the grain hardness locus in four independent lineages of polyploid wheat. Plant Physiol. 2007, 146, 200–212. [Google Scholar] [CrossRef]
- Zhang, S.; Zhao, J.; Zhang, H.; Fu, D.; Qiao, L.; Wu, B.; Li, X.; Hao, Y.; Zheng, X.; Liang, Z.; et al. Structural chromosome variations from Jinmai 47 and Jinmai 84 affected agronomic traits and drought tolerance of wheat. J. Integr. Agric. 2024; in press. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, S.; Lin, T.; Kan, S.; Yu, P. The DUF506 gene family in Triticum aestivum: Genome-wide identification and expression profiling under salt stress. Agronomy 2025, 15, 281. [Google Scholar] [CrossRef]
- Ma, J.; Wang, R.; Zhao, H.; Li, L.; Zeng, F.; Wang, Y.; Chen, M.; Chang, J.; He, G.; Yang, G.; et al. Genome-wide characterization of the VQ genes in Triticeae and their functionalization driven by polyploidization and gene duplication events in wheat. Int. J. Biol. Macromol. 2023, 243, 125264. [Google Scholar] [CrossRef]
- Zahaf, O.; Blanchet, S.; De Zélicourt, A.; Alunni, B.; Plet, J.; Laffont, C.; De Lorenzo, L.; Imbeaud, S.; Ichanté, J.; Diet, A.; et al. Comparative transcriptomic analysis of salt adaptation in roots of contrasting Medicago truncatula genotypes. Mol. Plant 2012, 5, 1068–1081. [Google Scholar] [CrossRef]
- Payne, C.T.; Zhang, F.; Lloyd, A.M. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 2000, 156, 1349–1362. [Google Scholar] [CrossRef]
- Masucci, J.D.; Rerie, W.G.; Foreman, D.R.; Zhang, M.; Galway, M.E.; Marks, M.D.; Schiefelbein, J.W. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 1996, 122, 1253–1260. [Google Scholar] [CrossRef]
- Wang, S.; Chen, J.G. Arabidopsis transient expression analysis reveals that activation of GLABRA2 may require concurrent binding of GLABRA1 and GLABRA3 to the promoter of GLABRA2. Plant Cell Physiol. 2008, 49, 1792–1804. [Google Scholar] [CrossRef]
- Wada, T.; Kurata, T.; Tominaga, R.; Koshino-Kimura, Y.; Tachibana, T.; Goto, K.; Marks, M.D.; Shimura, Y.; Okada, K. Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 2002, 129, 5409–5419. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.C.; Oppenheimer, D.G.; Lloyd, A.M.; Paparozzi, E.T.; Marks, M.D. Roles of the GLABROUS1 and TRANSPARENT TESTA GLABRA genes in Arabidopsis trichome development. Plant Cell 1994, 6, 1065–1076. [Google Scholar] [CrossRef] [PubMed]
- Marks, M.D. Molecular genetic analysis of trichome development in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 137–163. [Google Scholar] [CrossRef] [PubMed]
- Mei, F.; Chen, B.; Du, L.; Li, S.; Zhu, D.; Chen, N.; Zhang, Y.; Li, F.; Wang, Z.; Cheng, X.; et al. A gain-of-function allele of a DREB transcription factor gene ameliorates drought tolerance in wheat. Plant Cell 2022, 34, 4472–4494. [Google Scholar] [CrossRef]
- Merchuk-Ovnat, L.; Barak, V.; Fahima, T.; Ordon, F.; Lidzbarsky, G.; Krugman, T.; Saranga, Y. Ancestral QTL alleles from wild emmer wheat improve drought resistance and productivity in modern wheat cultivars. Front. Plant Sci. 2016, 7, 452. [Google Scholar] [CrossRef]

| Samples | Raw Bases (bp) | Raw Reads (bp) | Clean Bases (bp) | Clean Reads (bp) | Clean Bases (%) | Clean Reads (%) | Q30 1 (%) | GC (%) | Duplication 2 (%) |
|---|---|---|---|---|---|---|---|---|---|
| LSH_pool1+ | 7,354,933,800 | 49,032,892 | 7,125,150,346 | 48,695,506 | 96.88 | 99.31 | 96.9 | 52.5 | 4.1 |
| LSH_pool1− | 8,657,837,700 | 57,718,918 | 8,390,741,334 | 57,334,828 | 96.91 | 99.33 | 96.7 | 52.6 | 3.9 |
| LSH_pool2+ | 7,762,636,200 | 51,750,908 | 7,606,138,898 | 51,401,226 | 97.98 | 99.32 | 96.1 | 52.9 | 4.0 |
| LSH_pool2− | 7,286,962,200 | 48,579,748 | 7,046,164,016 | 48,241,024 | 96.70 | 99.30 | 97.0 | 52.9 | 4.1 |
| ZKLM1+ | 4,711,597,500 | 31,410,650 | 4,526,831,572 | 31,254,282 | 96.08 | 99.50 | 97.0 | 51.9 | 4.1 |
| SM830− | 4,432,805,400 | 29,552,036 | 4,250,976,330 | 29,393,964 | 95.90 | 99.47 | 96.9 | 52.3 | 3.5 |
| SM2262− | 4,359,853,500 | 29,065,690 | 4,211,924,814 | 28,922,050 | 96.61 | 99.51 | 96.7 | 51.1 | 3.9 |
| Samples | Mapping Rate 1 (%) | Average Depth 2 (×) | Coverage 3 ≥ 4× (%) | Coverage ≥ 10× (%) | Coverage ≥ 30× (%) | Capture Rate 4 (%) |
|---|---|---|---|---|---|---|
| LSH_pool1+ | 99.99 | 60.46 | 97.16 | 94.33 | 73.11 | 31.30 |
| LSH_pool1− | 99.99 | 69.97 | 97.23 | 94.59 | 77.69 | 31.15 |
| LSH_pool2+ | 99.99 | 59.74 | 97.26 | 94.46 | 72.89 | 31.12 |
| LSH_pool2− | 99.99 | 59.91 | 97.18 | 94.11 | 72.53 | 30.85 |
| ZKLM1+ | 99.99 | 37.62 | 94.65 | 87.59 | 48.52 | 28.53 |
| SM830− | 99.99 | 36.55 | 94.50 | 87.45 | 47.68 | 29.89 |
| SM2262− | 99.99 | 28.09 | 93.17 | 82.16 | 32.48 | 22.84 |
| Populations | QTLs/LD Block | Peak Intervals | Positions 1 (Mb) | References |
|---|---|---|---|---|
| C8 RILs | QLsh.saas-4D | Xbarc48—D_GB5Y7FA01AGCKH_224 | 504.32–511.67 | [8] |
| CC RILs | QLsh.saas-4D | Xbarc48–Xbarc1183 | 504.32–517.02 | [8] |
| Ae. tauschii accessions | LD block-trichomes-4DL | Between 530 kb LD block on 4DL | 502.84–503.26 | [15] |
| SM830/ZKLM1 F2 | QLsh.cwnu-4D | n.501803422–n.503905439 | 501.80–503.91 | This study |
| SM2262/ZKLM1 F2 | QLsh.cwnu-4D | n.501803422–n.503905382 | 501.80–503.91 | This study |
| Cultivars | Probabilities of Localization (Top 4) | Probabilities of Membrane Association Type | ||||||
|---|---|---|---|---|---|---|---|---|
| Cytoplasm | Nucleus | Mitochondrion | Peroxisome | Peripheral | Transmembrane | Lipid anchor | Soluble | |
| ZKLM1 | 0.2662 | 0.9365 | 0.1483 | 0.1795 | 0.3550 | 0.1420 | 0.0870 | 0.8170 |
| SM830 | 0.2820 | 0.8966 | 0.1186 | 0.3141 | 0.3080 | 0.1090 | 0.0720 | 0.8370 |
| CM42 | 0.2122 | 0.9478 | 0.1061 | 0.4360 | 0.2540 | 0.0950 | 0.0450 | 0.8630 |
| Gene/QTL | Species/Locus | Molecular Type and Function | References |
|---|---|---|---|
| TaSAIN1-4D | Wheat (4DL) | Salt-inducible nuclear protein; unique to Triticeae | This study |
| Hl1/Hl1th | Wheat (4BL)/Th. ponticum introgression | Dominant locus for leaf-blade/sheath pubescence | [16] |
| QLsh.saas-4D | Wheat (4DL) | QTL for hairy leaf sheath (LSH) | [8] |
| Hsh | Barley (4HL) | Gene controlling leaf sheath hairiness (Hsh) | [9] |
| Hp1 | Rye (5RL) | Hairy peduncle gene (Hp) | [9] |
| GL6/Mhl1 | Rice (Chr. 6)/Maize (Chr. 9) | Homologous loci regulating trichome density/length | [9] |
| Hg (WOX3-like) | Wheat (1AS) | WOX transcription factor | [9] |
| GL1 (R2R3-MYB) | Arabidopsis | Activator of trichome initiation (MBW complex) | [29] |
| GL3/EGL3 (bHLH) | Arabidopsis | Partners with GL1 and TTG1; activates GL2 | [30] |
| TTG1 (WD40) | Arabidopsis | WD40 scaffold protein | [31] |
| TRY/CPC (R3-MYBs) | Arabidopsis | Mobile negative regulators of trichome density | [32] |
| DREB family | Wheat (Various loci) | AP2/ERF transcription factors | [33] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, L.; He, J.; Shen, S.; Li, Y.; He, J.; Hu, X. Fine Mapping of a Major Locus for Leaf Sheath Hairiness in Wheat Identifies TaSAIN1-4D as a Candidate Gene. Genes 2025, 16, 1117. https://doi.org/10.3390/genes16091117
Wu L, He J, Shen S, Li Y, He J, Hu X. Fine Mapping of a Major Locus for Leaf Sheath Hairiness in Wheat Identifies TaSAIN1-4D as a Candidate Gene. Genes. 2025; 16(9):1117. https://doi.org/10.3390/genes16091117
Chicago/Turabian StyleWu, Lijuan, Jundong He, Shian Shen, Yulin Li, Jinbai He, and Xinkun Hu. 2025. "Fine Mapping of a Major Locus for Leaf Sheath Hairiness in Wheat Identifies TaSAIN1-4D as a Candidate Gene" Genes 16, no. 9: 1117. https://doi.org/10.3390/genes16091117
APA StyleWu, L., He, J., Shen, S., Li, Y., He, J., & Hu, X. (2025). Fine Mapping of a Major Locus for Leaf Sheath Hairiness in Wheat Identifies TaSAIN1-4D as a Candidate Gene. Genes, 16(9), 1117. https://doi.org/10.3390/genes16091117








