Novel KIAA0825 Variants Underlie Nonsyndromic Postaxial Polydactyly
Abstract
1. Introduction
2. Materials and Methods
2.1. Recruitment of Families
2.2. Blood Collection and DNA Extraction
2.3. Whole Exome Sequencing and Genotyping
3. Results
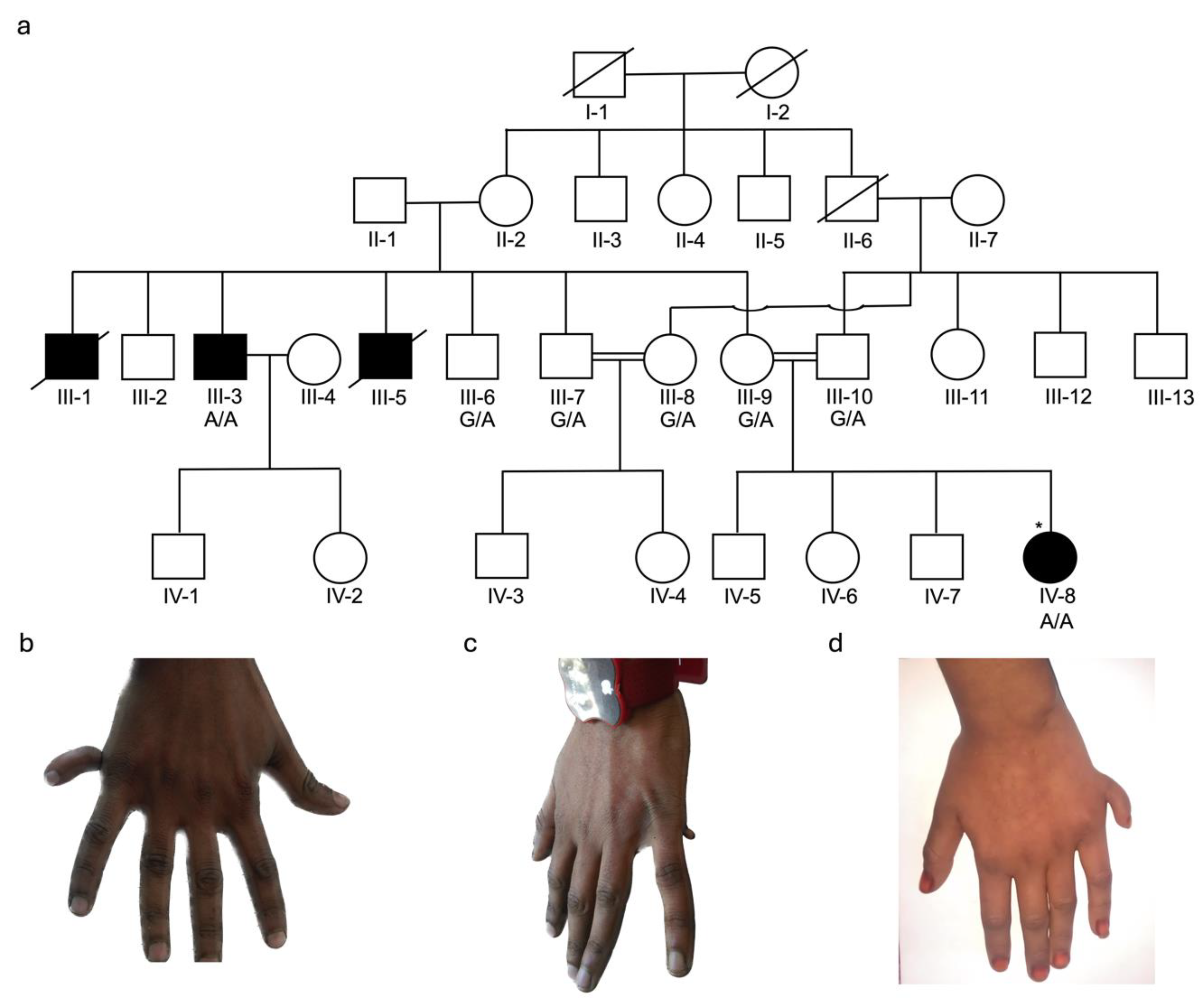
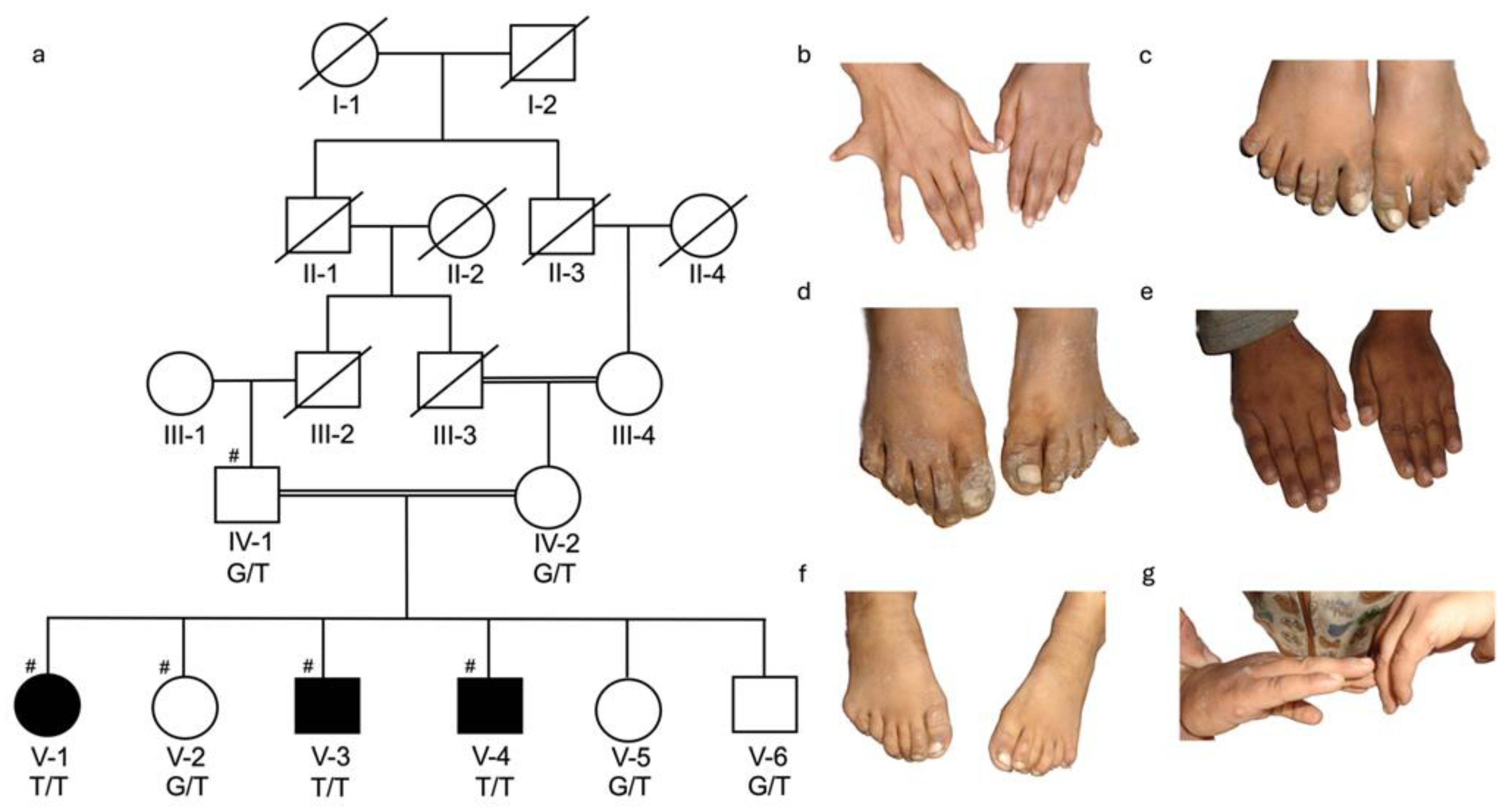
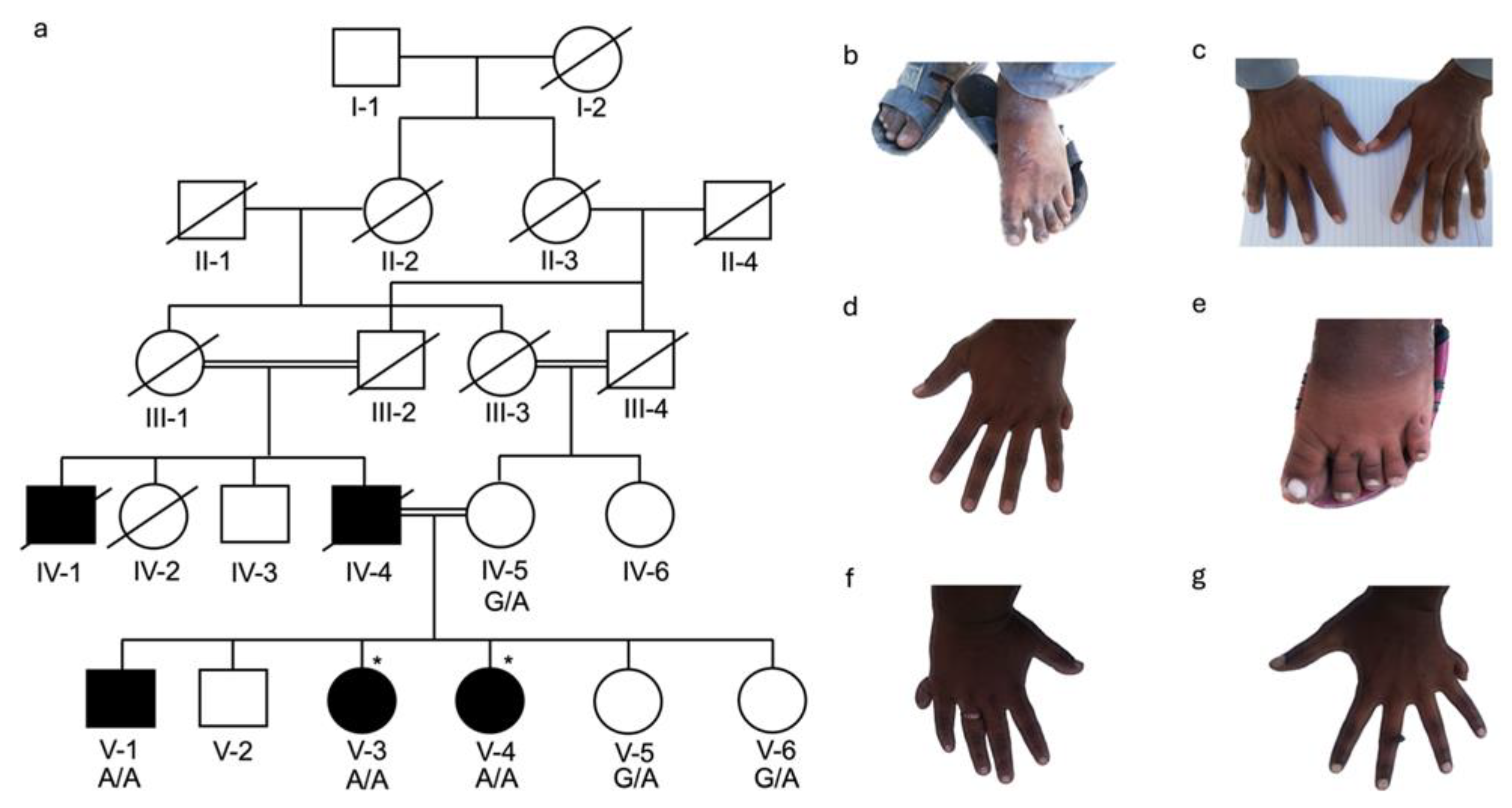
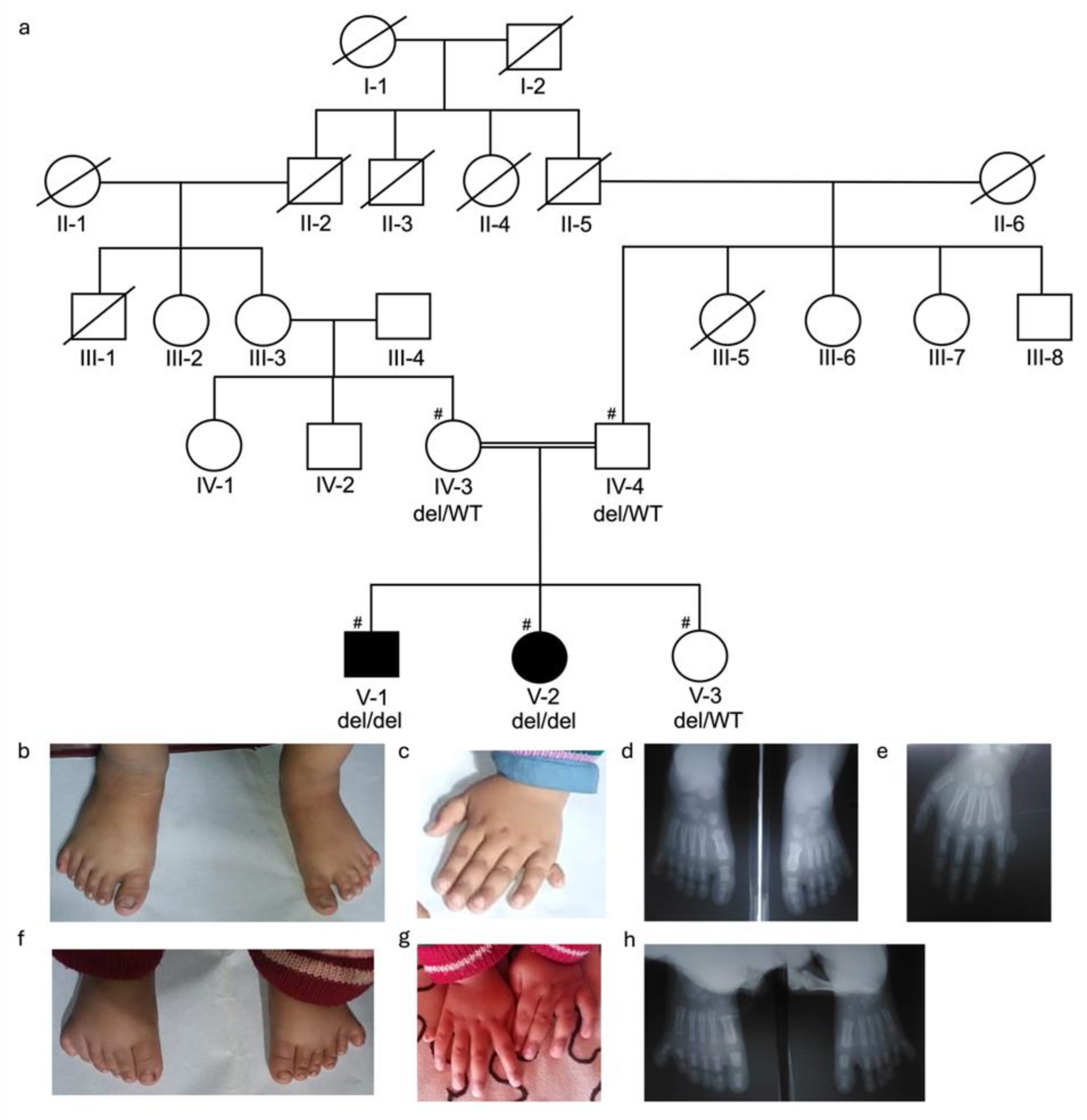
3.1. Clinical Description of Families
3.2. Identification of KIAA0825 Variants
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Biesecker, L.G. Polydactyly: How Many Disorders and How Many Genes? 2010 Update. Dev. Dyn. 2011, 240, 931–942. [Google Scholar] [CrossRef] [PubMed]
- Al Amin, A.S.M.; Carter, K.R. Polydactyly(Archived). In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Bouldin, C.M.; Harfe, B.D. Aberrant FGF Signaling, Independent of Ectopic Hedgehog Signaling, Initiates Preaxial Polydactyly in Dorking Chickens. Dev. Biol. 2009, 334, 133–141. [Google Scholar] [CrossRef]
- Perez-Lopez, L.M.; La Iglesia, D.G.; Cabrera-Gonzalez, M. Radial Polydactyly. What’s New? Curr. Pediatr. Rev. 2018, 14, 91–96. [Google Scholar] [CrossRef]
- Umair, M.; Ahmad, F.; Bilal, M.; Ahmad, W.; Alfadhel, M. Clinical Genetics of Polydactyly: An Updated Review. Front. Genet. 2018, 9, 447. [Google Scholar] [CrossRef]
- Malik, S.; Ullah, S.; Afzal, M.; Lal, K.; Haque, S. Clinical and Descriptive Genetic Study of Polydactyly: A Pakistani Experience of 313 Cases. Clin. Genet. 2014, 85, 482–486. [Google Scholar] [CrossRef]
- Umair, M.; Bilal, M.; Ali, R.H.; Alhaddad, B.; Ahmad, F.; Abdullah; Haack, T.B.; Alfadhel, M.; Ansar, M.; Meitinger, T.; et al. Whole-exome Sequencing Revealed a Nonsense Mutation in STKLD1 Causing Non-syndromic Pre-axial Polydactyly Type A Affecting Only Upper Limb. Clin. Genet. 2019, 96, 134–139. [Google Scholar] [CrossRef]
- Tilemis, F.-N.; Marinakis, N.M.; Kosma, K.; Fostira, F.; Traeger-Synodinos, J. Identification of a Novel IQCE Large Deletion through Copy Number Variant Analysis from Whole-exome Sequencing Data of a Patient with Postaxial Polydactyly Type A7. Mol. Syndromol. 2023, 14, 225–230. [Google Scholar] [CrossRef]
- Temtamy, S.A.; McKusick, V.A. The Genetics of Hand Malformations. Birth Defects Orig. Artic. Ser. 1978, 14, i–xviii. [Google Scholar] [CrossRef] [PubMed]
- Umair, M.; Palander, O.; Bilal, M.; Almuzzaini, B.; Alam, Q.; Ahmad, F.; Younus, M.; Khan, A.; Waqas, A.; Rafeeq, M.M.; et al. Biallelic Variant in DACH1, Encoding Dachshund Homolog 1, Defines a Novel Candidate Locus for Recessive Postaxial Polydactyly Type A. Genomics 2021, 113, 2495–2502. [Google Scholar] [CrossRef]
- Ahmed, H.; Akbari, H.; Emami, A.; Akbari, M.R. Genetic Overview of Syndactyly and Polydactyly. Plast. Reconstr. Surg. Glob. Open 2017, 5, e1549. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.K.; El-Harouni, A.A. Review of Literature: Genes Related to Postaxial Polydactyly. Front. Pediatr. 2015, 3, 8. [Google Scholar] [CrossRef]
- Barham, G.; Clarke, N.M.P. Genetic Regulation of Embryological Limb Development with Relation to Congenital Limb Deformity in Humans. J. Child. Orthop. 2008, 2, 1–9. [Google Scholar] [CrossRef]
- Stewart, T.A.; Bhat, R.; Newman, S.A. The Evolutionary Origin of Digit Patterning. EvoDevo 2017, 8, 21. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Yang, H.; Wang, K. Genomic Variant Annotation and Prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 2015, 10, 1556–1566. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The Mutational Constraint Spectrum Quantified from Variation in 141,456 Humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Seelow, D.; Schuelke, M.; Hildebrandt, F.; Nurnberg, P. HomozygosityMapper—An Interactive Approach to Homozygosity Mapping. Nucleic Acids Res. 2009, 37, W593–W599. [Google Scholar] [CrossRef] [PubMed]
- Lyles, C.R.; Lunn, M.R.; Obedin-Maliver, J.; Bibbins-Domingo, K. The New Era of Precision Population Health: Insights for the All of Us Research Program and Beyond. J. Transl. Med. 2018, 16, 211. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, M.H.; Qian, H.; Hou, Z.; Rosen, J.D.; Tapia, A.L.; Shan, Y.; Jain, D.; Argos, M.; Arnett, D.K.; Avery, C.; et al. Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium Whole Genome Sequences Improves Imputation Quality and Detection of Rare Variant Associations in Admixed African and Hispanic/Latino Populations. PLoS Genet. 2019, 15, e1008500. [Google Scholar] [CrossRef] [PubMed]
- Ullah, I.; Kakar, N.; Schrauwen, I.; Hussain, S.; Chakchouk, I.; Liaqat, K.; Acharya, A.; Wasif, N.; Santos-Cortez, R.L.P.; Khan, S.; et al. Variants in KIAA0825 Underlie Autosomal Recessive Postaxial Polydactyly. Hum. Genet. 2019, 138, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Hayat, A.; Umair, M.; Abbas, S.; Rauf, A.; Ahmad, F.; Ullah, S.; Ahmad, W.; Khan, B. Identification of a Novel Biallelic Missense Variant in the KIAA0825 Underlies Postaxial Polydactyly Type A. Genomics 2020, 112, 2729–2733. [Google Scholar] [CrossRef]
- Bilal, M.; Ahmad, W. A Frameshift Variant in KIAA0825 Causes Postaxial Polydactyly. Mol. Syndromol. 2021, 12, 20–24. [Google Scholar] [CrossRef]
- Ahmad, S.; Ali, M.Z.; Muzammal, M.; Khan, A.U.; Ikram, M.; Muurinen, M.; Hussain, S.; Loid, P.; Khan, M.A.; Mäkitie, O. Identification of GLI1 and KIAA0825 Variants in Two Families with Postaxial Polydactyly. Genes 2023, 14, 869. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Deng, S.; Zhu, F. Prenatal Detection of Novel Compound Heterozygous Splice Site Variants of the KIAA0825 Gene in a Fetus with Postaxial Polydactyly Type A. Genes 2022, 13, 1230. [Google Scholar] [CrossRef] [PubMed]

| Family | Type | Reference Genome | Transcript | Genomic Position (hg38) | cDNA Change | Amino Acid Change | gnomAD All | gnomAD South Asian | Classification 1 |
|---|---|---|---|---|---|---|---|---|---|
| BD375 | Nonsense | Hg19 | NM_001145678.3 | 5:94452997 | c.2319G>A | p.(Trp773*) | 2.03 × 10−5 | 3.75 × 10−4 | Pathogenic (PVS1, PP1, PM2) |
| BD650 | Nonsense | Hg38 | NM_001145678.3 | 5:94452997 | c.2319G>A | p.(Trp773*) | 2.03 × 10−5 | 3.75 × 10−4 | Pathogenic (PVS1, PP1, PM2) |
| BD551 | Missense | Hg38 | NM_001145678.3 | 5:94520248 | c.970G>T | p.(Val324Phe) | 5.05 × 10−6 | 1.17 × 10−5 | Likely pathogenic (PP1, PM2, PP3) |
| BDKA21 | In-frame deletion | Hg38 | NM_001145678.3 | 5:94403701 | c.2743_2754del | p.(Gln915-Val918del) | Absent | Absent | Likely pathogenic (PP1, PM2, PM4) |
| Study | Phenotype | Variant Type | Zygosity | cDNA | Amino Acid Change | Country of Origin |
|---|---|---|---|---|---|---|
| Ullah et al., 2019 [22] | PAPA, PAPB, and clinodactyly | Frameshift | Homozygous | c.591dupA | p.(Gln198Thrfs*21) | Pakistani |
| Ullah et al., 2019 [22] | PAPA, PAPB and mild camptodactyly | Nonsense | Homozygous | c.2173A>T | p.(Lys725*) | Pakistani |
| Hayat el at., 2020 [23] | PAPA and camptodactyly | Missense | Homozygous | c.50T>C | p.(Leu17Ser) | Pakistani |
| Bilal and Ahmad, 2021 [24] | PAPA and syndactyly | Frameshift deletion | Homozygous | c.143delG | p.(Cys48Serfs*28) | Pakistani |
| Yao et al., 2022 [26] | PAPA | Splice site | Compound heterozygous | c.-1-2A>T | − | Chinese |
| Yao et al., 2022 [26] | PAPA | Splice site | Compound heterozygous | c.2247-2A>G | − | Chinese |
| Ahmad et al., 2023 [25] | PAPA, syndactyly, and clinodactyly | Missense | Homozygous | c.3572C>T | p.(Pro1191Leu) | Pakistani |
| Present Study | PAPB, clinodactyly, brachydactyly, and whitish nails | Nonsense | Homozygous | c.2319G>A | p.(Trp773*) | Pakistani |
| Present Study | PAPA, PAPB, and dysplastic nails | Missense | Homozygous | c.970G>T | p.(Val324Phe) | Pakistani |
| Present Study | PAPA and PAPB | In-frame deletion | Homozygous | c.2743_2754del | p.(Gln915-Val918del) | Pakistani |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdullah; Bharadwaj, T.; Javed, S.; Khan, H.; Acharya, A.; Ji, W.; Umm-e-Kalsoom; Ali, H.; Schrauwen, I.; Ahmad, W.; et al. Novel KIAA0825 Variants Underlie Nonsyndromic Postaxial Polydactyly. Genes 2025, 16, 1118. https://doi.org/10.3390/genes16091118
Abdullah, Bharadwaj T, Javed S, Khan H, Acharya A, Ji W, Umm-e-Kalsoom, Ali H, Schrauwen I, Ahmad W, et al. Novel KIAA0825 Variants Underlie Nonsyndromic Postaxial Polydactyly. Genes. 2025; 16(9):1118. https://doi.org/10.3390/genes16091118
Chicago/Turabian StyleAbdullah, Thashi Bharadwaj, Saffia Javed, Hammal Khan, Anushree Acharya, Weizhen Ji, Umm-e-Kalsoom, Hamid Ali, Isabelle Schrauwen, Wasim Ahmad, and et al. 2025. "Novel KIAA0825 Variants Underlie Nonsyndromic Postaxial Polydactyly" Genes 16, no. 9: 1118. https://doi.org/10.3390/genes16091118
APA StyleAbdullah, Bharadwaj, T., Javed, S., Khan, H., Acharya, A., Ji, W., Umm-e-Kalsoom, Ali, H., Schrauwen, I., Ahmad, W., Lakhani, S. A., & Leal, S. M. (2025). Novel KIAA0825 Variants Underlie Nonsyndromic Postaxial Polydactyly. Genes, 16(9), 1118. https://doi.org/10.3390/genes16091118







