De Novo Transcriptome Sequencing and Profiling of Ovarian Development of Argas persicus Along the Trophogonic Cycle
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. RNA Isolation and Library Construction
2.3. Transcript Splicing and Functional Annotation
2.4. Differential Expression Analysis of Genes
2.5. Functional Enrichment Analysis
2.6. Screening of Candidate Genes Related to Ovarian Development
2.7. Quantitative Real-Time PCR (qPCR) for mRNA Quantitation
3. Results
3.1. Quality Analysis of RNA Sequencing
3.2. The Profiling of DEGs of A. persicus Ovaries
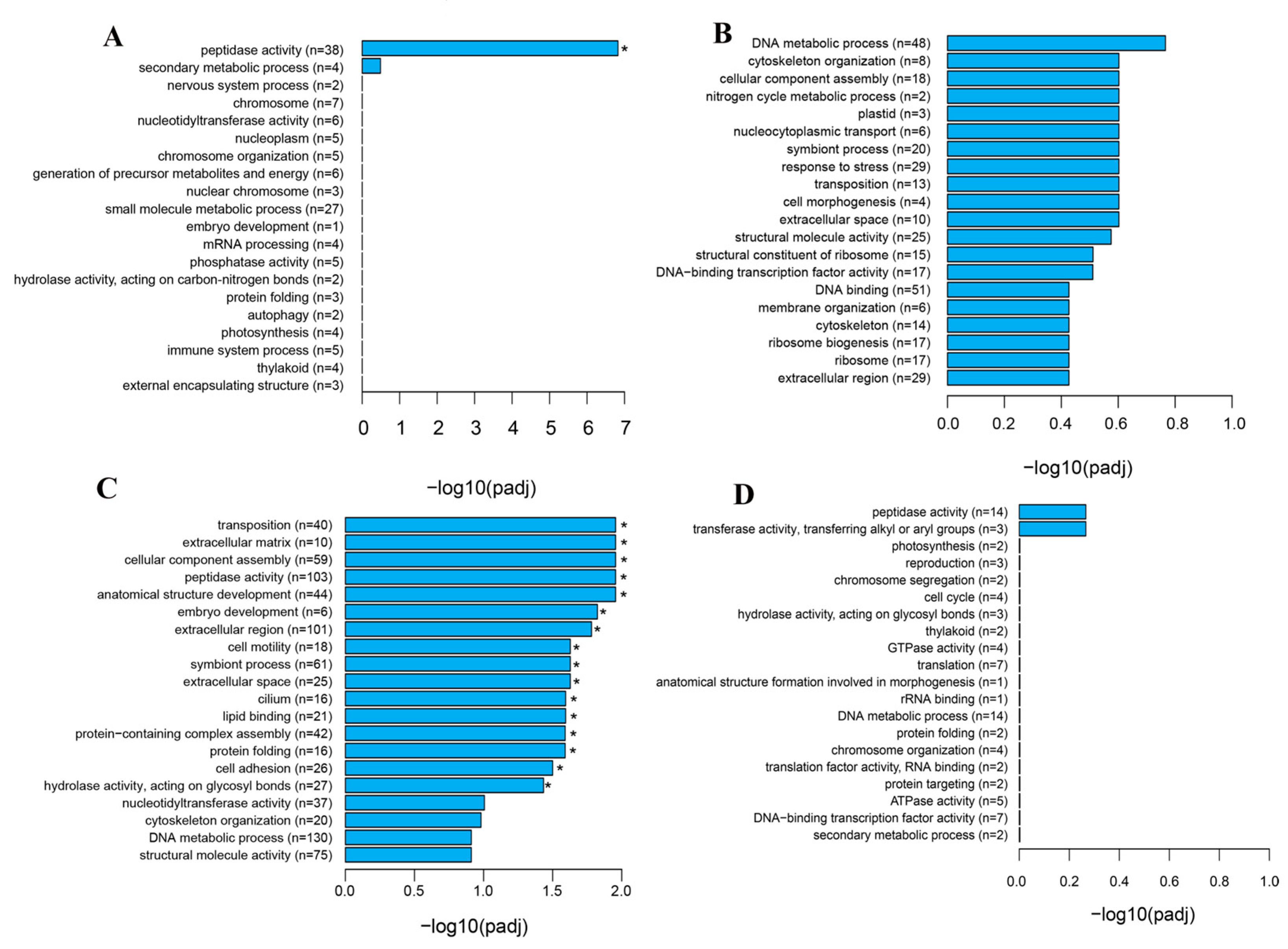
3.3. GO Analysis of DEGs
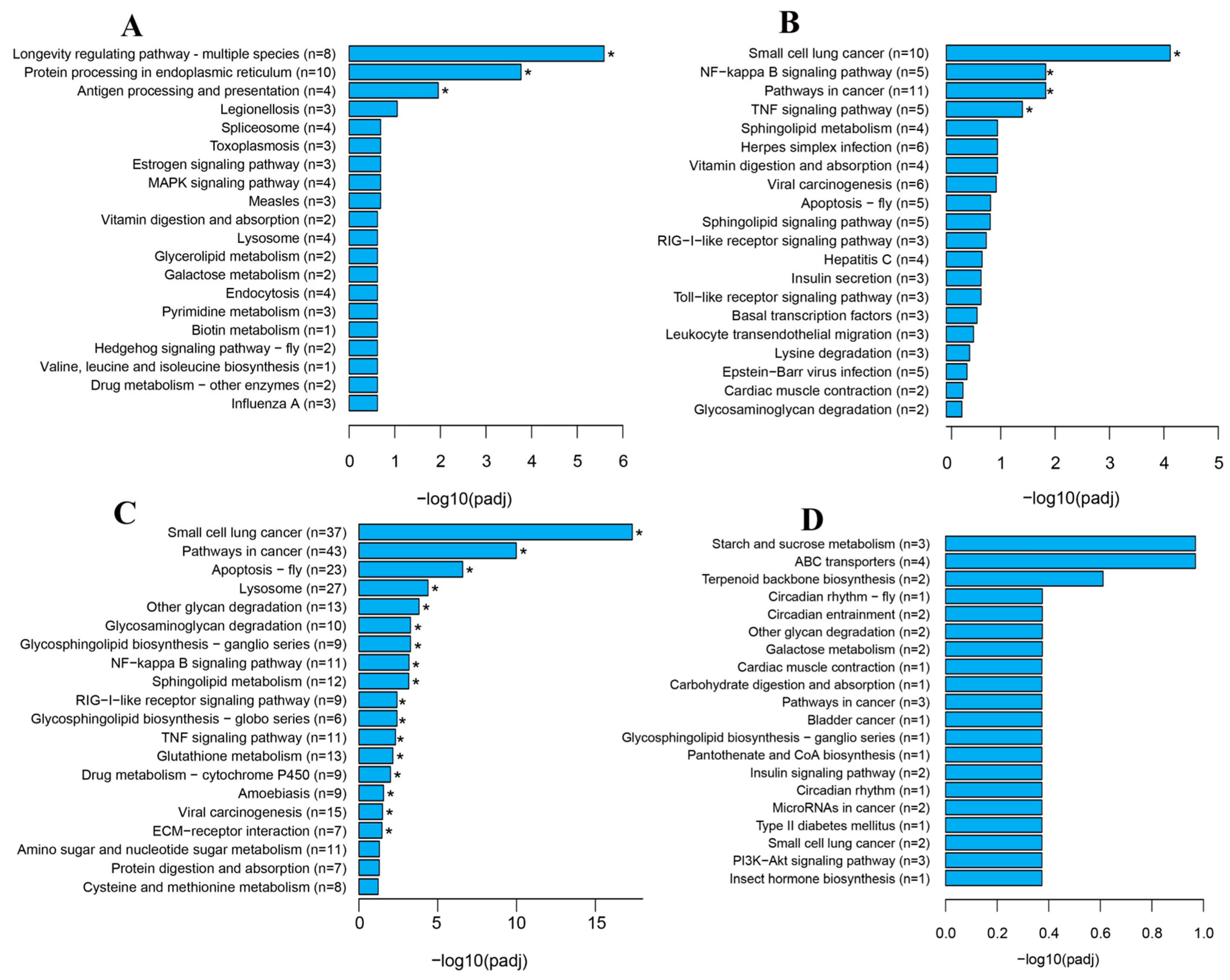
3.4. KEGG Analysis of DEGs
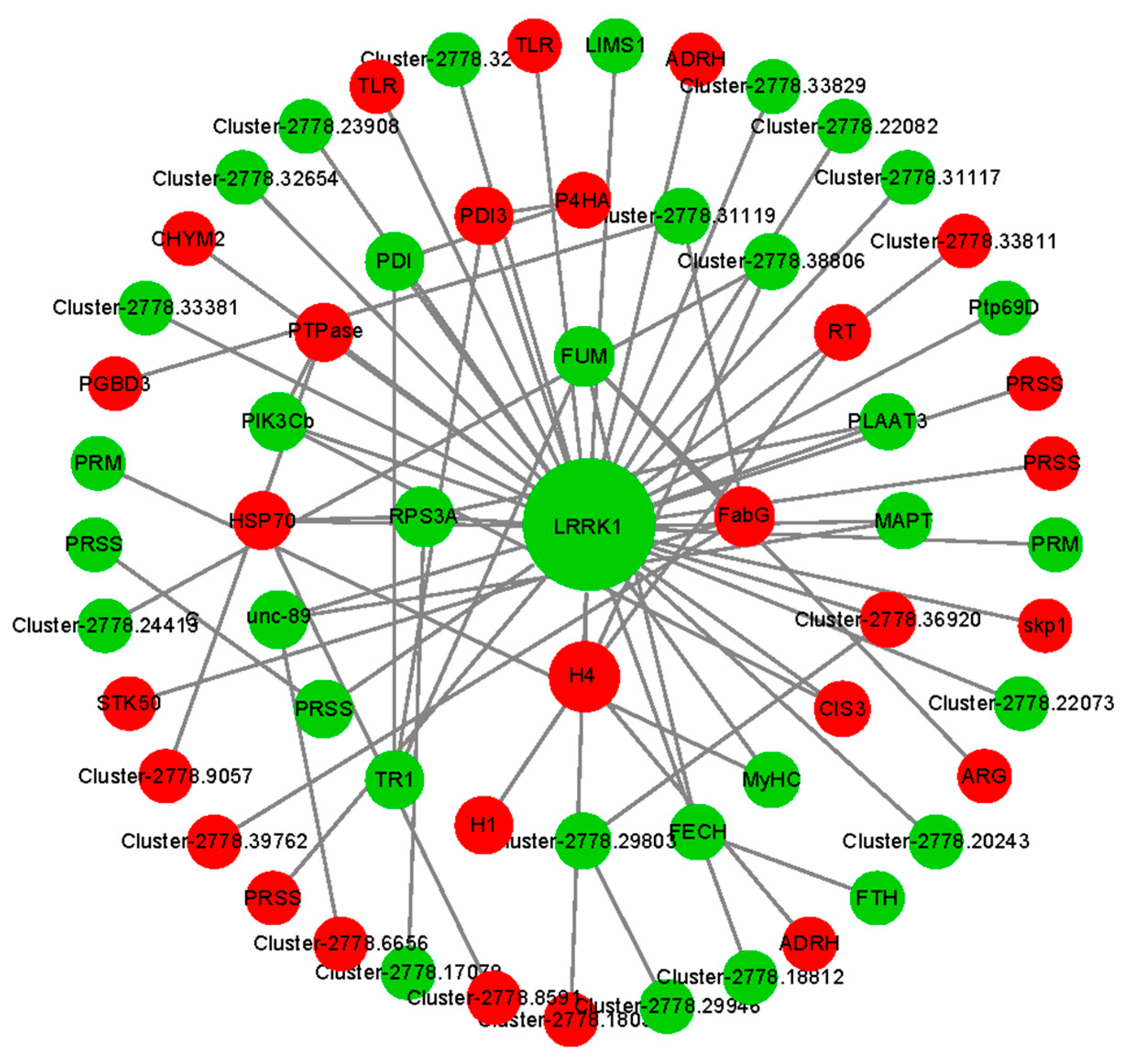
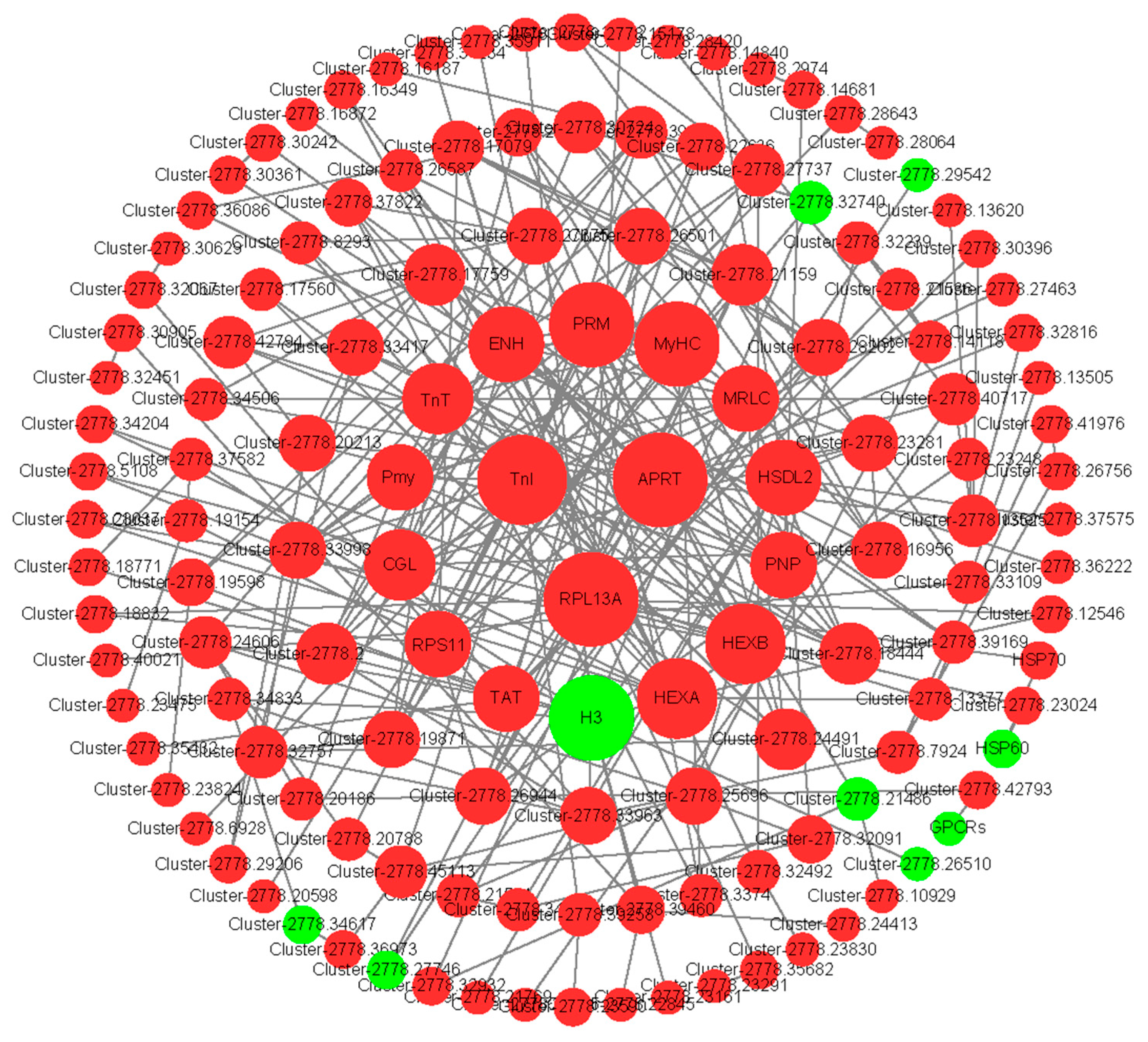
3.5. Candidate Genes Related to Ovarian Development
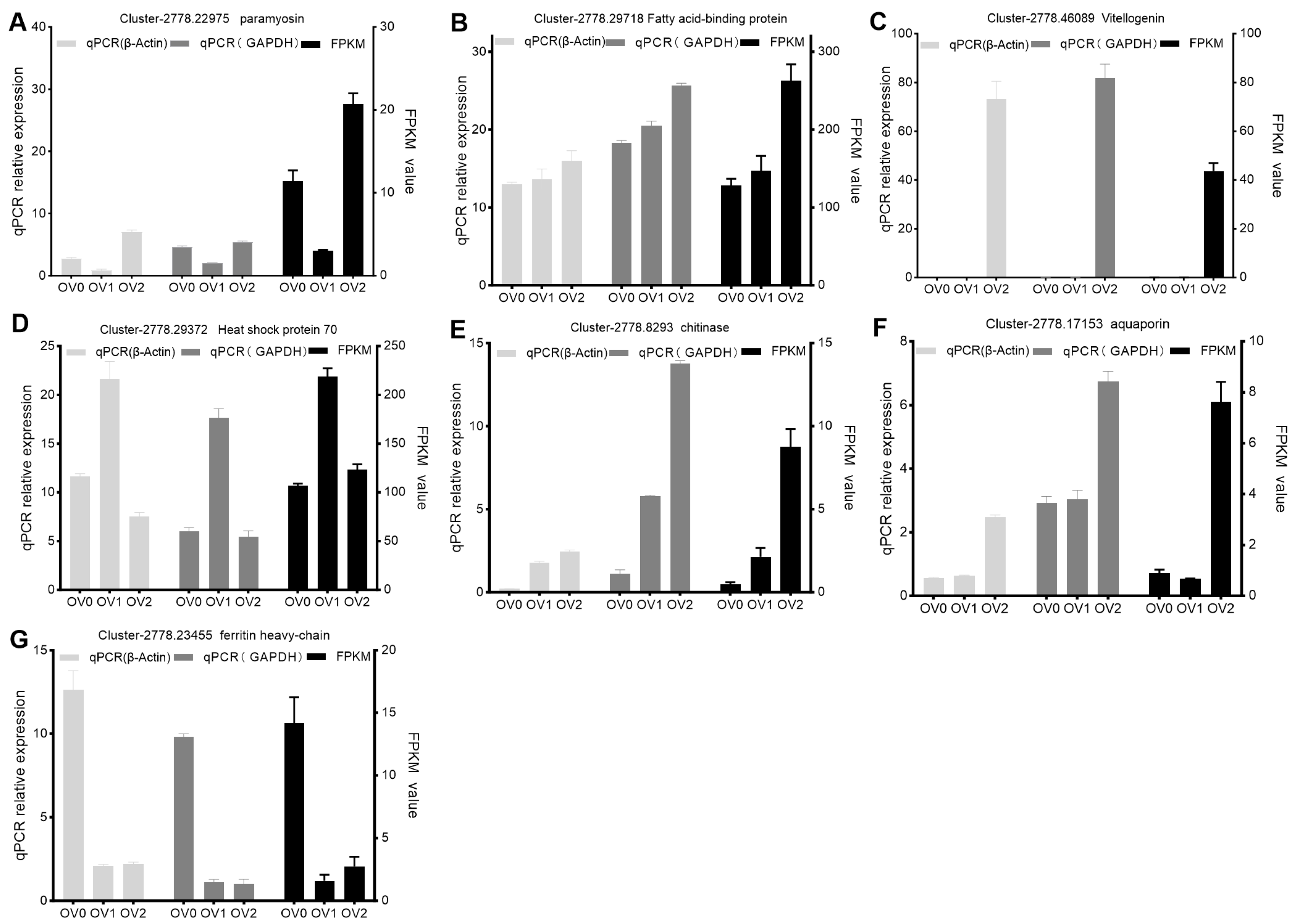
3.6. Verification of DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| A. persicus | Argas persicus |
| DEGs | differentially expressed genes |
| PPI | protein–protein interaction |
| LD | linear dichroism |
| HSP70 | heatshockprotein70 |
| PDI | protein disulfide isomerase |
| PDI3 | protein disulfide isomerase 3 |
| PRM | paramyosin |
| TnI | troponin I protein |
| HEX | beta-hexosaminidase |
| Vg | vitellogenin |
| Vn | vitellin |
| H. longicornis | Haemaphysalis longicornis |
References
- Kohls, G.M.; Hoogstraal, H.; Clifford, C.M.; Kaiser, M.N. The Subgenus Persicargas (Ixodoidea, Argasidae, Argas). 9. Redescription and New World Records of Argas (P.) persicus (Oken), and Resurrection, Redescription, and Records of A. (P.) radiatus Railliet, A. (P.) sanchezi Duges, and A. (P.) miniatus Koch, New World Ticks Misidentified as A. (P.) persicus. Ann. Entomol. Soc. Am. 1970, 63, 590–606. [Google Scholar] [CrossRef]
- Gothe, R. Paralysis in chickens caused by Argas (Persicargas) persicus larvae. I. The effect of the degree of engorgement and infection rate on clinical symptoms. Z. Fur Parasitenkd. 1971, 35, 298–307. [Google Scholar] [CrossRef]
- Zahid, H.; Muñoz-Leal, S.; Khan, M.Q.; Alouffi, A.S.; Labruna, M.B.; Ali, A. Life Cycle and Genetic Identification of Argas persicus Infesting Domestic Fowl in Khyber Pakhtunkhwa, Pakistan. Front. Vet. Sci. 2021, 8, 664731. [Google Scholar] [CrossRef]
- Duan, D.-Y.; Liu, Y.-K.; Liu, L.; Liu, G.-H.; Cheng, T.-Y. Microbiome analysis of the midguts of different developmental stages of Argas persicus in China. Ticks Tick-Borne Dis. 2022, 13, 101868. [Google Scholar] [CrossRef]
- Cutler, S.; Abdissa, A.; Adamu, H.; Tolosa, T.; Gashaw, A. Borrelia in Ethiopian ticks. Ticks Tick-Borne Dis. 2012, 3, 14–17. [Google Scholar] [CrossRef]
- Pader, V.; Buniak, J.N.; Abdissa, A.; Adamu, H.; Tolosa, T.; Gashaw, A.; Cutler, R.R.; Cutler, S.J. Candidatus Rickettsia hoogstraalii in Ethiopian Argas persicus ticks. Ticks Tick-Borne Dis. 2012, 3, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Denardi, S.E.; Bechara, G.H.; de Oliveira, P.R.; Nunes, É.T.; Saito, K.C.; Mathias, M.I.C. Morphological characterization of the ovary and vitellogenesis dynamics in the tick Amblyomma cajennense (Acari: Ixodidae). Vet. Parasitol. 2004, 125, 379–395. [Google Scholar] [CrossRef] [PubMed]
- Marzouk, A.S.; Swelim, H.H.; Ali, A.A.B. Ultrastructural changes induced by the entomopathogenic fungus Beauveria bassiana in the ovary of the tick Argas (Persicargas) persicus (Oken). Ticks Tick-Borne Dis. 2020, 11, 101507. [Google Scholar] [CrossRef] [PubMed]
- El Kammah, K.M.; Abdel Wahab, K.S. Argas (Persicargas) persicus life cycle under controlled and outdoor conditions. Acarologia 1980, 21, 163–172. [Google Scholar]
- Rodríguez, M.; Penichet, M.; Mouris, A.; Labarta, V.; Luaces, L.L.; Rubiera, R.; Cordovés, C.; Sánchez, P.; Ramos, E.; Soto, A.; et al. Control of Boophilus microplus populations in grazing cattle vaccinated with a recombinant Bm86 antigen preparation. Vet. Parasitol. 1995, 57, 339–349. [Google Scholar] [CrossRef]
- Fuente, J.D.; Rodríguez, M.; García-Garí, J.C. Immunological control of ticks through vaccination with Boophilus microplus gut antigens. Ann. N. Y. Acad. Sci. 2000, 916, 617–621. [Google Scholar] [CrossRef]
- Zhao, Y.; Qu, Z.-H.; Jiao, F.-C. De novo transcriptome sequencing and comparative profiling of the ovary in partially engorged and fully engorged Haemaphysalis flava ticks. Parasitol. Int. 2021, 83, 102344. [Google Scholar] [CrossRef]
- Vechtova, P.; Fussy, Z.; Cegan, R.; Sterba, J.; Erhart, J.; Benes, V.; Grubhoffer, L. Catalogue of stage-specific transcripts in Ixodes ricinus and their potential functions during the tick life-cycle. Parasites Vectors 2020, 13, 311. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wang, T.; Zhang, M.; Shi, X.; Zhang, M.; Wang, H.; Yang, X.; Yu, Z.; Liu, J. The Ovarian Development Genes of Bisexual and Parthenogenetic Haemaphysalis longicornis Evaluated by Transcriptomics and Proteomics. Front. Vet. Sci. 2021, 8, 783404. [Google Scholar] [CrossRef]
- Moreira, H.N.S.; Barcelos, R.M.; Vidigal, P.M.P.; Klein, R.C.; Montandon, C.E.; Maciel, T.E.F.; Carrizo, J.F.A.; de Lima, P.H.C.; Soares, A.C.; Martins, M.M.; et al. A deep insight into the whole transcriptome of midguts, ovaries and salivary glands of the Amblyomma sculptum tick. Parasitol. Int. 2017, 66, 64–73. [Google Scholar] [CrossRef]
- Tirloni, L.; Braz, G.; Nunes, R.D.; Gandara, A.C.P.; Vieira, L.R.; Assumpcao, T.C.; Sabadin, G.A.; da Silva, R.M.; Guizzo, M.G.; Machado, J.A.; et al. A physiologic overview of the organ-specific transcriptome of the cattle tick Rhipicephalus microplus. Sci. Rep. 2020, 10, 18296. [Google Scholar] [CrossRef]
- Santos, I.K.F.D.M.; Valenzuela, J.G.; Ribeiro, J.M.C.; DE Castro, M.; Costa, J.N.; Costa, A.M.; DA Silva, E.R.; Neto, O.B.R.; Rocha, C.; Daffre, S.; et al. Gene discovery in Boophilus microplus, the cattle tick: The transcriptomes of ovaries, salivary glands, and hemocytes. Ann. N. Y. Acad. Sci. 2004, 1026, 242–246. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.T.M.; Antoshechkin, I.; Hill, E.; Perry, M.W.; Olafson, P.U.; Saelao, P.; Lohmeyer, K.H.; Akbari, O.S. First transcriptome analysis of the winter tick (Dermacentor albipictus) reveals sex-specific expression patterns and potential targets for genetic control. G3 Genes Genomes Genet. 2025, 15, jkaf116. [Google Scholar] [CrossRef] [PubMed]
- Rio, D.C.; Ares, M., Jr.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, 2010, pdb.prot5439. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; A Simão, F.; Zdobnov, E.M.; Kelley, J. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Tang, Y.; Ghosal, S.; Roy, A. Nonparametric bayesian estimation of positive false discovery rates. Biometrics 2007, 63, 1126–1134. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. S4), S11. [Google Scholar] [CrossRef]
- Kim, T.K.; Waldman, J.; Ibanez-Carrasco, F.; Tirloni, L.; Waltero, C.; Calixo, C.; Braz, G.R.; Mulenga, A.; Junior, I.d.S.V.; Logullo, C. Stable internal reference genes for quantitative RT-PCR analyses in Rhipicephalus microplus during embryogenesis. Ticks Tick-Borne Dis. 2023, 14, 102251. [Google Scholar] [CrossRef]
- Chinzei, Y.; Minoura, H. Host immunoglobulin G titre and antibody activity in haemolymph of the tick, Ornithodoros moubata. Med. Vet. Entomol. 1987, 1, 409–416. [Google Scholar] [CrossRef]
- Galay, R.L.; Matsuo, T.; Hernandez, E.P.; Talactac, M.R.; Kusakisako, K.; Umemiya-Shirafuji, R.; Mochizuki, M.; Fujisaki, K.; Tanaka, T. Immunofluorescent detection in the ovary of host antibodies against a secretory ferritin injected into female Haemaphysalis longicornis ticks. Parasitol. Int. 2018, 67, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Tellam, R.; Kemp, D.; Riding, G.; Briscoe, S.; Smith, D.; Sharp, P.; Irving, D.; Willadsen, P. Reduced oviposition of Boophilus microplus feeding on sheep vaccinated with vitellin. Vet. Parasitol. 2002, 103, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liao, M.; Hatta, T.; Tanaka, M.; Xuan, X.; Fujisaki, K. Identification of a follistatin-related protein from the tick Haemaphysalis longicornis and its effect on tick oviposition. Gene 2006, 372, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Hussein, H.E.; Johnson, W.C.; Taus, N.S.; Suarez, C.E.; Scoles, G.A.; Ueti, M.W. Silencing expression of the Rhipicephalus microplus vitellogenin receptor gene blocks Babesia bovis transmission and interferes with oocyte maturation. Parasites Vectors 2019, 12, 7. [Google Scholar] [CrossRef]
- Domingues, L.N.; Bendele, K.G.; Bodine, D.M.; Halos, L.; Cutolo, A.A.; Liebstein, M.; Widener, J.; Figueiredo, M.; Moreno, Y.; Epe, C.; et al. A reverse vaccinology approach identified novel recombinant tick proteins with protective efficacy against Rhipicephalus microplus infestation. Ticks Tick-Borne Dis. 2024, 15, 102403. [Google Scholar] [CrossRef]
- Parthasarathi, B.C.; Kumar, B.; Bhure, S.K.; Sharma, A.K.; Manisha; Nagar, G.; Kumar, S.; Nandi, A.; Manjunathachar, H.V.; Chigure, G.M.; et al. Co-Immunization Efficacy of Recombinant Antigens against Rhipicephalus microplus and Hyalomma anatolicumTick Infestations. Pathogens 2023, 12, 433. [Google Scholar] [CrossRef]
- El Shoura, S.M.; Banaja, A.A.; Roshdy, M.A. Fine structure of the developing oocytes in adult Argas (Persicargas) arboreus (Ixodoidea: Argasidae). Exp. Appl. Acarol. 1989, 6, 143–156. [Google Scholar] [CrossRef]
- Le Gall, V.L.; Klafke, G.M.; Torres, T.T. Detoxification mechanisms involved in ivermectin resistance in the cattle tick, Rhipicephalus (Boophilus) microplus. Sci. Rep. 2018, 8, 12401. [Google Scholar] [CrossRef]
- Leyria, J.; Orchard, I.; Lange, A.B. The involvement of insulin/ToR signaling pathway in reproductive performance of Rhodnius prolixus. Insect Biochem. Mol. Biol. 2021, 130, 103526. [Google Scholar] [CrossRef]
- Valanne, S.; Wang, J.-H.; Rämet, M. The Drosophila Toll signaling pathway. J. Immunol. 2011, 186, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Franta, Z.; Frantová, H.; Konvičková, J.; Horn, M.; Sojka, D.; Mareš, M.; Kopáček, P. Dynamics of digestive proteolytic system during blood feeding of the hard tick Ixodes ricinus. Parasites Vectors 2010, 3, 119. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Pei, T.; Shi, X.; Nwanade, C.F.; Bing, Z.; Gao, Z.; Meng, J.; Li, L.; Liu, J. The functions of DNA methyltransferases during the feeding and development of Haemaphysalis longicornis are potentially associated with lysosome pathways. BMC Genom. 2024, 25, 1109. [Google Scholar] [CrossRef] [PubMed]
- Sabadin, G.A.; Parizi, L.F.; Kiio, I.; Xavier, M.A.; Matos, R.d.S.; Camargo-Mathias, M.I.; Githaka, N.W.; Nene, V.; Vaz, I.d.S. Effect of recombinant glutathione S-transferase as vaccine antigen against Rhipicephalus appendiculatus and Rhipicephalus sanguineus infestation. Vaccine 2017, 35, 6649–6656. [Google Scholar] [CrossRef]
- Ndawula, C.; Xavier, M.A.; Villavicencio, B.; Lopes, F.C.; Juliano, M.A.; Parizi, L.F.; Verli, H.; Vaz, I.d.S.; Ligabue-Braun, R. Prediction, mapping and validation of tick glutathione S-transferase B-cell epitopes. Ticks Tick-Borne Dis. 2020, 11, 101445. [Google Scholar] [CrossRef]
- Nagan, N.; A Zoeller, R. Plasmalogens: Biosynthesis and functions. Prog. Lipid Res. 2001, 40, 199–229. [Google Scholar] [CrossRef]
- Ohanian, J.; Ohanian, V. Sphingolipids in mammalian cell signalling. Cell. Mol. Life Sci. 2001, 58, 2053–2068. [Google Scholar] [CrossRef]
- Xavier, M.A.; Tirloni, L.; Pinto, A.F.M.; Diedrich, J.K.; Yates, J.R.; Mulenga, A.; Logullo, C.; Vaz, I.d.S.; Seixas, A.; Termignoni, C. A proteomic insight into vitellogenesis during tick ovary maturation. Sci. Rep. 2018, 8, 4698. [Google Scholar] [CrossRef]
- Oleaga, A.; Soriano, B.; Llorens, C.; Pérez-Sánchez, R.; Caimano, M.J. Sialotranscriptomics of the argasid tick Ornithodoros moubata along the trophogonic cycle. PLoS Neglected Trop. Dis. 2021, 15, e0009105. [Google Scholar] [CrossRef]
- Verma, J. Antiviral Mechanism of Serine Protease in Various Insects. In Trends in Insect Molecular Biology and Biotechnology; Kumar, D., Gong, C., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 143–161. [Google Scholar]
- LeMosy, E.K.; Leclerc, C.L.; Hashimoto, C. Biochemical defects of mutant nudel alleles causing early developmental arrest or dorsalization of the Drosophila embryo. Genetics 2000, 154, 247–257. [Google Scholar] [CrossRef]
- Zou, P.; Wu, L.; Wen, S.; Pei, Y.; Hu, Z.; Zuo, Y. Disruption of Spodoptera exigua serine protease 2 (Ser2) results in male sterility by CRISPR/Cas9 technology. Pest Manag. Sci. 2025, 81, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Jialaliding, Z.; Gu, J.; Merchant, A.; Zhang, Q.; Zhou, X. Knockout of ovary serine protease Leads to Ovary Deformation and Female Sterility in the Asian Corn Borer, Ostrinia furnacalis. Int. J. Mol. Sci. 2023, 24, 16311. [Google Scholar] [CrossRef] [PubMed]
- Asad, M.; Liao, J.; Chen, J.; Munir, F.; Pang, S.; Abbas, A.N.; Yang, G. Exploring the role of the ovary-serine protease gene in the female fertility of the diamondback moth using CRISPR/Cas9. Pest Manag. Sci. 2024, 80, 3194–3206. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Bi, H.; Wang, Y.; Li, X.; Xu, J.; Liu, Z.; He, L.; Li, K.; Huang, Y. Disruption of the ovarian serine protease (Osp) gene causes female sterility in Bombyx mori and Spodoptera litura. Pest Manag. Sci. 2020, 76, 1245–1255. [Google Scholar] [CrossRef]
- Xu, X.; Wang, Y.; Chen, J.; Du, X.; Yao, L.; Xu, J.; Zhang, Y.; Huang, Y.; Wang, Y. Mutation of Serine protease 1 Induces Male Sterility in Bombyx mori. Front. Physiol. 2022, 13, 828859. [Google Scholar] [CrossRef] [PubMed]
- Anisuzzaman; Islam, M.K.; Alim, M.A.; Miyoshi, T.; Hatta, T.; Yamaji, K.; Matsumoto, Y.; Fujisaki, K.; Tsuji, N. Longistatin is an unconventional serine protease and induces protective immunity against tick infestation. Mol. Biochem. Parasitol. 2012, 182, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Costa, G.C.; Silva, F.A.; Manzato, V.M.; Torquato, R.J.S.; Gonzalez, Y.G.; Parizi, L.F.; Junior, I.d.S.V.; Tanaka, A.S. A multiepitope chimeric antigen from Rhipicephalus microplus-secreted salivary proteins elicits anti-tick protective antibodies in rabbit. Vet. Parasitol. 2023, 318, 109932. [Google Scholar] [CrossRef]
- Camillo, L.d.M.B.; Ferreira, G.C.; Duran, A.F.A.; da Silva, F.R.S.; Garcia, W.; Scott, A.L.; Sasaki, S.D. Structural modelling and thermostability of a serine protease inhibitor belonging to the Kunitz-BPTI family from the Rhipicephalus microplus tick. Biochimie 2021, 181, 226–233. [Google Scholar] [CrossRef]
- Xu, Z.; Yan, Y.; Cao, J.; Zhou, Y.; Zhang, H.; Xu, Q.; Zhou, J. A family of serine protease inhibitors (serpins) and its expression profiles in the ovaries of Rhipicephalus haemaphysaloides. Infect. Genet. Evol. 2020, 84, 104346. [Google Scholar] [CrossRef]
- Liu, X.Y.; de la Fuente, J.; Cote, M.; Galindo, R.C.; Moutailler, S.; Vayssier-Taussat, M.; Bonnet, S.I.; Vinetz, J.M. IrSPI, a tick serine protease inhibitor involved in tick feeding and Bartonella henselae infection. PLoS Neglected Trop. Dis. 2014, 8, e2993. [Google Scholar] [CrossRef]
- Lu, J.; Wang, K.; Gao, Z.; Zhang, S.; Li, H.; Shi, Y.; Song, X.; Liu, J.; Yu, Z.; Yang, X. Doenitin-1: A novel Kunitz family protein with versatile functions during feeding and reproduction of the tick Haemaphysalis doenitzi. Front. Vet. Sci. 2022, 9, 872244. [Google Scholar] [CrossRef]
- Chinzei, Y. Quantitative changes of vitellogenin and vitellin in adult female ticks, Ornithodoros moubata, during vitellogenesis. Mie Med. J. 1983, 27, 117–127. [Google Scholar]
- Logullo, C.; Moraes, J.; Dansa-Petretski, M.; Vaz, I.; Masuda, A.; Sorgine, M.; Braz, G.; Masuda, H.; Oliveira, P. Binding and storage of heme by vitellin from the cattle tick, Boophilus microplus. Insect Biochem. Mol. Biol. 2002, 32, 1805–1811. [Google Scholar] [CrossRef]
- Kuniyori, M.; Sato, N.; Yokoyama, N.; Kawazu, S.-I.; Xuan, X.; Suzuki, H.; Fujisaki, K.; Umemiya-Shirafuji, R. Vitellogenin-2 Accumulation in the Fat Body and Hemolymph of Babesia-Infected Haemaphysalis longicornis Ticks. Front. Cell. Infect. Microbiol. 2022, 12, 908142. [Google Scholar] [CrossRef] [PubMed]
- He, X.-M.; Liu, L.; Cheng, T.-Y. HSC70 from Haemaphysalis flava (Acari: Ixodidae) exerts anticoagulation activity in vitro. Ticks Tick-Borne Dis. 2019, 10, 170–175. [Google Scholar] [CrossRef]
- Wang, F.; Wang, Y.; Wang, G.; Zhang, H.; Kuang, C.; Zhou, Y.; Cao, J.; Zhou, J. Ovary Proteome Analysis Reveals RH36 Regulates Reproduction via Vitellin Uptake Mediated by HSP70 Protein in Hard Ticks. Front. Cell. Infect. Microbiol. 2020, 10, 93. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Shu, J.; Zhang, Q. Expression of the Tribolium castaneum (Coleoptera: Tenebrionidae) hsp83 gene and its relation to oogenesis during ovarian maturation. J. Genet. Genom. 2010, 37, 513–522. [Google Scholar] [CrossRef]
- Myllyharju, J. Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol. 2003, 22, 15–24. [Google Scholar] [CrossRef]
- Winter, A.D.; McCormack, G.; Page, A.P. Protein disulfide isomerase activity is essential for viability and extracellular matrix formation in the nematode Caenorhabditis elegans. Dev. Biol. 2007, 308, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Cui, Y.; Namarra, U.; Tian, X.; Rivas-Giorgi, F.; Fikrig, E.; Norris, S.J. Dual roles for a tick protein disulfide isomerase during the life cycle of the Lyme disease agent. mBio 2024, 15, e0175424. [Google Scholar] [CrossRef]
- Elfvin, M.; Levine, R.J.; Dewey, M.M. Paramyosin in invertebrate muscles. I. Identification and localization. J. Cell Biol. 1976, 71, 261–272. [Google Scholar] [CrossRef]
- Yang, J.; Yang, Y.; Gu, Y.; Li, Q.; Wei, J.; Wang, S.; Boireau, P.; Zhu, X. Identification and characterization of a full-length cDNA encoding paramyosin of Trichinella spiralis. Biochem. Biophys. Res. Commun. 2008, 365, 528–533. [Google Scholar] [CrossRef]
- McManus, D.P.; Wong, J.Y.; Zhou, J.; Cai, C.; Zeng, Q.; Smyth, D.; Li, Y.; Kalinna, B.H.; Duke, M.J.; Yi, X. Recombinant paramyosin (rec-Sj-97) tested for immunogenicity and vaccine efficacy against Schistosoma japonicum in mice and water buffaloes. Vaccine 2001, 20, 870–878. [Google Scholar] [CrossRef]
- Gazarian, K.G.; Solis, C.F.; Gazarian, T.G.; Rowley, M.; Laclette, J.P. Synthetic peptide-targeted selection of phage display mimotopes highlights immunogenic features of α-helical vs non-helical epitopes of Taenia solium paramyosin: Implications for parasite- and host-protective roles of the protein. Peptides 2012, 34, 232–241. [Google Scholar] [CrossRef]
- Nisbet, A.J.; Huntley, J.F.; MacKELLAR, A.; Sparks, N.; Mcdevitt, R. A house dust mite allergen homologue from poultry red mite Dermanyssus gallinae (De Geer). Parasite Immunol. 2006, 28, 401–405. [Google Scholar] [CrossRef]
- Wu, P.-X.; Cui, X.-J.; Cao, M.-X.; Lv, L.-H.; Dong, H.-M.; Xiao, S.-W.; Liu, J.-Z.; Hu, Y.-H. Evaluation on two types of paramyosin vaccines for the control of Haemaphysalis longicornis infestations in rabbits. Parasites Vectors 2021, 14, 309. [Google Scholar] [CrossRef]
- Fukumoto, S.; Sakaguchi, T.; You, M.; Xuan, X.; Fujisaki, K. Tick troponin I-like molecule is a potent inhibitor for angiogenesis. Microvasc. Res. 2006, 71, 218–221. [Google Scholar] [CrossRef]
- You, M.-J. Immunization effect of recombinant P27/30 protein expressed in Escherichia coli against the hard tick Haemaphysalis longicornis (Acari: Ixodidae) in rabbits. Korean J. Parasitol. 2004, 42, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Pasini, M.E.; Intra, J.; Gomulski, L.M.; Calvenzani, V.; Petroni, K.; Briani, F.; Perotti, M.E. Identification and expression profiling of Ceratitis capitata genes coding for β-hexosaminidases. Gene 2011, 473, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, H.; Liu, F.; Wu, Q.; Shen, X.; Yang, Q. Structural determinants of an insect beta-N-Acetyl-D-hexosaminidase specialized as a chitinolytic enzyme. J. Biol. Chem. 2011, 286, 4049–4058. [Google Scholar] [CrossRef]
- Del Pino, F.A.; Brandelli, A.; Gonzales, J.C.; Henriques, J.A.; Dewes, H. Effect of antibodies against beta-N-acetylhexosaminidase on reproductive efficiency of the bovine tick Boophilus microplus. Vet. Parasitol. 1998, 79, 247–255. [Google Scholar] [CrossRef] [PubMed]

| Gene ID | Gene | Nucleotide Sequence (5′–3′) | Size (bp) |
|---|---|---|---|
| Cluster-2778.22975 | paramyosin | F: CAACCGCCGCATTCACGAGTAC | 123 |
| R: CGTTGGCACTTGGCTTCCTCCT | |||
| Cluster-2778.29718 | Fatty acid-binding protein | F: TAGGATGGGGCGGAAATGTG | 125 |
| R: CTTCATCGTCCACTGGTCCC | |||
| Cluster-2778.46089 | Vitellogenin | F: TGGCAGCAACTCCTCCGTCAAC | 194 |
| R: TGGTCAGCACAAGTGGCGACAA | |||
| Cluster-2778.29372 | Heat shock protein 70 | F: CGACATGGACGCCAACGGTATC | 139 |
| R: CTGTTCAGCCTCCTTCAGCATCC | |||
| Cluster-2778.8293 | chitinase | F: GCTCTGAAATCGAGCGCAAG | 138 |
| R: CAGACAGCGAAGTGTTTGCC | |||
| Cluster-2778.17153 | aquaporin | F: CGCCCTGGTGTACCTCATTT | 106 |
| R: GGGGTACGTAGCGAAGATGG | |||
| Cluster-2778.23455 | ferritin heavy-chain | F: AGACCCTGGATGGAGATGACT | 97 |
| R: CAAGGTCAACCACAGAAGAGC | |||
| Reference 1 | β-Actin | F: AGAGCAAGCGTGGCATCCTGA | 109 |
| R: CGCAGCTCGTTGTAGAAGGTGT | |||
| Reference 2 | GAPDH | F: ATGAAGCCTGCCCAGATTCC | 122 |
| R: ACCACCTTTTTGGCTCCTCC |
| Summary | Number of Genes | Percentage (%) |
|---|---|---|
| Statistics of transcriptome assembly | ||
| Total transcripts | 163,186 | |
| Total Unigenes | 63,500 | |
| Longest Unigene (bp) | 22,653 | |
| Shortest Unigene (bp) | 301 | |
| N50 transcript length (bp) | 2111 | |
| N50 Unigene length (bp) | 1693 | |
| GC_pct | 51.83–54.37 (mean = 53.61) | |
| Statistics of transcriptome annotation | ||
| Annotated in NR | 24,796 | 39.04 |
| Annotated in NT | 9353 | 14.72 |
| Annotated in KEGG | 10,887 | 17.14 |
| Annotated in SwissProt | 16,807 | 26.46 |
| Annotated in PFAM | 21,555 | 33.94 |
| Annotated in GO | 21,552 | 33.94 |
| Annotated in KOG/COG | 8855 | 13.94 |
| Annotated in all databases | 3305 | 5.2 |
| Annotated in at least one database | 32,440 | 51.08 |
| Total number of CDS | 29,695 | 46.76 |
| Number of transdecoder-predicted CDS | 9859 | 33.2 |
| Number of CDS searched in databases | 19,836 | 66.8 |
| GeneID | FPKM (OV0) | FPKM (OV1) | log2FC | padj | NR Description |
|---|---|---|---|---|---|
| Cluster-2778.40968 | 0 | 7.98 | 8.0112 | 1.58 × 10−10 | serine protease |
| Cluster-2778.8195 | 0 | 2.653 | 7.5078 | 4.66 × 10−9 | -- |
| Cluster-2778.10352 | 0.073 | 9.8267 | 7.1624 | 9.35 × 10−9 | serine protease |
| Cluster-2778.41816 | 0 | 8.8467 | 6.9324 | 2.55 × 10−7 | -- |
| Cluster-2778.6648 | 0 | 2.61 | 6.8712 | 2.67 × 10−7 | -- |
| Cluster-2778.7580 | 0.03 | 3.66 | 6.8066 | 7.71 × 10−8 | serine protease |
| Cluster-2778.40652 | 0 | 2.4067 | 6.7153 | 1.29 × 10−6 | -- |
| Cluster-2778.38220 | 0 | 2.33 | 6.461 | 3.51 × 10−6 | -- |
| Cluster-2778.9289 | 0.22 | 19.067 | 6.4352 | 7.43 × 10−113 | Na(+)/citrate cotransporter |
| Cluster-2778.8272 | 0 | 1.68 | 6.3875 | 4.76 × 10−6 | -- |
| Cluster-2778.37873 | 0.403 | 0 | −5.7259 | 0.0002864 | -- |
| Cluster-2778.15372 | 0.983 | 0 | −5.7556 | 0.00052655 | tcb1 |
| Cluster-2778.38261 | 2.22 | 0 | −5.9901 | 9.37 × 10−5 | -- |
| Cluster-2778.11877 | 2.247 | 0 | −6.3043 | 9.11 × 10−6 | -- |
| Cluster-2778.38351 | 1.59 | 0 | −6.4119 | 9.02 × 10−6 | cubilin |
| Cluster-2778.17280 | 2.02 | 0 | −6.5729 | 1.98 × 10−6 | MPV17L |
| Cluster-2778.15450 | 1.703 | 0 | −6.626 | 1.45 × 10−6 | |
| Cluster-2778.22920 | 3.443 | 0 | −6.6583 | 1.61 × 10−6 | -- |
| Cluster-2778.21097 | 6.763 | 0.0567 | −6.7136 | 1.46 × 10−7 | Allergen |
| Cluster-13619.0 | 0.823 | 0 | −6.7309 | 1.21 × 10−6 | -- |
| GeneID | FPKM (OV1) | FPKM (OV2) | log2FC | padj | NR Description |
|---|---|---|---|---|---|
| Cluster-2778.43242 | 0.07 | 646.643 | 13.163 | 2.44 × 10−90 | -- |
| Cluster-2778.11888 | 0.593 | 1205.44 | 10.98 | 0 | -- |
| Cluster-2778.42853 | 0.13 | 244.063 | 10.93 | 1.30 × 10−120 | -- |
| Cluster-2778.23188 | 0.043 | 70.457 | 10.619 | 1.97 × 10−36 | -- |
| Cluster-2778.18139 | 0.237 | 266.783 | 10.146 | 1.50 × 10−49 | -- |
| Cluster-2778.9849 | 0 | 208.253 | 9.998 | 3.62 × 10−16 | -- |
| Cluster-2778.46089 | 0.05 | 43.693 | 9.9447 | 1.30 × 10−126 | vitellogenin |
| Cluster-2778.31631 | 1.27 | 1224.01 | 9.9275 | 0 | -- |
| Cluster-2778.1351 | 0 | 29.927 | 9.924 | 4.18 × 10−16 | monotonin |
| Cluster-2778.1477 | 0 | 65.35 | 9.6753 | 1.31 × 10−15 | Kunitz-type serine protease inhibitors |
| Cluster-2778.40730 | 1.21 | 0 | −5.6285 | 0.00024 | -- |
| Cluster-2778.8690 | 1.837 | 0 | −5.6349 | 0.000274 | -- |
| Cluster-2778.36369 | 1.627 | 0 | −5.7533 | 0.000128 | -- |
| Cluster-2797.0 | 3.157 | 0 | −5.7766 | 0.000137 | -- |
| Cluster-2987.0 | 1.253 | 0 | −5.8111 | 0.000108 | -- |
| Cluster-2778.41221 | 1.487 | 0 | −5.8152 | 0.000175 | -- |
| Cluster-2778.20830 | 4.417 | 0 | −5.9664 | 5.37 × 10−5 | -- |
| Cluster-2778.40843 | 1.293 | 0 | −6.1418 | 2.58 × 10−5 | -- |
| Cluster-2778.40652 | 2.407 | 0 | −6.7034 | 1.59 × 10−6 | -- |
| Cluster-2778.7580 | 3.66 | 0.033 | −6.7942 | 8.86 × 10−8 | serine protease |
| Gene ID | Name | Score | FPKM | log2FC | Annotation | |
|---|---|---|---|---|---|---|
| OV0 | OV1 | |||||
| Cluster-2778.209 | LRRK1 | 39 | 2.2 | 0.057 | −5.0273 | leucine-rich repeat serine/threonine-protein kinase 1 |
| Cluster-2778.34485 | RPS3A | 5 | 14.78 | 6.96 | −1.0684 | 40S ribosomal protein S3a |
| Cluster-2778.38419 | H4 | 5 | 0.91 | 2.33 | 1.3937 | Histone 4 |
| Cluster-2778.13663 | FabG | 4 | 3.57 | 10.79 | 1.6286 | 3-oxoacyl-[acyl carrier protein] reductase |
| Cluster-2778.24606 | FUM | 4 | 2.34 | 0.62 | −1.8773 | fumarase |
| Cluster-2778.21121 | PIK3Cb | 4 | 1.77 | 0.49 | −1.755 | phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform |
| Cluster-2778.34617 | PDI3 | 3 | 5.23 | 10.557 | 1.0391 | protein disulfide isomerase 3 |
| Cluster-2778.26261 | PDI | 3 | 126.82 | 60.43 | −1.0714 | protein disulfide isomerase |
| Cluster-2778.24796 | PTEN | 3 | 2.83 | 9 | 1.6946 | phosphatidylinositol triP phosphatase |
| Cluster-2778.29372 | HSP70 | 3 | 106.83 | 218.94 | 1.0625 | heat shock protein 70 |
| Gene ID | Name | Score | FPKM | log2FC | Annotation | |
|---|---|---|---|---|---|---|
| OV1 | OV2 | |||||
| Cluster-2778.23724 | APRT | 14 | 13.57 | 34.25 | 1.3188 | adenine phosphoribosyltransferase |
| Cluster-2778.29659 | RPL13A | 14 | 437.09 | 876.26 | 1.0599 | ribosomal protein L13A |
| Cluster-2778.34298 | TnI | 13 | 3.31 | 15.87 | 2.2441 | troponin I protein |
| Cluster-2778.22975 | PRM | 12 | 3.93 | 27.59 | 2.7926 | paramyosin |
| Cluster-2778.30915 | MyHC | 12 | 2.64 | 16.04 | 2.5853 | myosin heavy chain |
| Cluster-2778.38419 | H3 | 12 | 2.3 | 1.1 | −1.1013 | Histone 3 |
| Cluster-2778.17397 | HEX (HEXB) | 11 | 1.38 | 8.19 | 2.5592 | beta-hexosaminidase subunit beta |
| Cluster-2778.32843 | HEX (HEXA) | 11 | 6.13 | 29.88 | 2.2908 | beta-hexosaminidase subunit alpha |
| Cluster-2778.33259 | ENH | 10 | 2.53 | 6.92 | 1.4376 | adaptor protein enigma |
| Cluster-2778.26644 | HSDL2 | 10 | 1.55 | 3.37 | 1.1192 | hydroxysteroid dehydrogenase protein 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, F.; Duan, D.; Meng, J.; Cheng, T. De Novo Transcriptome Sequencing and Profiling of Ovarian Development of Argas persicus Along the Trophogonic Cycle. Genes 2025, 16, 1107. https://doi.org/10.3390/genes16091107
Yan F, Duan D, Meng J, Cheng T. De Novo Transcriptome Sequencing and Profiling of Ovarian Development of Argas persicus Along the Trophogonic Cycle. Genes. 2025; 16(9):1107. https://doi.org/10.3390/genes16091107
Chicago/Turabian StyleYan, Fen, Deyong Duan, Jinzhu Meng, and Tianyin Cheng. 2025. "De Novo Transcriptome Sequencing and Profiling of Ovarian Development of Argas persicus Along the Trophogonic Cycle" Genes 16, no. 9: 1107. https://doi.org/10.3390/genes16091107
APA StyleYan, F., Duan, D., Meng, J., & Cheng, T. (2025). De Novo Transcriptome Sequencing and Profiling of Ovarian Development of Argas persicus Along the Trophogonic Cycle. Genes, 16(9), 1107. https://doi.org/10.3390/genes16091107





